Abstract
Abstract 1833
Poster Board I-859
Individualization of therapy for multiple myeloma (MM), based on gene expression profiling, has not yet been achieved.
We previously described a “metagene” genomic model that synergizes with standard clinical staging to robustly prognosticate in MM (ASCO 2009 meeting, oral abstract # 8521; submitted for publication). In the current work we queried the Connectivity Map (Lamb J et al., Science 313(5795):1929-35, 2006); http://www.broadinstitute.org/cmap) using the metagene model, to elicit novel therapies that are likely to mitigate the poor-prognosis clinical phenotype we can identify with that model. We then employed gene expression profiling, the metagene model, and probit binary regression analysis to classify 12 untreated MM cell lines as having a similar or dissimilar molecular phenotype to poor-prognosis patients. Lastly, we performed MTT tetrazolium viability assays to survey, in triplicate, the sensitivity of those cell lines to the drug identified as well as melphalan, which we used as a conventional therapeutic control.
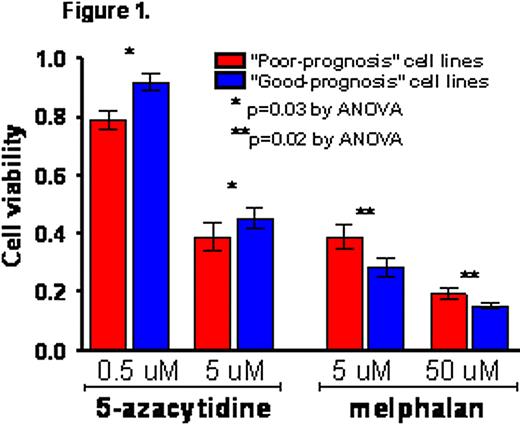
We used Connectivity Map to analyze the metagene model, which we had previously discovered and validated in silico using three patient cohorts with prospectively collected clinical follow-up data (n=624 total patients). In doing so, we elicited 5-azacytidine as a likely agent to reverse the poor-prognosis phenotype. Using the metagene model, we then characterized 8 untreated MM cell lines as having a “good prognosis” molecular phenotype (i.e., a gene expression profile dissimilar to poor-prognosis patients according to the metagene model), and 4 cell lines as having a “poor prognosis” molecular phenotype (i.e., similar to poor-prognosis patients based on the metagene model). “Poor-prognosis” cell lines were susceptible to viability reduction by 5-azacytidine in a dose-independent manner and, importantly, at a physiologically achievable dose in humans (5 μM), whereas ‘good-prognosis‘ cell lines were comparatively less susceptible (p=0.03 by repeated measures 2-way ANOVA). Conversely, melphalan, which we used as a clinically relevant control that was not elicited by our analysis of the metagene model as likely to be effective in poor-prognosis patients, reduced the viability of “good-prognosis” cell lines more than that of “poor-prognosis” cell lines (fig. 1; p=0.02).
Computational analysis of the extensively validated and clinically relevant metagene prognostic model indicates, and in vitro assays confirm, that 5-azacytidine is more effective in ameliorating a poor-prognosis phenotype than melphalan in a multiple myeloma pre-clinical model. This provides proof-of-concept that gene expression profiling may 1) reveal novel and effective therapeutic approaches for identifiable high-risk subgroups of patients, and 2) enable clinicians to decide prospectively which conventional agents, such as melphalan, are unlikely to be effective in certain patients' myeloma. Clinical trials are needed to study such individualized approaches to the treatment of this molecularly heterogeneous disease.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal