Abstract
Recent studies support the notion that there is an intricate relationship between hematopoiesis and bone homeostasis in normal steady states. Using mice undergoing chronic inflammatory arthritis, we investigated the relationship between hematopoiesis and bone homeostasis in pathologic conditions. We demonstrate that mice undergoing chronic inflammatory arthritis displayed osteoporosis resulting from a severe defect in osteoblast function. Despite the defective osteoblast function, however, the hematopoietic stem cells from these mice exhibited normal properties in either long-term repopulation or cell cycling. Therefore, the bone-forming capacity of osteoblasts is distinct from their ability to maintain hematopoietic stem cells in chronic inflammatory conditions.
Introduction
Under normal physiologic conditions, hematopoietic stem cells (HSCs) residing within the specialized bone marrow (BM) niche maintain a balance between self-renewal and differentiation and provide continuous supply of circulating mature immune cells with a limited life span. An intricate relationship exits between hematopoiesis and bone homeostasis. As such, osteoblasts serve as an HSC niche, whereas osteoclasts mediate HSCs and progenitor egress from the BM.1,2 Specifically, an increase in osteoblast number and/or activation through conditional Alk3 deletion or parathyroid hormone administration augments the HSC frequency in BM.3,4 Conversely, ablation of osteoblasts results in a decrease in absolute number of phenotypic primitive hematopoietic progenitors.5
Rheumatoid arthritis (RA) is a chronic systemic inflammatory autoimmune disease of unknown etiology afflicting 1% of the population. It leads to destruction of cartilage and bone at multiple joints with a distal to proximal preference. RA is also attended by systemic osteoporosis. However, the mechanisms of RA-associated osteoporosis are less appreciated than how joints are destroyed. The KRNxNOD (herein K/BxN) mouse model of inflammatory arthritis recapitulates many of the features of human RA.6,7 These mice were generated fortuitously when mice transgenic for a T-cell receptor recognizing an epitope of bovine RNase (C57BL/6.KRN, herein KRN) were bred onto an NOD background.8 They developed spontaneous chronic and severely destructive arthritis with 100% penetrance that resembled human RA.8 KRN with a C57BL/6 line congenic for the NOD MHC H-2g7 (C57BL/6.H-2g7; herein G7) was used to distinguish the contribution of MHC from non-MHC NOD-derived genes to disease development. The KRNxC57BL/6.H-2g7 (herein KRNxG7) offspring all develop overt joint swelling and the histologic hallmarks of arthritis of K/BxN mice, indicating that H-2g7 is sufficient for RA development.8
Using a KRNxG7 mouse model, we investigated the relationship between HSCs and bone homeostasis in chronic inflammatory conditions. We demonstrate that, similar to patients with RA, mice with inflammatory arthritis develop osteoporosis. However, unlike the osteolyisis of inflamed joints, which reflects accelerated osteoclast activity, the systemic bone loss of arthritic mice is the result of arrested osteoblast function. This conclusion is consistent with the decrease in generation of mature osteoclasts in vivo. Unexpectedly, the osteoblast deficiency in bone formation did not affect the long-term repopulating potential of HSCs in these arthritic mice. Collectively, we provide evidence that marrow HSCs can be maintained in the absence of functional osteoblasts in chronic inflammatory environments.
Materials
Mice
KRN (T-cell receptor transgenic) mice on a C57BL/6 background were crossed with G7 (I-Ag7) to generate KRNxG7 mice. C57BL/6J (CD45.2 allele) and B6.SJL-PtprcaPep3b/BoyJ (CD45.1 allele) mice were obtained from The Jackson Laboratory. All animals were housed in accordance with National Institutes of Health and American Association for Accreditation of Laboratory Animal Care regulations, and animal protocols were reviewed and approved by the Washington University animal studies committee.
Cell preparation and flow cytometric analyses
BM cells were prepared by vigorously flushing femur and tibia 6 to 8 times. Peripheral blood (PB) was obtained by retro-orbital collection. Spleen cells were prepared by gently crushing the tissue and filtering through a 40-μm cell strainer (BD Falcon). Liver cells were obtained by gently crushing the tissue and filtering through a 70-μm cell strainer (BD Falcon). All collected cells were treated with red blood cell lysis buffer (Roche Diagnostics) before analyses.
Fluorescence-activated cell sorter (FACS) analyses were performed as described previously.9 For KSL analyses, we used fluorescein isothiocyanate (FITC)–conjugated antibodies against CD4, CD8, Mac-1, Gr-1, Ter119, and B220 (lineage marker antibodies, BD Biosciences or eBioscience), phycoerythrin (PE)–conjugated anti–Sca-1, peridinin chlorophyll protein (PerCP)/Cy5.5-conjugated anti-CD45, and allophycocyanin (APC)–conjugated anti–c-Kit (eBioscience). For some experiments, anti-CD45 APC-Alexa 750 (eBioscience) or anti–c-Kit PerCP/Cy5.5 (BioLegend) was used. SLAM analyses were performed using anti-CD150 PE (BioLegend), anti-CD48 FITC (eBioscience), and anti-CD41 FITC (BD Biosciences). Mature lineage analyses were performed with FITC- and PE-conjugated antibodies to the lineage markers as well as Alexa 647–conjugated anti–Mac-1 (BD Biosciences) and APC-conjugated F4/80 (eBioscience).
Lymphoid primed multipotential progenitor (Kit+Sca1+Lin−Flk2hiCD34+) and common lymphoid progenitor (CLP; Lin−Flk2+IL-7Rα+) analyses were performed with the following fluorophore-conjugated antibodies: α-Sca1-FITC, α-IL-7Rα-biotin, α-cKit-APC-Alexa 750 (eBioscience), α-Flk2-PE, α-CD34-Alexa647, α-lineage-APC cocktail, streptavidin-PerCP/Cy5.5 (BD Biosciences), and α-lineage-biotin cocktail (Miltenyi Biotec). PreproB analyses were performed with α-IgM-FITC, α-CD43-PE, α-NK1.1 PerCP/Cy5.5, α-CD11c PE-Cy7, α-B220-biotin (BD Bioscience), and α-CD19-Alexa647 and streptavidin APC-eFluor 708 (eBioscience). Other antibodies used for B-cell precursor analysis were α-B220-PerCP/Cy5.5 (eBioscience) and α-AA4.1-FITC (BD Bioscience). Cells were analyzed using a Facscalibur (4-color), a FACScan adapted for 5-color analysis, or FACScanto (6-color) and data analyzed with CellQuest (BD Biosciences) or FlowJo software (TreeStar).
BrdU labeling
KRNxG7 and control mice (6 to 8 weeks old) were injected with a single dose (1 mg per 6 g of body mass) of sterile-filtered 5-bromodeoxyuridine (BrdU; Sigma-Aldrich) dissolved in phosphate-buffered saline. Mice were killed 2 to 3 hours later and BM cells harvested as described earlier. Harvested BM cells were subjected to lineage-cell depletion by magnetic separation using the lineage cell depletion kit (Miltenyi Biotec). Lineage-depleted cells were stained with α-cKit-FITC (eBioscience), α-Sca1-PE, and streptavidin-PerCP-Cy5.5 (BD Biosciences PharMingen). Cells were subsequently fixed and intracellularly stained with APC-conjugated α-BrdU antibody using the APC-BrdU flow kit (BD Biosciences PharMingen). Cells were analyzed using the BD FACScalibur and data analyzed using BD CellQuest software.
Ki-67/Hoechst analysis
Lin+ cells were depleted from BM by magnetic sorting as described in “Cell preparation and flow cytometric analyses” using biotin-conjugated anti-Lin antibodies (Miltenyi Biotec). Cells were surface stained with α-Sca1 PE, streptavidin PerCP/Cy5.5 (BD Biosciences), and α-cKit APC (eBioscience). Ki-67/Hoechst staining has previously been used to assess KSL cycling, and our method was adapted from this previous study.10 Surface-stained cells were fixed and permeabilized using BD Cytofix/Cytoperm buffer followed by intracellular staining with α-Ki-67 FITC for 30 minutes followed by a 5-minute Hoechst incubation (20 μg/mL). Five-color flow cytometry was performed with a MoFlo (Dako North America), which has UV excitation capability. Data analysis was performed with Summit or FlowJo software. Doublets were excluded in the gated populations that were analyzed for Ki-67 expression. The Ki-67− population was defined based on staining a control population of cells with an FITC-conjugated isotype control antibody (BD Biosciences).
Progenitor assay
Cells from BM and spleen were replated in Methocult M3434 (StemCell Technologies). Colonies were counted 7 to 10 days later.
Cell transplantation
For serial BM transplantation, lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipients were injected (intravenously) with unfractionated 106 BM cells from 6-week-old KRNxG7 (CD45.2xCD45.2) or B6xG7 (CD45.2xCD45.2) mice (5 recipients for each group). PB samples were analyzed for CD45.1 and CD45.2 every 4 weeks. Seven months after transplantation, BM suspensions were prepared from primary recipients, and 106 nucleated cells were injected into new lethally irradiated B6xG7 (CD45.1xCD45.2) recipient mice (8 for control and 9 for KRNxG7). The tertiary transplantation was performed 7 months after secondary transplantation (7 for control and 9 for KRNxG7). The recipients of serial BM transplantation were subjected to lineage analyses for donor contributions 6 or 7 months after transplantations.
Competitive repopulation assay has been described previously.11 Briefly, 6-week-old B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) BM cells, 2 × 105, were mixed with 2 × 105 B6xG7 (CD45.1xCD45.2) competitor BM cells and injected (intravenously) into lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipient mice. PB samples were collected retro-orbitally 3 and 5 months after transplantation and analyzed for CD45.1 and CD45.2.
Lineage-negative (Lin−) spleen cells from 6-week-old B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) mice were isolated using MACS Lineage Cell Depletion Kit (Miltenyi Biotec). A total of 105 sorted Lin− cells were injected into lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipients. Reconstitution of donor-derived cells (CD45.2) was monitored by staining retro-orbitally obtained PB cells with monoclonal antibodies against CD45.2 and CD45.1 (eBioscience) followed by FACS analysis.
Serum TRAP5b activity and serum osteocalcin activity
Blood was collected retro-orbitally under anesthesia before sacrifice. The serum TRAP5b activities of 6-week-old G7 and KRNxG7 mice were measured by MouseTRAP Assay ELISA kit (Immunodiagnostic Systems Inc). Serum osteocalcin levels of 6-week-old G7 and KRNxG7 mice were measured by the Mouse Osteocalcin ELISA kit (Biomedical Technologies Inc).
Histology and histomorphometry
The tibiae of 6-week-old B6xG7 and KRNxG7 mice were fixed with 70% ethanol followed by plastic embedding and Goldner staining, or with 10% neutral buffered formalin followed by the decalcification in 14% ethylenediaminetetraacetic acid for 4 to 5 days, paraffin embedding, and TRAP staining. Calcein (Sigma-Aldrich; 7.5 mg/kg, intraperitoneally) was injected on days 7 and 12. Mice were killed on day 14. Osteoclastic and osteoblastic perimeters were measured and analyzed using Osteomeasure (OsteoMetrics) in a blinded fashion.
μCT
The trabecular volume in the distal femoral metaphysic was measured using a Scanco μCT40 scanner (Scanco Medical AG). A threshold of 300 was used for evaluation of all scans. Thirty slices were analyzed, starting with the first slice in which condyles and primary spongiosa were no longer visible.
Quantitative RT-PCR
RNA preparation and cDNA synthesis were previously described.12 Primer sequences used in this study are provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Statistical analyses
Statistical significance was assessed by 2-tailed Student t test. Values of P less than .05 were considered statistically significant.
Results
KRNxG7 mice are osteoporotic because of diminished bone formation
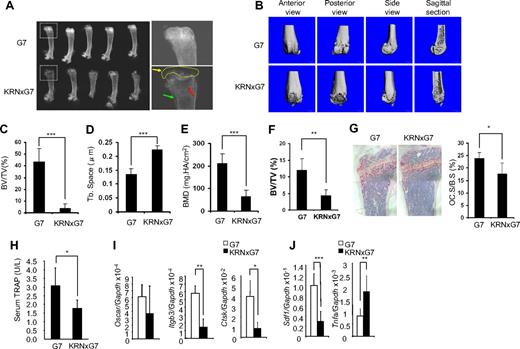
K/BxN and KRNxG7 mice develop arthritic symptoms, including ankle swelling shortly after 3 weeks of age.8 The ankle thickness increases up to 5 to 6 weeks of age, reaching a maximum of 4 to 5 mm and remaining constant at a slightly lower level thereafter.8 Typically, 6-week-old KRNxG7 mice in C57BL/6 genetic background were used in this study, as they show overt inflammation at this time point. As expected, KRNxG7 mice develop rheumatoid joint pannus and lysis of periarticular bone (Figure 1A-B). Because human inflammatory arthritis is also attended by systemic bone loss, we asked whether the same holds true in this murine model. Radiographs of KRNxG7 tibiae showed destruction of epiphyseal bone as well as metaphyseal demineralization. Histomorphometric and μCT analysis of the same bones established a marked reduction of trabecular bone volume and consequently increased trabecular spacing (Figure 1C-D,F). A dual-energy X-ray absorptiometry analysis exhibited decreased bone mineral density in arthritic mice (Figure 1E). Despite the profound metaphyseal osteoporosis, however, the number of mature resorptive cells was decreased in the marrow of endosteal bone (Figure 1G). This observation was confirmed by diminished serum levels of the global osteoclast marker TRAP5b (Figure 1H) and impaired expression of osteoclast specific genes in whole BM (Figure 1I).
Severe joint destruction and osteoporosis in KRN/G7 mice. (A) Radiographs of femurs of 6-week-old G7 and KRNxG7 mice. (Right panels) Higher magnification images of boxed areas in the left panels. Yellow arrow and dashed circle in lower right panel denote destroyed articular surface and secondary ossification center, respectively. Red arrow points to trabecular bone region. Green arrow indicates microfractures. (B) Representative 3-dimensional reconstruction of the femur by μCT. (C) The percentage of trabecular bone volume/tissue volume (BV/TV) determined by μCT. (D) Trabecular separation determined by μCT (Tb. space). (E) Dual-energy X-ray absorptiometry determined bone mineral density (BMD). Data are presented as mean ± SD; n = 5 in each group of mice. (F) Histomorphometric determination of BV/TV. Trabecular bone volume normalized to total marrow space (BV/TV). (G) TRAP (red reaction product) stained histologic sections of G7 and KRNxG7 tibia. Data are expressed as percentage trabecular bone surface covered by osteoclasts; n = 5. (H) Global osteoclast number, in vivo, was quantified by serum TRAP5b enzyme-linked immunosorbent assay. (I) Oscar, integrin β3, and cathepsin K expression was analyzed by quantitative polymerase chain reaction (PCR) with RNA from G7 and KRNxG7 BM. Shown is the mean expression ± SD for each gene normalized to GAPDH; n = 3. (J) SDF1 and TNF-α expression was analyzed by quantitative reverse-transcribed (RT)–PCR with RNA from G7 and KRNxG7 BM. Shown is the mean expression ± SD for each gene normalized to GAPDH; n = 3. *P < .05. **P < .01. ***P < .001.
Severe joint destruction and osteoporosis in KRN/G7 mice. (A) Radiographs of femurs of 6-week-old G7 and KRNxG7 mice. (Right panels) Higher magnification images of boxed areas in the left panels. Yellow arrow and dashed circle in lower right panel denote destroyed articular surface and secondary ossification center, respectively. Red arrow points to trabecular bone region. Green arrow indicates microfractures. (B) Representative 3-dimensional reconstruction of the femur by μCT. (C) The percentage of trabecular bone volume/tissue volume (BV/TV) determined by μCT. (D) Trabecular separation determined by μCT (Tb. space). (E) Dual-energy X-ray absorptiometry determined bone mineral density (BMD). Data are presented as mean ± SD; n = 5 in each group of mice. (F) Histomorphometric determination of BV/TV. Trabecular bone volume normalized to total marrow space (BV/TV). (G) TRAP (red reaction product) stained histologic sections of G7 and KRNxG7 tibia. Data are expressed as percentage trabecular bone surface covered by osteoclasts; n = 5. (H) Global osteoclast number, in vivo, was quantified by serum TRAP5b enzyme-linked immunosorbent assay. (I) Oscar, integrin β3, and cathepsin K expression was analyzed by quantitative polymerase chain reaction (PCR) with RNA from G7 and KRNxG7 BM. Shown is the mean expression ± SD for each gene normalized to GAPDH; n = 3. (J) SDF1 and TNF-α expression was analyzed by quantitative reverse-transcribed (RT)–PCR with RNA from G7 and KRNxG7 BM. Shown is the mean expression ± SD for each gene normalized to GAPDH; n = 3. *P < .05. **P < .01. ***P < .001.
The chemokine SDF-1 plays a critical role in osteoclastogenesis by promoting osteoclast differentiation and the cell's longevity.13,14 In addition, inhibition of BM SDF-1 expression promotes osteoclast progenitor cell mobilization to the periphery.15 We found that SDF-1 BM mRNA levels were decreased in KRNxG7 mice; thus, its suppression probably mediates, at least in part, the noted in vivo arrest of terminal osteoclast differentiation (Figure 1J). Because tumor necrosis factor-α (TNF-α), which accelerates osteoclast progenitor mobilization in inflammatory erosive arthritis, also suppresses SDF-1 expression,15 we posited that TNF-α level is increased in KRNxG7 mice. We indeed found that TNF-α mRNA and protein are increased in KRNxG7 BM and serum, respectively (Figure 1J and data not shown). Thus, although a direct link between TNF-α and SDF-1 in KRNxG7 mice needs to be established, in face of suppressed Sdf-1 expression, osteoclast progenitor cells most probably do not readily assume the full resorptive phenotype but are mobilized to the periphery and migrate to the inflamed joint, which they degrade on maturation.
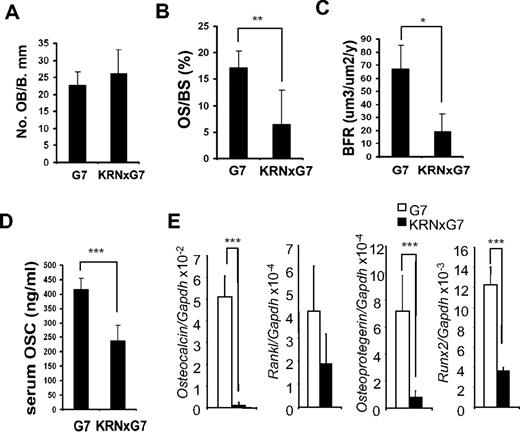
Osteoporosis may reflect stimulated osteoclast or diminished osteoblast activity. KRNxG7 mice have reduced marrow osteoclasts in face of systemic osteoporosis, suggesting that the paucity of bone extant in these animals reflects suppressed bone formation. To address this issue, we first histomorphometrically determined the number of trabecular osteoblasts/millimeter bone surface, which we found indistinguishable in KRNxG7 and G7 mice (Figure 2A). Moreover, in vitro osteoblast formation as well as bone nodule formation assessed by alkaline phosphatase activity and mineralization assays, respectively, were indistinguishable between KRNxG7 and controls (supplemental Figure 1A-B). However, the percentage of metaphyseal bone surface covered by osteoid was reduced in the arthritic mice, suggesting that the bone-synthesizing population was diminished (Figure 2B). This posture was confirmed by dynamic histomorphometry, which established that the rate of metaphyseal bone formation is less than one-third of control (Figure 2C). Similarly, serum osteocalcin as well as Osteocalcin mRNA, a marker of global bone formation, was reduced in KRNxG7 mice (Figure 2D-E). In addition, mRNA expression of osteoblast specific genes, receptor activator of nuclear factor-κB ligand (Rankl), Osteoprotegerin, and Runx2 were markedly diminished (Figure 2E). Thus, the systemic osteoporosis attending the inflammatory arthritis of KRNxG7 mice reflects diminished bone formation and not accelerated bone resorption.
Impaired bone formation rate in KRN/G7 mice. (A) Osteoblast number/millimeter bone perimeter (No. OB/B.mm). (B) Percentage trabecular surface covered by osteoid (OS.S/B.mm). (C) Bone formation rate (BFR) histomorphometrically quantitated from double calcein labeled tibia. (D) In vivo bone formation was quantified by serum osteocalcin (Osc) level at 6 weeks of age (n = 5). (E) RNA of KRN and KRNxG7 BM was analyzed for gene expression of osteoblast markers, receptor activator of nuclear factor-κB ligand (Rankl), Osteoprotegerin (Opg), Runx2, and Osteocalcin by quantitative RT-PCR. Shown is the mean expression ± SD for each gene normalized to GAPDH. *P < .05. **P < .01. ***P < .001.
Impaired bone formation rate in KRN/G7 mice. (A) Osteoblast number/millimeter bone perimeter (No. OB/B.mm). (B) Percentage trabecular surface covered by osteoid (OS.S/B.mm). (C) Bone formation rate (BFR) histomorphometrically quantitated from double calcein labeled tibia. (D) In vivo bone formation was quantified by serum osteocalcin (Osc) level at 6 weeks of age (n = 5). (E) RNA of KRN and KRNxG7 BM was analyzed for gene expression of osteoblast markers, receptor activator of nuclear factor-κB ligand (Rankl), Osteoprotegerin (Opg), Runx2, and Osteocalcin by quantitative RT-PCR. Shown is the mean expression ± SD for each gene normalized to GAPDH. *P < .05. **P < .01. ***P < .001.
Systemic increase in Gr1+ cells and decrease in B220+ cells accompanied by impaired KRNxG7 marrow B lymphopoiesis
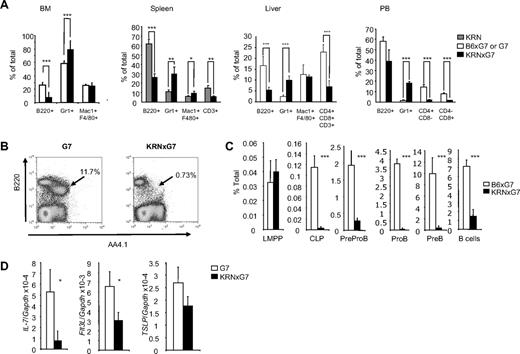
Our data so far show that osteoblasts are functionally defective in KRNxG7 mice. In addition to bone formation, osteoblasts have been reported to play crucial roles in hematopoiesis by providing a niche to maintain HSCs and supporting B lymphopoiesis.3-5,16-18 We noticed that KRNxG7 BM cellularity was higher (∼ 50% more) compared with that of controls irrespective of the method of BM collection (supplemental Figure 2A). To investigate the specific hematopoietic changes occurring in chronic inflammation, we examined mature hematopoietic cell lineages in BM, spleen, liver, and PB (Figure 3A). There was an increase in myeloid cells, specifically Gr1+ cells, in all KRNxG7 tissues analyzed. Myeloid cells, including neutrophils, are abundant in the joint inflammation of human RA patients21 and are critical for the disease, as depletion of neutrophils or macrophages ameliorates inflammatory joint disease in a serum transfer model of RA.22,23 In KRNxG7 mice, T cells (detected by CD3, CD4, or CD8) bearing T-cell receptor transgene undergo negative selection.8 Thus, as expected, T cells were reduced.
Characterization of mature cells and B-cell development defect in KRNxG7 mice. (A) Systemic increase in myeloid cells and decrease in lymphoid cells. BM, spleen, liver, and PB cells were harvested from 6- to 8-week-old KRNxG7 and control (KRN, G7, or B6xG7) mice, stained for the indicated lineage markers, and analyzed by flow cytometry. B220 is a B-cell marker, Gr1 stains granulocytes and monocyte populations, Mac1 and F480 combination stains macrophages, and CD3, CD4, and CD8 stain T cells. Shown is the mean for 3 to 11 mice analyzed per strain for each tissue. (B) FACS plot for B-cell precursors in BM. BM cells from KRNxG7 or G7 controls are stained for B220 and AA4.1 (a marker for B-cell precursors). B-cell precursors (B220loAA4.1+; arrow) are virtually depleted in KRNxG7 BM, whereas most of the residual cells are B220hi and IgM+ (IgM staining not shown). (C) Systematic analysis of BM B-cell development. FACS determined frequency of various B-cell precursors from earliest (left) to the latest (right) are shown. Lymphoid primed multipotential progenitor (Kit+Sca1+Lin−Flk2hiCD34+), CLP (Lin−Flk2+IL-7Rα+), PreproB (B220+IgM−CD19−CD43+NK1.1−CD11c−), ProB (B220+IgM−CD19+CD43+), PreB (B220+IgM−CD19+CD43−), and B cells (B220+IgM+). We confirmed that Lin−Flk2+IL-7Rα+ CLPs were almost predominantly KitloSca1lo (data not shown) as previously reported.19,20 (D) Expression of various B lymphopoiesis promoting cytokines in whole marrow. Expression was determined by quantitative real-time PCR, followed by normalization to GAPDH. IL-7, Flt3-L (ligand for Flk2), thymic stromal lymphopoietin (TSLP). *P < .05. **P < .01. ***P < .001.
Characterization of mature cells and B-cell development defect in KRNxG7 mice. (A) Systemic increase in myeloid cells and decrease in lymphoid cells. BM, spleen, liver, and PB cells were harvested from 6- to 8-week-old KRNxG7 and control (KRN, G7, or B6xG7) mice, stained for the indicated lineage markers, and analyzed by flow cytometry. B220 is a B-cell marker, Gr1 stains granulocytes and monocyte populations, Mac1 and F480 combination stains macrophages, and CD3, CD4, and CD8 stain T cells. Shown is the mean for 3 to 11 mice analyzed per strain for each tissue. (B) FACS plot for B-cell precursors in BM. BM cells from KRNxG7 or G7 controls are stained for B220 and AA4.1 (a marker for B-cell precursors). B-cell precursors (B220loAA4.1+; arrow) are virtually depleted in KRNxG7 BM, whereas most of the residual cells are B220hi and IgM+ (IgM staining not shown). (C) Systematic analysis of BM B-cell development. FACS determined frequency of various B-cell precursors from earliest (left) to the latest (right) are shown. Lymphoid primed multipotential progenitor (Kit+Sca1+Lin−Flk2hiCD34+), CLP (Lin−Flk2+IL-7Rα+), PreproB (B220+IgM−CD19−CD43+NK1.1−CD11c−), ProB (B220+IgM−CD19+CD43+), PreB (B220+IgM−CD19+CD43−), and B cells (B220+IgM+). We confirmed that Lin−Flk2+IL-7Rα+ CLPs were almost predominantly KitloSca1lo (data not shown) as previously reported.19,20 (D) Expression of various B lymphopoiesis promoting cytokines in whole marrow. Expression was determined by quantitative real-time PCR, followed by normalization to GAPDH. IL-7, Flt3-L (ligand for Flk2), thymic stromal lymphopoietin (TSLP). *P < .05. **P < .01. ***P < .001.
A decrease was also seen in B220+ cells in KRNxG7 mice for all tissues analyzed (Figure 3A), and this indeed reflects a decrease in B-lineage cells, not merely a decrease in B220 expression (supplemental Figure 3A). To determine whether defective marrow B lymphopoiesis in KRNxG7 mice could at least in part explain the diminution in B220+ frequency, we examined the frequency of marrow B-cell precursors. We found that a majority of the residual B220+ cells in KRNxG7 marrow were B cells (B220hiIgM+), with almost complete depletion of B-cell precursors (B220loAA4.1+; Figure 3B; and data not shown).24,25 Further analysis revealed that not only were B cell–committed, pre-proB, proB, and preB precursors absent but CLPs19 were also absent from KRNxG7 marrow (Figure 3C; supplemental Figure 3B). Analysis of whole BM gene expression also revealed a down-regulation in several marrow B lymphopoiesis promoting factors, including SDF-1, interleukin-7 (IL-7), and Fms-like tyrosine kinase 3 ligand (Flt3-L; Figures 1J, 3D). Therefore, KRNxG7 mice have impaired marrow B lymphopoiesis attending the defective osteoblasts.
The frequency of c-Kit+Sca1+Lin− cells is not changed in KRNxG7 BM
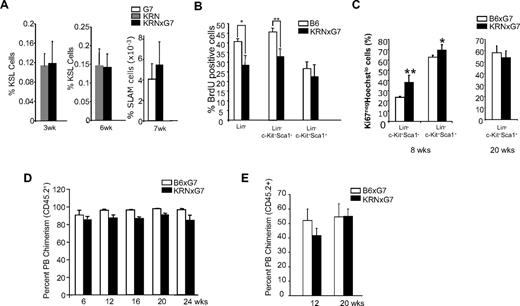
Based on the current understanding that endosteal osteoblasts serve as a HSC niche and maintain the quiescence of the HSCs,3,4,16 the impairment of osteoblast bone-forming capacity in KRNxG7 mice raised the possibility that its role in the HSC niche was also compromised. To determine whether HSC and progenitor cell homeostasis was affected in the absence of functional osteoblasts in chronic inflammatory arthritic environments, we subjected KRNxG7 and control BM, spleen, liver, and PB cells to CD45 (pan hematopoietic marker), c-Kit, Sca-1, and Lin marker staining. The frequency of HSC-enriched KSL cells in the BM was similar between the control and KRNxG7 mice when examined at 3 weeks of age, just before the onset of joint swelling (Figure 4A). There was no increase in BM cellularity at this age (not shown). The KSL frequency was also similar at 6 weeks of age, when all KRNxG7 mice show overt arthritis, irrespective of the method of marrow isolation (Figure 4A; supplemental Figure 2B). The frequency of CD150+CD48−CD41− (SLAM) cells, also enriched for HSCs,26 in the BM was also similar (Figure 4A). As the total BM cellularity was increased (supplemental Figure 2A), there was a net increase in absolute KSL number in KRNxG7 BM despite osteoblast deficiency (supplemental Figure 2C).
Hematopoietic stem cells are not impaired in KRNxG7 mice. (A) Immunophenotypic analyses of HSC containing populations. BM from 3-, 6-, and 7-week-old KRN, B6xG7, and KRNxG7 mice were subjected to FACS analyses for cKit, Sca1, and lineage or CD150, CD48, and CD41 markers. The frequency ± SD of KSL or SLAM is shown on the y-axis (n ≥ 4). (B) Cell-cycle analysis of BM subpopulations. Six- to 8-week-old mice were injected with a single dose of BrdU proportionate to body mass for 2 to 3 hours before death. Different BM fractions were analyzed for BrdU incorporation by flow cytometry. Shown is the mean ± SD of BrdU+ cells for each population for 2 independent experiments (n = 4 or 5 total mice). (C) Quiescent fraction analysis of BM KSL cells. BM cells from KRNxG7 and B6xG7 mice (n = 4 or 5) of different ages were lineage depleted, surface stained for c-Kit and Sca-1, and subjected to intracellular staining for Ki-67 and Hoechst (supplemental Figure 5). Quiescent cells do not express Ki-67 (Ki-67−) and stain low for Hoechst because of their 2N DNA content (vs 4N DNA content of S/G2/M phase cells). Ki-67−Hoechstlow (quiescent cell) fraction of the stem cell–enriched Lin−c-Kit+Sca1+ (KSL) population and the non–stem cell–enriched Lin−cKit+Sca1− population are shown. As expected, KSL cells are more quiescent than Lin−cKit+Sca1− progenitors. However, there is no appreciable difference in quiescent fraction in KRNxG7 KSL cells compared with B6xG7 KSL cells. (D) B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) BM was transplanted into lethally irradiated B6xG7 (CD45.1xCD45.2) recipients. PB was analyzed for donor contribution (CD45.2) every 6 weeks after BM transplantation for 6 months. The percentage ± SD of CD45.2+ chimerism is shown on the y-axis (n = 4/genotype). (E) B6xG7 (CD45.2+) or KRNxG7 (CD45.2+) BM cells (2 × 105) were mixed with B6xG7 (CD45.1+; CD45.2+) competitor BM cells (2 × 105) and injected intravenously into lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipient mice. PB CD45.2+ cells of were analyzed 3 months and 5 months after transplantation. Data represent the average percentages of PB chimerism ± SEM. *P < .05. **P < .01.
Hematopoietic stem cells are not impaired in KRNxG7 mice. (A) Immunophenotypic analyses of HSC containing populations. BM from 3-, 6-, and 7-week-old KRN, B6xG7, and KRNxG7 mice were subjected to FACS analyses for cKit, Sca1, and lineage or CD150, CD48, and CD41 markers. The frequency ± SD of KSL or SLAM is shown on the y-axis (n ≥ 4). (B) Cell-cycle analysis of BM subpopulations. Six- to 8-week-old mice were injected with a single dose of BrdU proportionate to body mass for 2 to 3 hours before death. Different BM fractions were analyzed for BrdU incorporation by flow cytometry. Shown is the mean ± SD of BrdU+ cells for each population for 2 independent experiments (n = 4 or 5 total mice). (C) Quiescent fraction analysis of BM KSL cells. BM cells from KRNxG7 and B6xG7 mice (n = 4 or 5) of different ages were lineage depleted, surface stained for c-Kit and Sca-1, and subjected to intracellular staining for Ki-67 and Hoechst (supplemental Figure 5). Quiescent cells do not express Ki-67 (Ki-67−) and stain low for Hoechst because of their 2N DNA content (vs 4N DNA content of S/G2/M phase cells). Ki-67−Hoechstlow (quiescent cell) fraction of the stem cell–enriched Lin−c-Kit+Sca1+ (KSL) population and the non–stem cell–enriched Lin−cKit+Sca1− population are shown. As expected, KSL cells are more quiescent than Lin−cKit+Sca1− progenitors. However, there is no appreciable difference in quiescent fraction in KRNxG7 KSL cells compared with B6xG7 KSL cells. (D) B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) BM was transplanted into lethally irradiated B6xG7 (CD45.1xCD45.2) recipients. PB was analyzed for donor contribution (CD45.2) every 6 weeks after BM transplantation for 6 months. The percentage ± SD of CD45.2+ chimerism is shown on the y-axis (n = 4/genotype). (E) B6xG7 (CD45.2+) or KRNxG7 (CD45.2+) BM cells (2 × 105) were mixed with B6xG7 (CD45.1+; CD45.2+) competitor BM cells (2 × 105) and injected intravenously into lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipient mice. PB CD45.2+ cells of were analyzed 3 months and 5 months after transplantation. Data represent the average percentages of PB chimerism ± SEM. *P < .05. **P < .01.
c-Kit+Sca1+Lin− cells in KRNxG7 BM cycle normally
It has been suggested that quiescence and restricted proliferation of HSCs are important in maintaining stem cell properties and that osteoblasts maintain HSCs by promoting their quiescence.16,27,28 We therefore next investigated whether KRNxG7 HSCs displayed altered proliferation and/or cell cycling. To this end, KRNxG7 arthritic as well as B6 control mice were subjected to BrdU incorporation and cell-cycle analyses. Specifically, mice were injected with a single dose of BrdU, killed 2 to 3 hours later and BM cells were harvested and subjected to BrdU staining. There was no difference in BrdU labeling in KSL cell populations between control and KRNxG7 BM at 6 to 8 weeks, nor at earlier or later time points (Figure 4B; supplemental Figure 4). We were also unable to detect a decrease in quiescence of KSL cells assessed by Ki-67 and Hoechst staining (Ki-67−Hoechstlow, Figure 4C; supplemental Figure 5). For unknown reasons, however, more mature progenitor fractions (ie, Lin−c-Kit+Sca1− or Lin−) had reduced BrdU+ fraction and increased Ki-67−Hoechstlow, suggesting an overall decrease in cycling (Figure 4B-C). We also compared the expression of several cell-cycle regulators, including the “stemness” gene Bmi in arthritic and control KSL cells, but failed to detect any differences for most genes (supplemental Figure 6). Expression of the cell-cycle inhibitors, p21 and p27 (supplemental Figure 6), was decreased in arthritic KSL cells; however, it has previously been shown that complete deletion of these genes does not alter HSC function and/or pool size or cycling.29,30 KRNxG7 KSL cells did not show any significant differences in annexin V staining pattern from controls (not shown), suggesting that the KRNxG7 KSL cell survival/longevity is not changed. Collectively, these data suggest that KRNxG7 KSL cell cycle is unaltered.
The long-term repopulating potential of KRNxG7 hematopoietic stem cells is not impaired
To assess whether the properties of HSCs are altered in the defective bone-forming osteoblast environments, lethally irradiated B6xG7 (CD45.1xCD45.2) recipients were transplanted with 106 whole BM cells from 6-week-old KRNxG7 (CD45.2xCD45.2) mice. Age-matched B6xG7 (CD45.2xCD45.2) BM cells served as controls. The donor contribution from KRNxG7 (CD45.2xCD45.2) BM was similar to that from B6xG7 (CD45.2xCD45.2) donor cells (Figure 4D). As expected, all blood cell lineages of donor origin were found as evidenced by lineage analyses of PB at 7 months after transplantation, although T-cell generation was deficient because of negative selection (supplemental Figure 7).
Subsequent secondary and tertiary transplantation studies indicated that KRNxG7 HSCs displayed no obvious defects in self-renewal potential (supplemental Figure 7). To rule out the possibility that the long-term repopulation potential of HSCs is diminished in older animals, BM transplantation from 4-month-old KRNxG7 mice was performed. Again, there was no compromise in hematopoietic reconstitution potential of KRNxG7 HSCs from old animals (supplemental Figure 8).
To further confirm that HSC function was not compromised in these mice, we next subjected KRNxG7 HSCs to competitive repopulation studies.31 Specifically, B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) BM cells were mixed with an equal number of B6xG7 (CD45.1xCD45.2) competitor BM cells (2 × 105 cells each) and injected into lethally irradiated (1000 cGy) B6xG7 (CD45.1xCD45.2) recipient mice. PB cells were collected and analyzed for CD45.1 and CD45.2. The contribution from KRNxG7 BM cells was indistinguishable from that of the controls (Figure 4E). Collectively, we conclude that, despite the severe defects in osteoblast bone-forming function in KRNxG7 mice, KRNxG7 BM HSC cycling was similar to controls and long-term repopulating potential of these HSCs was not impaired.
c-Kit+Sca1+Lin− cells are maintained in KRNxG7 spleen
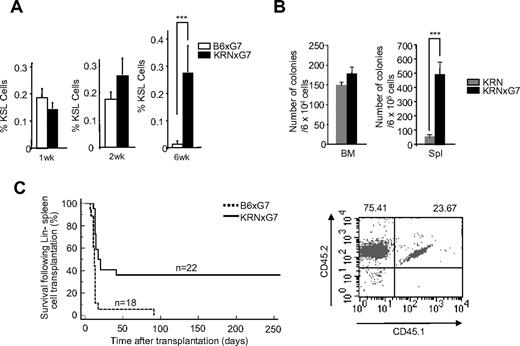
We noticed that most of the KRNxG7 mice had splenomegaly with no obvious hepatomegaly (supplemental Figure 9A-B; and data not shown). Intriguingly, the frequency of KSL cells as well as absolute KSL number was much greater in KRNxG7 spleen compared with B6xG7 spleen at 6 weeks of age (Figure 5A; supplemental Figure 9C; and data not shown). Similar changes in the KSL frequency also occurred in K/BxN spleen (data not shown). To confirm that the apparently high frequency of phenotypic hematopoietic stem/progenitors in the spleen of arthritic mice indeed correlated with an actual increase in functional hematopoietic stem/progenitor cells, we performed hematopoietic replating and transplantation studies. Specifically, unfractionated BM or spleen cells from 6-week-old KRNxG7 and KRN controls were cultured in methylcellulose hematopoietic replating medium. KRNxG7 splenocytes generated a substantially higher number of hematopoietic colonies (Figure 5B). We further transplanted sorted Lin− cells (105 cells/mouse) from KRNxG7 (CD45.2xCD45.2) and control B6xG7 (CD45.2xCD45.2) spleens into lethally irradiated B6xG7 (CD45.1xCD45.2) mice. A majority of the mice that received Lin− cells from control spleen died within 2 weeks, with 100% dying within 3 months. However, a significant number of mice that received Lin− cells from KRNxG7 spleen still survived past 6 months after transplantation (Figure 5C left). A high donor chimerism was obvious when recipients of KRNxG7 splenic Lin− cells were analyzed 3 months after transplantation (Figure 5C right). These replating and transplantation studies corroborate FACS analyses showing that functional hematopoietic progenitors are particularly abundant in KRNxG7 spleen.
Characterization of HSCs and progenitors in KRNxG7 spleen. (A) Spleen from 1-, 2-, and 6-week-old B6xG7, KRN, and KRNxG7 mice were subjected to FACS analyses for cKit, Sca1, and lineage markers. The frequency ± SD of CD45+KSL− is shown on the y-axis (n ≥ 5). (B) BM and spleen cells from 6-week-old KRN and KRNxG7 mice were subjected to hematopoietic replating assay (n = 3). Hematopoietic colonies were counted 7 to 10 days after replating. (C) Lin− spleen cells from B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) were transplanted into lethally irradiated B6xG7 (CD45.1xCD45.2) mice. Survival rate of the recipients is shown on the left. CD45.2+ cells were analyzed 3 months after transplantation. Representative FACS data are shown on the right. ***P < .001.
Characterization of HSCs and progenitors in KRNxG7 spleen. (A) Spleen from 1-, 2-, and 6-week-old B6xG7, KRN, and KRNxG7 mice were subjected to FACS analyses for cKit, Sca1, and lineage markers. The frequency ± SD of CD45+KSL− is shown on the y-axis (n ≥ 5). (B) BM and spleen cells from 6-week-old KRN and KRNxG7 mice were subjected to hematopoietic replating assay (n = 3). Hematopoietic colonies were counted 7 to 10 days after replating. (C) Lin− spleen cells from B6xG7 (CD45.2xCD45.2) or KRNxG7 (CD45.2xCD45.2) were transplanted into lethally irradiated B6xG7 (CD45.1xCD45.2) mice. Survival rate of the recipients is shown on the left. CD45.2+ cells were analyzed 3 months after transplantation. Representative FACS data are shown on the right. ***P < .001.
The high frequency of hematopoietic stem and/or progenitor cells in the KRNxG7 spleen raised a possibility that hematopoietic stem/progenitor cells readily mobilized into the periphery in chronic inflammation. However, KSL cells in the PB and other organs, such as the liver, of KRNxG7 mice were hardly detectable and not significantly different from controls (data not shown). Moreover, the KSL frequency in B6xG7 and KRNxG7 spleen was similar in young mice (1-2 weeks of age). Whereas KSL cells were maintained in KRNxG7 spleen (6 weeks), they decreased greatly in age-matched mice (Figure 5A). We suggest that HSCs are sustained in the KRNxG7 spleen, although the possibility that hematopoietic stem/progenitor cells are continuously mobilized at very low levels to the spleen in chronic inflammatory environments cannot be ruled out at this time.
Discussion
Inflammatory bone loss is associated with several chronic diseases in humans, including RA.32 Such human inflammatory joint disease is generally complicated by systemic osteoporosis, which is particularly severe in KRNxG7 mice. Although focal destruction of bone, within the rheumatoid joint, is the product of aggressive osteoclast recruitment, whether its attendant osteoporosis is the product of accelerated resorption or attenuated formation is less clear. Given the abundance of systemic inflammatory cytokines, the pathogensis of rheumatoid-associated osteoporosis has been assumed to be primarily osteoclastic. We demonstrate, however, that the number of endosteal osteoclasts and confirmatory markers of bone resorption are diminished. Although the osteoblast number per bone surface was similar, the absolute osteoblast number was reduced in KRNxG7 mice because of decreased trabecular bone volume. Given that osteoblast bone-forming activity is also ablated, at least in the KRNxG7 model of RA, the attendant osteoporosis reflects retarded osteogenesis.
Our results extend a growing body of work, examining the relationship between bone homeostasis and hematopoiesis, to a disease model. Our findings that there was a net increase in absolute number of BM HSCs in osteoblast-deficient KRNxG7 mice compared with controls seem to be at odds with the current view of the role of osteoblasts as HSC niche cells. Specifically, transgenic mice expressing a constitutively active PPR (PTH/PTHrP receptors) under the control of the type 1 (I) collagen promoter (col1-caPPR) stimulated osteoblast and increased their number and stromal cells from these mice supported HSCs in culture. These transgenic mice as well as parathyroid hormone treatment of wild-type mice also increased the frequency of KSL cells as well as functional HSCs.3 Moreover, poly I:C treatment of Mx1-Cre+Bmpr1a fx/fx mice resulted in approximately 2-fold increase in the percentage of KSL cells, which correlated with an increase in endosteal osteoblast number.4 Conversely, ablation of osteoblasts by ganciclovir treatment of transgenic mice expressing herpesvirus thymidine kinase (TK) gene under the 2.3 kb of the rat collagen α1 type I promoter (Col2.3ΔTK) resulted in a decrease in absolute number of KSL cells, although the frequency was increased in these mice because of reduced BM cellularity.5 These previous studies will predict that HSCs in KRNxG7 bones would be reduced, which is contrary to what we observe.
We suggest several possible explanations for the apparent discrepancy. First, the remaining osteoblasts in the KRNxG7 mice, although reduced in numbers, may be sufficient to support the HSC maintenance. Second, osteoblast lineage cells are multifunctional. For example, they not only manufacture bone but stimulate osteoclastogenesis via expression of RANKL and macrophage colony-stimulating factor. Indeed, there are circumstances of disassociation of these events as seen in multiple myeloma in which bone formation is arrested but osteoclastogenesis is exuberant, presumably reflecting altered Wnt signaling.33 It is therefore possible that KRNxG7 osteoblasts still support the HSC maintenance, although their bone-forming ability is impaired. In this regard, it is important to note that Zhang et al determined histologically that HSCs in BMPR1a conditional knockout mice were located adjacent to spindle-shaped bone lining cells that express N-cadherin.4 Bone-lining cells are classically regarded as quiescent nonfunctional osteoblasts34 or immature osteoblasts4 and have distinguished morphology from the cuboidal osteoblasts that are responsible for bone formation.34 Lymperi et al have shown that increasing total osteoblasts without increasing N-cadherin+ osteoblasts enhances bone formation without increasing HSCs, highlighting the dissociation between bone formation and HSC maintenance at the cellular level.35
Even though HSCs were maintained normally in KRNxG7, B cells were greatly reduced in these mice. Previous studies have shown that conditional ablation of osteoblasts in Col2.3ΔTK transgenic mice resulted in defects in B lymphopoiesis.5,17 Furthermore, it has been shown that a cell-autonomous defect of Gsα signaling in osteoblasts also impairs marrow B-cell development.18 Our results are more similar to the effects of osteoblast depletion using the Col2.3 TK transgenic system. We find that, similar to Zhu et al,17 preproB, proB and preB precursors are all depleted in KRNxG7 mice in contrast with Wu et al,18 which showed preproB cells are intact. This more severe B-cell depletion in KRNxG7 mice is also reflected by gene expression analysis, which reveals a decrease in not just IL-7 as obtained by Wu et al18 but also of SDF-1 (which is not changed in Wu et al18 ) and of Flt3-L. Intriguingly CLPs are also absent from arthritic BM, suggesting that importance of osteoblasts in marrow B lympophopoiesis occurs higher up in the developmental hierarchy than previously determined, before B-cell commitment. It is possible that the decreased cycling of Kit+Sca−Lin− cells in KRNxG7 BM is the result of undefined progenitors in the lymphoid lineage. Previous studies have suggested that immunization, infection, and inflammatory cytokines can mobilize B-cell precursors into the periphery.36-38 However, it is at the moment unclear if and where B lymphopoiesis might be relocated to in the KRNxG7 mice.
Taken together, osteoblast determinants involved in bone formation versus B lymphopoiesis versus hematopoietic stem supporting activity could be distinct and uncoupled. Alternatively, although osteoblasts may be obligatory for B-cell development and bone formation, additional HSC niche cells, such as endothelial or reticular cells,39,40 could compensate for the osteoblast defects in maintaining HSC integrity in inflammatory environments of KRNxG7 mice. To this end, previous studies demonstrate that endothelial cells of hematopoietic tissues, such as BM or extramedullary organs, express cell surface molecules, including E-selectin and VCAM-1. Intriguingly, these molecules are not expressed in quiescent endothelium of nonhematopoietic tissues but become up-regulated in inflammation.41,42 Thus, it will be particularly important to know whether the accumulation of HSCs and progenitors that we see in the spleens of arthritic mice is associated with changes in the endothelial niche in chronic inflammation. Moreover, studies distinguishing the requirements of osteoblast versus endothelial niche in HSC maintenance in normal versus pathologic conditions should be addressed thoroughly in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR055923-01, F.L.; AR032788 and AR046523, S.L.T.), and the National Heart, Lung, and Blood Institute (HL63736 and HL55337, K.C.).
National Institutes of Health
Authorship
Contribution: C.P., Y.D.M., and K.A.O. characterized hematopoietic phenotype; H.Z., C.P., and Y.D.M. characterized bone phenotype; X.T. performed in vitro osteoblast culture and experiments; P.M.A. provided K/BxN and KRNxG7 mice; P.M.A., F.L., and S.L.T. discussed and edited the manuscript; and C.P., Y.D.M., K.A.O., H.Z., S.L.T., and K.C. planned the project, interpreted the data, prepared the figures, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kyunghee Choi, Department of Pathology and Immunology, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO 63110; e-mail: kchoi@pathology.wustl.edu.
References
Author notes
Y.D.M. and C.P. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal