In this issue of Blood, Hoogenkamp and colleagues describe chromatin modifications associated with activation of the hematopoietic transcription factor Pu.1 gene in the earliest stages of blood cell production in differentiating ES cells. Surprisingly, the Runx1 transcription factor is a major but transient player required only in this opening act.
Pu.1 is a well-known pivotal factor in adult hematopoiesis. Complete deficiency of Pu.1 results in late gestational lethality, with an absence of fetal liver granulocytes, macrophages, and B lymphocytes.1,2 The Runx1 transcription factor is an upstream regulator of Pu.1 and is required for Pu.1 expression.3 Pu.1, in turn, is pivotal for the expression of the colony-stimulating factor 1 receptor (Csfr1), which is required for macrophage differentiation. In the adult bone marrow, hematopoietic stem cells, common lymphocyte progenitors (CLPs), and common myeloid progenitors (CMPs) all express Pu.1. Varying levels of Pu.1 in CMPs influence cell fate decisions,4 with high-expressing CMPs efficient in granulocyte/macrophage differentiation and low-expressing CMPs committed to the megakaryocyte/erythroid lineage. Pu.1 is down-regulated in the transition of CLP to the B-lymphoid lineage and is extinguished in the T-lymphoid lineage. While the Pu.1 cis regulatory elements have been extensively characterized in vivo,3,5 due to limiting numbers of hematopoietic progenitors available in these models, no studies until now have been able to examine the epigenetic changes accompanying the onset of Pu.1 expression. In this issue of Blood, the report by Hoogenkamp et al is the first to correlate chromatin changes in Pu.1 regulatory elements with hematopoietic commitment.6
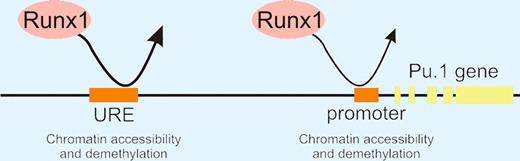
“Hit and run” model of Runx1 action in the unfolding of chromatin at Pu.1 regulatory regions in early hematopoietic development.
“Hit and run” model of Runx1 action in the unfolding of chromatin at Pu.1 regulatory regions in early hematopoietic development.
By focusing on the Pu.1 gene in readily available embryonic stem (ES) cells undergoing hematopoietic commitment, the authors were able to identify the stage at which the regulatory regions of Pu.1 assume an open chromatin configuration. DnaseI analysis on chromatin obtained from an ES cell differentiation series showed that the Pu.1 promoter and the upstream regulatory element (URE) were beginning to become accessible just prior to the hemangioblast stage and before the onset of Pu.1 expression. Chromatin in the Pu.1 promoter and URE was found to reach a similar unfolded structure in hemangioblasts as found in macrophages. Moreover, a selective demethylation (another indication of chromatin reorganization) was found to occur within the Pu.1 promoter and URE in a majority of cells in the hemangioblast population. Neither of the assays showed priming of the Csfr1 regulatory regions in hemangioblast chromatin. Thus, the Pu.1 gene assumes an open chromatin configuration early in development. Of note, this is all occurring in the absence of elevated levels of active or bivalent chromatin marks (Histone 3 [H3] protein methylated at lysine 27 or lysine 4). As shown by others for key early developmental genes in ES cells,7 chromatin domains containing a predominance of H3 lysine 27 methylation indicate negative regulation or silencing of gene transcription, whereas H3 lysine 4 methylation indicates positive regulation or activation of gene transcription. It will be interesting in the future to determine whether other histone marks are associated with events leading to the expression of the Pu.1 gene.
Most interestingly, the authors show that Runx1 plays a critical role in chromatin unfolding around the Pu.1 regulatory elements. Unlike wild-type ES cells, they found that Dnase1 accessibility at the Pu.1 regulatory elements did not change in Runx1−/− ES cell differentiation cultures. Curiously, DMS footprinting (which detects stable interactions) showed no protection at the Pu.1 URE in Runx1+/+ hemangioblasts. ChIp studies on Runx1−/− ES cells engineered to express Runx1 upon Dox exposure (iRunx ES cells) suggested that Runx1 only transiently interacts with Pu.1 regulatory elements during the hemangioblast stage. Using an elegant method in which Runx1−/− ES cells were engineered with a Dox-inducible Runx1-Dam methylase fusion gene, the authors show that indeed a transient (“hit-and-run”) interaction occurred. The Runx1-Dam protein left its identifiable mark on the Pu.1 URE and promoter, thus confirming an interaction. All together, these studies show that Runx1 is the major player contributing to the opening of the chromatin surrounding the Pu.1 gene, licensing it for future expression. In this opening act, Runx1 does not assemble into a detectable stable transcription factor complex, but instead it takes the lead to begin the gradual changes in chromatin structure that, upon reaching a threshold, trigger Pu.1 expression and a dramatic program of hematopoietic lineage differentiation.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal