Abstract
Previous studies in the mouse have shown that high levels of α-globin gene expression in late erythropoiesis depend on long-range, physical interactions between remote upstream regulatory elements and the globin promoters. Using quantitative chromosome conformation capture (q3C), we have now analyzed all interactions between 4 such elements lying 10 to 50 kb upstream of the human α cluster and their interactions with the α-globin promoter. All of these elements interact with the α-globin gene in an erythroid-specific manner. These results were confirmed in a mouse model of human α globin expression in which the human cluster replaces the mouse cluster in situ (humanized mouse). We have also shown that expression and all of the long-range interactions depend largely on just one of these elements; removal of the previously characterized major regulatory element (called HS −40) results in loss of all the interactions and α-globin expression. Reinsertion of this element at an ectopic location restores both expression and the intralocus interactions. In contrast to other more complex systems involving multiple upstream elements and promoters, analysis of the human α-globin cluster during erythropoiesis provides a simple and tractable model to understand the mechanisms underlying long-range gene regulation.
Introduction
Analysis of the human globin gene clusters and their associated mutations that cause thalassemia have elucidated many of the general principles underlying the regulation of mammalian gene expression. In many gene clusters there is evidence that remote regulatory elements are essential for high-level, tissue-specific expression and that these elements physically interact with their cognate promoters. These findings pose the general questions of how such sequences interact with their promoters and how they influence gene expression. In this study, we have evaluated the human α-globin cluster as a model to understand the mechanisms underlying long-range regulation of gene expression.
In all mammals studied to date, the α-globin genes appear to be regulated by one or more of 4 remote, multispecies conserved sequences (MCS-R1 to R4) corresponding to erythroid-specific DNase1 hypersensitive sites (DHSs) lying upstream of the cluster.1 In the mouse α-globin cluster, we have previously shown that several upstream elements physically interact with the α-globin promoters late in erythropoiesis as the α-globin genes are activated.2 However, orthologous upstream elements may play different roles in different species and the mouse does not appear to be an ideal model for the human α gene cluster. Deletion of the most highly conserved, orthologous element (MCS-R2) leads to almost complete down-regulation of human α gene expression but to only a modest reduction (∼50%) in mouse α-globin synthesis.3 This suggests that other upstream elements may interact and/or enhance α-globin expression independent of MCS-R2 in the mouse,3 possibly including a hypersensitive site (HS −12) that has no human equivalent and may be rodent specific.4
A better model is provided by a “humanized” mouse, in which 87 kb of the mouse α cluster is replaced in situ with 117 kb of the human cluster. This replaces the mouse genes and regulatory elements with sequences that contain the human elements and genes in their normal relative positions. The human MCS-R1 to -R4 correspond to 4 DHSs (HS −48, HS −40, HS −33, HS −10; Figure 1A), of which MCS-R2 (HS −40) appears to be the major remote, distal enhancer of adult α gene expression.5 The influence (if any) of the other elements (HS −48, HS −33, and HS −10) is not clear.1 Here we have assessed in detail the physical interactions of these elements with each other and with the α-globin promoter and shown similar interactions in both normal primary human erythroblasts and erythroblasts from the humanized mouse. Furthermore, we demonstrate that in the absence of HS −40 in the mouse model, in which there is no α gene expression, none of the remaining upstream elements interacts with the α-globin promoter. When HS −40 is reinserted at an ectopic site 3′ to the α genes, both the interactions and expression are reinstated. Therefore, in the human α-globin cluster both physical interaction and enhancement of expression appear to depend predominantly on a single upstream element (MCS-R2/HS −40). This provides a relatively simple model for addressing the general question of how remote upstream regulatory elements interact with their cognate promoters in vivo.
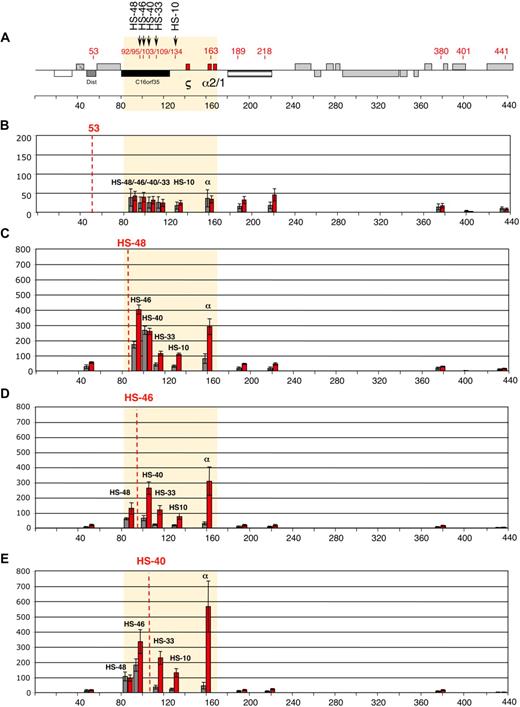
Intrachromosomal interactions, between the human α-globin genes and conserved upstream elements. (A) Chromosomal organization of the human α-globin locus annotated as previously described.23 Red numbers indicate the points (coordinates) analyzed by q3C. q3C assays were performed using HindIII-digested, fixed chromatin from primary T lymphocytes (gray bars) and primary erythroblasts (red bars), generating restriction fragments of 3 to 5 kb. This allowed us to use q3C to evaluate all interactions between the α-globin genes and a variety of points along the α-globin cluster, including all the remote upstream elements. Twelve restriction fragments located within (HS −48, HS −46, HS −40, HS −33, and HS −10 and the α-globin genes) or outside (fragments 53, 189, 218, 380, 401, and 441) the α-globin locus were chosen to determine the locus conformation (A). The shaded area corresponds to the region containing all sequences that interact with the α-globin genes (B-J). The bar chart (y-axis) shows the enrichment of PCR product (%) normalized to the enrichment within the human Ercc3 gene (= 100%). This provides an internal, genomic control for the cross-linking procedure and any general changes in nuclear or chromatin structure.24 These independent graphs represent a measure of the association between the points indicated (x-axis) to the anchor points (B) “fragment 53,” (C) HS −48, (D) HS −46, (E) HS −40, (F) HS −33, (G) HS −10, (H) α-globin genes, (I) “fragment 189,” and (J) “fragment 380.” For each new anchor point, a new Taqman probe and reverse primer were designed and thus results can be compared only within, but not between, each set of graphs. The resolution of this 3C assay can be estimated by comparing “positive” and “negative” restriction fragments located at a similar distance to their target. For example, the locus position 189 is located 26 kb downstream of α-globin and does not interact, whereas HS −10, located 26 kb upstream of α-globin, shows a 3-fold enrichment in erythroid cells (H). Data shown represent the average of a least 3 independent experiments using Taqman/real-time PCR. Error bars denote SEM. Each PCR was performed several times and averaged. Signals were normalized to the total amount of DNA used, estimated with an amplicon located within a HindIII fragment (Table 1). Coordinates of the points analyzed are indicated on the x-axis.
Intrachromosomal interactions, between the human α-globin genes and conserved upstream elements. (A) Chromosomal organization of the human α-globin locus annotated as previously described.23 Red numbers indicate the points (coordinates) analyzed by q3C. q3C assays were performed using HindIII-digested, fixed chromatin from primary T lymphocytes (gray bars) and primary erythroblasts (red bars), generating restriction fragments of 3 to 5 kb. This allowed us to use q3C to evaluate all interactions between the α-globin genes and a variety of points along the α-globin cluster, including all the remote upstream elements. Twelve restriction fragments located within (HS −48, HS −46, HS −40, HS −33, and HS −10 and the α-globin genes) or outside (fragments 53, 189, 218, 380, 401, and 441) the α-globin locus were chosen to determine the locus conformation (A). The shaded area corresponds to the region containing all sequences that interact with the α-globin genes (B-J). The bar chart (y-axis) shows the enrichment of PCR product (%) normalized to the enrichment within the human Ercc3 gene (= 100%). This provides an internal, genomic control for the cross-linking procedure and any general changes in nuclear or chromatin structure.24 These independent graphs represent a measure of the association between the points indicated (x-axis) to the anchor points (B) “fragment 53,” (C) HS −48, (D) HS −46, (E) HS −40, (F) HS −33, (G) HS −10, (H) α-globin genes, (I) “fragment 189,” and (J) “fragment 380.” For each new anchor point, a new Taqman probe and reverse primer were designed and thus results can be compared only within, but not between, each set of graphs. The resolution of this 3C assay can be estimated by comparing “positive” and “negative” restriction fragments located at a similar distance to their target. For example, the locus position 189 is located 26 kb downstream of α-globin and does not interact, whereas HS −10, located 26 kb upstream of α-globin, shows a 3-fold enrichment in erythroid cells (H). Data shown represent the average of a least 3 independent experiments using Taqman/real-time PCR. Error bars denote SEM. Each PCR was performed several times and averaged. Signals were normalized to the total amount of DNA used, estimated with an amplicon located within a HindIII fragment (Table 1). Coordinates of the points analyzed are indicated on the x-axis.
Methods
Primary cells and cell lines
Human erythroid progenitors were derived from peripheral blood mononuclear cells of healthy subjects and grown in 2-step cultures.6,7 Human activated T lymphocytes were obtained from buffy coats by culture for 3 days in RPMI/20% fetal calf serum in the presence of 2% phytohemagglutinin M and 20 U/mL interleukin-2. Mouse primary erythroid cells were obtained from the spleens of acetylphenylhydrazine-treated mice,8 purified for Ter119-positive cells by auto–magnetic-activated cell sorting; Ter119-negative cells from the same source served as a nonerythroid control.
Chromosome conformation capture
The chromosome conformation capture assay (3C) was performed as described previously using the same set of controls.2 Human and mouse primers and probes used are shown in Table 1.
Oligonucleotide primers and probes (q3C)
| Designation . | Sequence . |
|---|---|
| Human primers | |
| 53 H3 P | TTTCTGGAGGAGGACACAACTGATTTCCC |
| HS48 H3 P | CAGGCCAAACATAGCAACACCATCCC |
| HS46 H3 P | AGTCTAGTCCCCTCAGCTTTTAGCAT |
| HS40 H3 P | CCTGATTGCTAAAGCCTTTTCACAGCTGG |
| HS33 H3 P | CAAGATGTGCAAGCCCACTGTGAT |
| HS10 H3 P | TGGTCTTTTGCGTTTGGTGACAGTCAA |
| a2 H3 P | CGGGAGCGATCTGGGTCGAGG |
| 189 H3 P | TCTCTCGGATCTGAATGAACCCCAAGTG |
| 380 H3 P | TCAATGCAAAGAAAACCATAGCCAATT |
| 153 H3 R | CACTATATTGGAAGACCATGCAACTC |
| 53 H3 A F | AAAGAAGGATGTGTCCAGCTCATC |
| HS48 H3 A R | AAGTGGACTGTGAAAGAAATAAGGAAA |
| HS46 H3 A R | TCACAGGTCCTGTCTAGTTCTAAGGTT |
| HS40 H3 A R | CATAAAGACACCACATGCACTAGCT |
| HS33 H3 A R | CTGGTATTTCTGCCATTTGGAAA |
| HS10 H3 A F | AACAGATGCCAGCCAAACAGA |
| 189 H3 A R | ACCTTGGTCTCCATGACAATGAC |
| 380 H3 A R | AGTTCGTTTTCCAGCAGTTTGACTA |
| a2 H3 A R | GAGAGGAAGGCGCCATCTC |
| 53 H3 F | AATCAGTTGTGTCCTCCTCCAGAA |
| HS48 H3 R | AATAAGGAAACTGGGATGGTGTTG |
| HS46 H3 R | GATGCTTTCTCAGATGCTAAAAGCT |
| HS40 H3 F | AGCTGTGAAAAGGCTTTAGCAATC |
| 109 H3 F | CCCTTATCCCAAACACTTAAACTTTC |
| HS33 H3 R | CTGGTATTTCTGCCATTTGGAAA |
| HS10 H3 F | GACTGTCACCAAACGCAAAAGAC |
| a2 H3 R | GCCGCTCACCTTGAAGTTGA |
| 189 H3 R | ACTTCGGTGGCAAGTTACACTTG |
| 218 H3 R | ACAGTCTTCGATAGAGTTTGAAAGCTT |
| 380 H3 R | TCTTTGCATTGATTCTTGTTAAGCTT |
| 401 H3 F | GCGATGACGCGGCACTT |
| 441 H3 F | AAACAGCCAAGTGGATACATTCAA |
| xpb H3 F | CGGTGAGGTGAGTTTGTGGAAT |
| xpb H3 R | AGGATCTCTGTTTAATGGAAAAGCTT |
| xpb H3 P | AAGGATGAAGGCGTGATCCGACTCTG |
| hPr/Ex1 F | GGGCCGGCACTCTTCTG |
| hPr/Ex1 R | GGCCTTGACGTTGGTCTTGT |
| hPr/Ex1 P | CCCACAGACTCAGAGAGAACCCACCATG |
| Mouse primers | |
| 24 H3 F | ATGGGACAATGGAGGGCTAAG |
| 55 H3 R | TCTATTAGGGATGGCACTTTACGAA |
| 72 H3 R | CCCCTGTGGGAAAGACAGAAA |
| 78 H3 F | AGACAAGGAAATGGAAGAGACATCA |
| HS31 H3 F | AGCTGTGCCTGGCAGATGAC |
| HS29 H3 F | AGGAAGAAGAGGTTCAACAAGCAT |
| HS26 H3 R | GAATCTCCATCTCCAAGGG |
| Zeta H3 F | ACAACGTGGAGAGGACTGAGAGAT |
| 174 H3 F | AATGGTTGCCTTTTAAAGACATTTGT |
| a2 H3 R | GTGGCCATGAGCCAAAGAAT |
| a2 H3 P | CCTCCAGGTACCATAAATCTCAGACTTG |
| xpb H3 F | CTTCTGCTACAAAGCGCTGACTT |
| xpb H3 R | TGCCCTCCCTGAAAATAAGGA |
| xpb H3 P | TGCACCCTGCTTTAGTGGCCTTGTAGG |
| mPr/Ex1Fw | TGGAGGGCATATAAGTGCTACTTG |
| mPr/EX1Rev | TGCTTTTGTCTTCCCCAGAGA |
| mPr/Ex1P | TGCAGGTCCAAGACACTTCTGATTCTGACA |
| Designation . | Sequence . |
|---|---|
| Human primers | |
| 53 H3 P | TTTCTGGAGGAGGACACAACTGATTTCCC |
| HS48 H3 P | CAGGCCAAACATAGCAACACCATCCC |
| HS46 H3 P | AGTCTAGTCCCCTCAGCTTTTAGCAT |
| HS40 H3 P | CCTGATTGCTAAAGCCTTTTCACAGCTGG |
| HS33 H3 P | CAAGATGTGCAAGCCCACTGTGAT |
| HS10 H3 P | TGGTCTTTTGCGTTTGGTGACAGTCAA |
| a2 H3 P | CGGGAGCGATCTGGGTCGAGG |
| 189 H3 P | TCTCTCGGATCTGAATGAACCCCAAGTG |
| 380 H3 P | TCAATGCAAAGAAAACCATAGCCAATT |
| 153 H3 R | CACTATATTGGAAGACCATGCAACTC |
| 53 H3 A F | AAAGAAGGATGTGTCCAGCTCATC |
| HS48 H3 A R | AAGTGGACTGTGAAAGAAATAAGGAAA |
| HS46 H3 A R | TCACAGGTCCTGTCTAGTTCTAAGGTT |
| HS40 H3 A R | CATAAAGACACCACATGCACTAGCT |
| HS33 H3 A R | CTGGTATTTCTGCCATTTGGAAA |
| HS10 H3 A F | AACAGATGCCAGCCAAACAGA |
| 189 H3 A R | ACCTTGGTCTCCATGACAATGAC |
| 380 H3 A R | AGTTCGTTTTCCAGCAGTTTGACTA |
| a2 H3 A R | GAGAGGAAGGCGCCATCTC |
| 53 H3 F | AATCAGTTGTGTCCTCCTCCAGAA |
| HS48 H3 R | AATAAGGAAACTGGGATGGTGTTG |
| HS46 H3 R | GATGCTTTCTCAGATGCTAAAAGCT |
| HS40 H3 F | AGCTGTGAAAAGGCTTTAGCAATC |
| 109 H3 F | CCCTTATCCCAAACACTTAAACTTTC |
| HS33 H3 R | CTGGTATTTCTGCCATTTGGAAA |
| HS10 H3 F | GACTGTCACCAAACGCAAAAGAC |
| a2 H3 R | GCCGCTCACCTTGAAGTTGA |
| 189 H3 R | ACTTCGGTGGCAAGTTACACTTG |
| 218 H3 R | ACAGTCTTCGATAGAGTTTGAAAGCTT |
| 380 H3 R | TCTTTGCATTGATTCTTGTTAAGCTT |
| 401 H3 F | GCGATGACGCGGCACTT |
| 441 H3 F | AAACAGCCAAGTGGATACATTCAA |
| xpb H3 F | CGGTGAGGTGAGTTTGTGGAAT |
| xpb H3 R | AGGATCTCTGTTTAATGGAAAAGCTT |
| xpb H3 P | AAGGATGAAGGCGTGATCCGACTCTG |
| hPr/Ex1 F | GGGCCGGCACTCTTCTG |
| hPr/Ex1 R | GGCCTTGACGTTGGTCTTGT |
| hPr/Ex1 P | CCCACAGACTCAGAGAGAACCCACCATG |
| Mouse primers | |
| 24 H3 F | ATGGGACAATGGAGGGCTAAG |
| 55 H3 R | TCTATTAGGGATGGCACTTTACGAA |
| 72 H3 R | CCCCTGTGGGAAAGACAGAAA |
| 78 H3 F | AGACAAGGAAATGGAAGAGACATCA |
| HS31 H3 F | AGCTGTGCCTGGCAGATGAC |
| HS29 H3 F | AGGAAGAAGAGGTTCAACAAGCAT |
| HS26 H3 R | GAATCTCCATCTCCAAGGG |
| Zeta H3 F | ACAACGTGGAGAGGACTGAGAGAT |
| 174 H3 F | AATGGTTGCCTTTTAAAGACATTTGT |
| a2 H3 R | GTGGCCATGAGCCAAAGAAT |
| a2 H3 P | CCTCCAGGTACCATAAATCTCAGACTTG |
| xpb H3 F | CTTCTGCTACAAAGCGCTGACTT |
| xpb H3 R | TGCCCTCCCTGAAAATAAGGA |
| xpb H3 P | TGCACCCTGCTTTAGTGGCCTTGTAGG |
| mPr/Ex1Fw | TGGAGGGCATATAAGTGCTACTTG |
| mPr/EX1Rev | TGCTTTTGTCTTCCCCAGAGA |
| mPr/Ex1P | TGCAGGTCCAAGACACTTCTGATTCTGACA |
Oligonucleotide sequences used in real-time PCR.
P indicates probe; R, reverse; A, anchor; and F, forward.
ES cell genetic manipulation and generation of humanized mouse models
The generation and characterization of the humanized mouse model with functional human α-globin genes and a second model with HS −40 deleted have been previously described.9 To create a mouse model in which the HS −40 was positioned at an ectopic location, the previously described BAC clone containing the human α-globin regulatory domain with the HS −40 deletion (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article) was further engineered by recombineering in Escherichia coli DY380 cells. This enabled the insertion of a cassette consisting of a PGK-zeocyin resistance gene (flanked by frt-F5 recombination sites) linked to a 1.1-kb HS −40 sequence into a position downstream of the α-globin genes (sequence position 169470 mapped on HG18). This newly modified BAC clone was then co-electroporated with a Cre expression plasmid into the double-targeted mouse embryonic stem (ES) cell line 2/H-1 with loxP and lox511 sites at the Dist (c16orf8) and theta (HbQ) genes. Cre recombination, as previously described,9 results in genomic replacement of the mouse sequence by human BAC sequence between positions delineated by loxP and lox511 sites and reconstruction of a hypoxanthine phosphoribosyl transferase (Hprt) minigene to allow selection of cells in medium containing hypoxanthine, aminopterin, and thymidine (HAT) (supplemental Figure 1B). From one electroporation, a single HAT-resistant clone was obtained that was also resistant to zeocyin and puromycin as expected. Cells of this HAT-resistant clone were subsequently electroporated with a FLPe expression vector to excise the Hprt, puromycin, and zeocyin resistance genes via their flanking frt, frt-F3, and frt-F5 sites, respectively (supplemental Figure 1B). Hprt-negative cells were selected in 6-thioguanine and resistant clones recovered at high frequency that were also puromycin sensitive and zeocyin sensitive. One such clone was checked to confirm the expected DNA sequence rearrangements. This was subsequently used to generate chimeras. Male chimeras were backcrossed to C57BL/6 females and agouti offspring genotyped to confirm germline transmission.
Genomic DNA analysis across the entire humanized insert and RNase protection assays were performed as previously described.9 In addition, the 3′HS −40 construct was also analyzed by sequencing and by polymerase chain reaction (PCR) using an amplicon spanning the junction of insertion (primer sequences: Fw, GCAATATCCGCTACAGCCAAAT; Rev, GAGCTCTTGGGCAGAGATATGG; probe, CGGATCCCCCAACAAGAAAACCAGC). Study protocols received ethical approval from Oxford University Ethical Review Panel and all animal work was carried out under Home Office License.
Results
3C analysis of regulatory elements and promoters in the human α-globin cluster
Here we have analyzed, for the first time, all interactions between the human α-globin regulatory elements and the α-globin promoters in primary human erythroid and nonerythroid cells using a quantitative 3C assay (q3C).2 HindIII restriction fragments containing HS −48, HS −46, HS −40, HS −33, HS −10, and the α genes, or regions outside the α-globin locus, were chosen to determine the locus conformation (Figure 1A). We initially evaluated q3C between the α genes and various fragments along the α-globin cluster (Figure 1H), demonstrating that the human α genes specifically interact with the upstream elements in erythroid cells. They also interact with HS −46, which is not an erythroid element but corresponds to a constitutive HS that binds CTCF in all tested cell types (Marco de Gobbi, unpublished data, October 2009). These data demonstrate that despite differences in the functioning and positions of these elements in human and mouse clusters, the overall architecture of the interactions is maintained in the 2 species.2
To examine these interactions in detail, each restriction fragment was used as a bait (anchor fragment) in q3C experiments, using a unique Taqman probe for each fragment. In total, 120 primer combinations have been used across 400 kb of DNA encompassing the α-globin gene cluster (Figure 1A). Regions located outside the globin locus do not interact with the α cluster either in erythroblasts or T lymphocytes (Figure 1B,I-J). By contrast, all fragments containing the remote regulatory elements and the α-globin genes come into close spatial proximity with each other in erythroblasts, whereas such interactions are relatively weak (or absent) in T lymphocytes (Figure 1C-H). Not all elements interact equally. For example, little interaction was seen between HS −40 and HS −48 in erythroid cells (Figure 1C,E).
3C analysis of regulatory elements and promoters in the humanized mouse
Having determined the interactions within the cluster in normal human erythroblasts, we determined whether these were replicated in the humanized mouse (Figure 2A). Expression of the human α genes is approximately 50% that of the mouse genes in these mice.9 Using q3C, these interactions were analyzed in erythroid cells from normal humanized mice, both in homozygotes (supplemental Figure 2) and heterozygotes (Figure 2C). This showed exactly the same pattern as seen in primary human erythroblasts (compare Figure 2C with Figure 1H). The control mouse cluster shows the expected interactions between the upstream elements with the promoters (Figure 2D) as seen previously.2 (Note that HS −10 could not be analyzed because of a SNP located at a HindIII site in these mice.) This humanized mouse therefore provides a suitable model to study this aspect of human α-globin regulation.
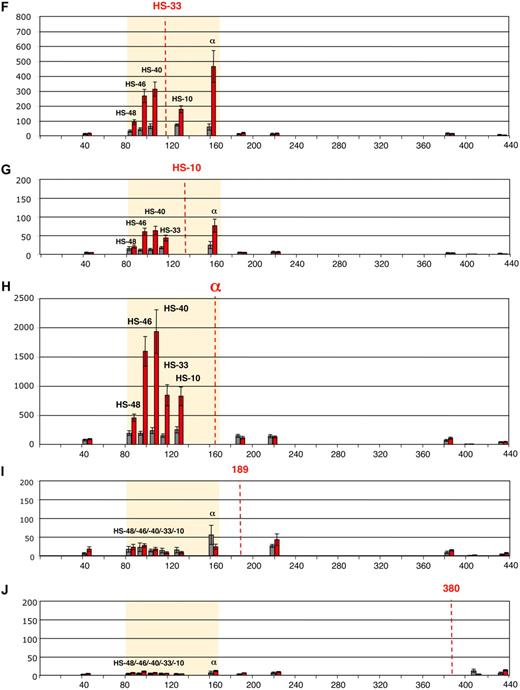
Deletion or ectopic reinsertion of HS −40 influences the interactions between the remote HS and the α-globin genes. Intrachromosomal interactions involving human remote regulatory sequences and the human α-globin genes in normal, ΔHS −40, and 3′HS −40 humanized alleles. (A) Chromosomal organization of the humanized α-globin locus. (B) Chromosomal organization of the mouse α-globin locus. Red and blue numbers indicate, respectively, the points (coordinates) of the human and mouse primers analyzed by q3C. q3C assays were performed using HindIII-digested, fixed chromatin from primary Ter119− (blue graph bars) and Ter119+ (red graph bars) cells from spleens obtained from phenylhydrazine-treated mice. The bar chart (y-axis in panels C-H) shows the enrichment of PCR product (%) normalized to the enrichment within the mouse Ercc3 gene (= 100%), and thus explains the scale difference compared with Figure 1, normalized on human Ercc3. These independent graphs represent a measure of the association between the remote regulatory sequences (x-axis) to the α-globin genes (anchor fragment) in normal-humanized locus (C), endogenous mouse locus (D), ΔHS −40 humanized locus (E), endogenous mouse locus (F), 3′HS −40 humanized locus (G), and endogenous mouse locus (H). Results obtained with endogenous mouse locus are in agreement with those observed previously.2 Data are represented as in Figure 1. To the left are images representing the proposed configurations interpreted from these data.
Deletion or ectopic reinsertion of HS −40 influences the interactions between the remote HS and the α-globin genes. Intrachromosomal interactions involving human remote regulatory sequences and the human α-globin genes in normal, ΔHS −40, and 3′HS −40 humanized alleles. (A) Chromosomal organization of the humanized α-globin locus. (B) Chromosomal organization of the mouse α-globin locus. Red and blue numbers indicate, respectively, the points (coordinates) of the human and mouse primers analyzed by q3C. q3C assays were performed using HindIII-digested, fixed chromatin from primary Ter119− (blue graph bars) and Ter119+ (red graph bars) cells from spleens obtained from phenylhydrazine-treated mice. The bar chart (y-axis in panels C-H) shows the enrichment of PCR product (%) normalized to the enrichment within the mouse Ercc3 gene (= 100%), and thus explains the scale difference compared with Figure 1, normalized on human Ercc3. These independent graphs represent a measure of the association between the remote regulatory sequences (x-axis) to the α-globin genes (anchor fragment) in normal-humanized locus (C), endogenous mouse locus (D), ΔHS −40 humanized locus (E), endogenous mouse locus (F), 3′HS −40 humanized locus (G), and endogenous mouse locus (H). Results obtained with endogenous mouse locus are in agreement with those observed previously.2 Data are represented as in Figure 1. To the left are images representing the proposed configurations interpreted from these data.
Interactions when HS −40 is deleted
We next studied humanized mice (heterozygotes) in which HS −40 had been deleted (ΔHS −40). In these mice, α-globin expression is severely down-regulated to less than 2% of mouse α gene expression9 (see also Figure 5). Given such changes in gene expression (which may also be associated with changes in chromatin accessibility), it was important to assess the cleavage of chromatin by endonuclease (HindIII) at critical sites because changes in the 3C assay might reflect changes in endonuclease accessibility rather than looping. Using a quantitative PCR assay, we demonstrated equal HindIII cleavage efficiency at critical sites in normal cells and cells from which HS −40 had been deleted (supplemental Figure 3).
The chromosome conformation assumed in the absence of HS −40 is shown in Figure 2E. In the absence of HS −40, no stable interactions occur between the remaining upstream elements (HS −48, HS −46, and HS −33) and the α genes. This indicates that HS −40 plays a crucial role in mediating the long-range interactions between the upstream elements and the α promoter. To ensure that the changes in chromosome looping were not simply due to differences in the stages of erythroid maturation between experiments, this was assessed by morphology and fluorescence-activated cell sorting analysis (Figure 3). As a further control, we have shown that interactions at the endogenous mouse α-globin cluster (the other allele in humanized mouse heterozygotes) are intact in all situations (Figure 2F).
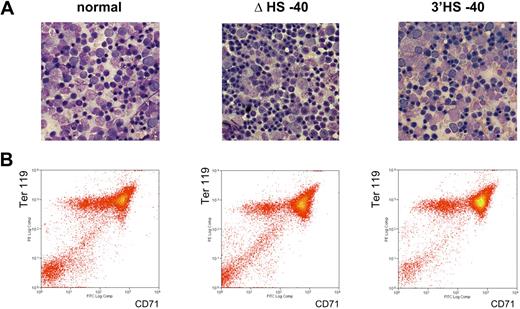
Erythroid cells analyzed in this study are all at similar stages of differentiation. (A) May-Grünwald Giemsa (MGG)–stained touch preparations from the spleens of phenylhydrazine-treated adult mice. Images were captured using a Nikon eclipse E600 microscope with a Nikon DXM1200C digital camera using NIS elements BR2.30 SP4 imaging software (all from Nikon UK). Original magnification 60×/1.40 NA oil objective (Nikon Japan) for all panels. (B) Flow cytometric analyses of mouse primary erythroblasts. Ter119+ erythroid cells were auto–magnetic-activated cell sorting–purified from the spleen of phenylhydrazine-treated adult mice and restained with anti-CD71 to show the equivalent maturation stages in all mice examined.
Erythroid cells analyzed in this study are all at similar stages of differentiation. (A) May-Grünwald Giemsa (MGG)–stained touch preparations from the spleens of phenylhydrazine-treated adult mice. Images were captured using a Nikon eclipse E600 microscope with a Nikon DXM1200C digital camera using NIS elements BR2.30 SP4 imaging software (all from Nikon UK). Original magnification 60×/1.40 NA oil objective (Nikon Japan) for all panels. (B) Flow cytometric analyses of mouse primary erythroblasts. Ter119+ erythroid cells were auto–magnetic-activated cell sorting–purified from the spleen of phenylhydrazine-treated adult mice and restained with anti-CD71 to show the equivalent maturation stages in all mice examined.
The effect of reinserting HS −40 at an ectopic site
Removal of HS −40 abolished all interactions. To determine whether these interactions could be restored by HS −40 alone, a humanized mouse was generated from ES cells with HS −40 reinserted into the α cluster, but at an ectopic site, 3′ to the α genes (3′HS −40, Figure 4A). To accomplish this, a previously described9 BAC clone, containing the human α-globin regulatory domain from which HS −40 had been deleted, was further modified by recombineering to insert a 1.1-kb HS −40 sequence linked to this new position in the same chromosomal orientation as found at its natural site (Figure 4A). This newly modified BAC clone was then used for Cre recombinase–mediated genomic replacement of the mouse α-globin synteny region using the ES cell line as previously described.9 An ES clone was obtained after selection with the correct replacement and this was subsequently modified by a final round of recombination using FLP recombinase to remove all selection marker cassettes (Figure 4 and supplemental Figure 1B). An ES cell clone now containing HS −40 at the ectopic position with minimal extraneous DNA sequences was checked extensively to confirm the correct sequence rearrangements were present, and used to establish the new humanized line (3′HS −40).
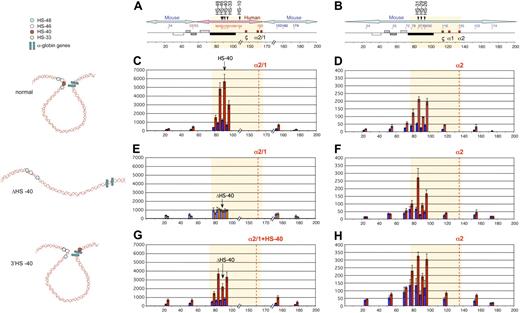
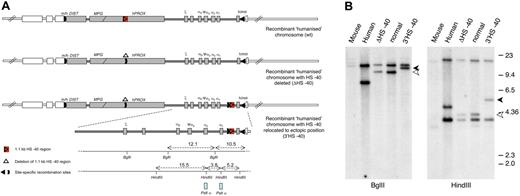
Humanized chromosomes used to study effect of ectopic reinsertion of HS −40. (A) An outline of the 3 recombinant humanized chromosomes analyzed in this study: (top) humanized chromosome with intact normal sequence in which HS −40 is located within the C16orf35 gene (referred to as normal humanized chromosome); (middle) humanized chromosome with HS −40 deleted from the C16orf35 gene (referred to as ΔHS −40 chromosome); (bottom) humanized chromosome with HS −40 deleted from the C16orf35 gene and relocated to an ectopic position (referred to as 3′HS −40 chromosome). Human genes are represented by gray filled rectangles; mouse genes, by open rectangles. Human and mouse sequences have boundaries in Dist and theta genes indicated by m/h and h/m. Human/mouse boundaries and positions of deletion and reinsertion of HS −40 correspond to the position of site-specific recombination sites (key) used to engineer these rearrangements. For a more detailed explanation of the chromosome engineering steps involved, see supplemental Figure 1A and B. (B) Southern blot analyses of the HS −40 reinsertion site with a PstI α probe that hybridizes with the α-globin genes. Genomic DNA was digested with BglII and HindIII and run on a 0.65% agarose gel. From left to right: genomic DNA from mouse, human, humanized mice lacking HS −40, normal humanized mouse, and with the reinsertion of HS −40 at the 3′ end of the cluster. Arrowheads show a size increase (1.1 kb) in the genomic DNA fragment of mice with the reinsertion of HS −40 between the α-globin gene and the θ gene. The normal 9.4 kb (open arrowhead) BglII fragment is increased to 10.5 kb (closed arrowhead) and the normal 4.1 kb (open arrowhead) HindIII fragment is increased to 5.2 kb (closed arrowhead). A small restriction map is represented at the bottom of panel A.
Humanized chromosomes used to study effect of ectopic reinsertion of HS −40. (A) An outline of the 3 recombinant humanized chromosomes analyzed in this study: (top) humanized chromosome with intact normal sequence in which HS −40 is located within the C16orf35 gene (referred to as normal humanized chromosome); (middle) humanized chromosome with HS −40 deleted from the C16orf35 gene (referred to as ΔHS −40 chromosome); (bottom) humanized chromosome with HS −40 deleted from the C16orf35 gene and relocated to an ectopic position (referred to as 3′HS −40 chromosome). Human genes are represented by gray filled rectangles; mouse genes, by open rectangles. Human and mouse sequences have boundaries in Dist and theta genes indicated by m/h and h/m. Human/mouse boundaries and positions of deletion and reinsertion of HS −40 correspond to the position of site-specific recombination sites (key) used to engineer these rearrangements. For a more detailed explanation of the chromosome engineering steps involved, see supplemental Figure 1A and B. (B) Southern blot analyses of the HS −40 reinsertion site with a PstI α probe that hybridizes with the α-globin genes. Genomic DNA was digested with BglII and HindIII and run on a 0.65% agarose gel. From left to right: genomic DNA from mouse, human, humanized mice lacking HS −40, normal humanized mouse, and with the reinsertion of HS −40 at the 3′ end of the cluster. Arrowheads show a size increase (1.1 kb) in the genomic DNA fragment of mice with the reinsertion of HS −40 between the α-globin gene and the θ gene. The normal 9.4 kb (open arrowhead) BglII fragment is increased to 10.5 kb (closed arrowhead) and the normal 4.1 kb (open arrowhead) HindIII fragment is increased to 5.2 kb (closed arrowhead). A small restriction map is represented at the bottom of panel A.
The humanized locus was analyzed by Southern blot (Figure 4B), as previously described,9 with probes spanning the entire humanized locus and its insertion sites. Junctions at the HS −40 reinsertion sites were checked by PCR amplification and subsequent sequence analysis.
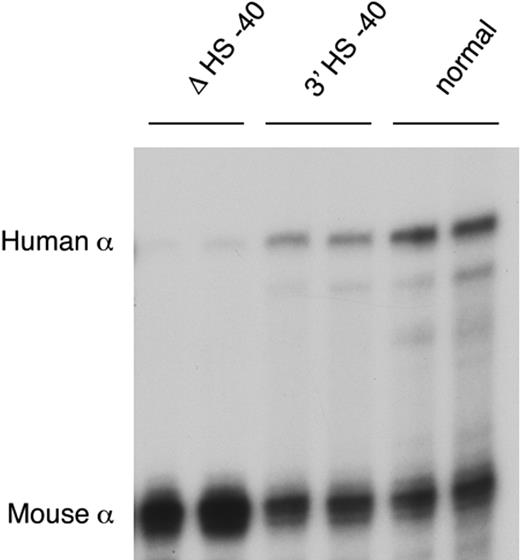
After reinsertion, α-globin RNA transcription occurs to approximately 50% of that seen when HS −40 is in its normal position (Figure 5). As HS −40 now lies in the same HindIII restriction fragment as the α genes themselves (Figure 4B), the HS −40/promoter interaction could not be examined. But, significantly, all the remaining upstream sequences again interacted normally with the anchor fragment (Figure 2G). Thus, in the presence of HS −40, albeit in a different position, the other upstream elements can once more interact with the α gene promoters and expression occurs, although this appears to be at suboptimal levels. Again, the endogenous mouse cluster is unaffected (Figure 2H).
α-globin expression in humanized mice. RNase protection assay of adult blood RNA from heterozygotes of lines ΔHS −40, 3′HS −40, and normal humanized mouse line.
α-globin expression in humanized mice. RNase protection assay of adult blood RNA from heterozygotes of lines ΔHS −40, 3′HS −40, and normal humanized mouse line.
Discussion
This study represents an important step toward understanding how the previously characterized remote upstream control elements (MCS-R1 to -R4) interact with the α-globin promoters to control gene expression. This in turn provides a good model for understanding, in general, how distal elements control mammalian gene expression. Our observations show that in humanized erythroid cells the physical interactions between all of the upstream elements and the α-globin promoters depend largely on a single element (MCS-R2), a 1.1-kb HpaI fragment containing HS −40 (Figure 4A). Deletion of this remote upstream element results in almost complete lack of expression (< 2% of normal) and of all the intrachromosomal interactions. However, when MCS-R2 is reinserted, now downstream of the α cluster, it restores both of these features (Figures 2–3x).
At present, the mechanism by which remote regulatory elements and their cognate promoters interact is still not clear, although several models (looping, tracking, facilitated tracking, and linking) have been proposed.10 Many observations have supported the idea that chromatin loops result from the stabilization of random interactions between cis-elements in chromatin. The observations made here are also most consistent with a looping model in which multiprotein complexes at the upstream element (MCS-R2) and the α gene promoters collide, and their interactions become stabilized, in erythroid cells (Figure 2). This in turn might nucleate interactions with other upstream elements (MCS-R1/HS −48 and MCS-R3/HS −33). Certainly, the establishment of these interactions is not entirely dependent on the position of HS −40, which activates α-globin from its natural location (upstream) and from its relocated position (downstream) of the cluster.
The true role of the other MCS-R elements is not clear. To date, there are no natural deletions that remove these elements without also removing HS −40.5 Furthermore, we and others have observed no major effect of these elements on expression in transgenic experiments.11-13 Nevertheless, because interactions with HS −48 and HS −33 are present when MCS-R2 is reinserted, these sites may yet be relevant to the control of α-globin gene expression. For example, they may in some way facilitate the interaction between HS −40 and the α-globin genes. It is therefore interesting to note that when MCS-R2 is reinserted downstream of the cluster, human α-globin expression is suboptimal (∼ 50% of that seen when MCS-R2 is located upstream), suggesting that its normal chromosomal position may be important for fully efficient activation. Together, these observations emphasize the need to analyze, in greater detail, the role of each individual regulatory element, and the importance of the position and orientation of MCS-R2 with respect to the α-globin cluster.
The MCS-R2 fragment contains binding sites for the GATA-1/SCL complex, NF-E2, and Sp/XKLF.2,14 It has been established that in the β-globin locus various transcription factors (GATA-1, EKLF), cofactors (Idb-1, a component of the GATA/SCL complex), and chromatin remodeling complex (BRG1, a component of the Swi/Snf complex) may mediate physical interactions between erythroid promoters and upstream elements.15-18 However, simple binding and interaction of GATA-1 or ldb-1 could not fully explain this phenomenon at the α-globin locus because these factors are recruited during the early phases of erythropoiesis, before looping occurs.14,19 However, Sp/XKLF factors are recruited, at the upstream elements and promoter late in erythropoiesis, when looping occurs.2 But, again, it is unlikely that a single member of this family could mediate looping because many of the upstream elements are bound by Sp/XKLF factors and yet these upstream elements do not interact with the α-promoters in the absence of HS −40. Furthermore, a previously described knockout of EKLF apparently had little effect on α-globin expression.20 It has also been suggested that PolII may play a role as a bridging factor between upstream elements and their cognate promoters.21 However, HS −48 still binds PolII in the absence of HS −40 and yet this element does not appear to interact with or facilitate significant recruitment of PolII to the α-globin promoters on its own. Furthermore, looping at the β-globin cluster persists in the absence of PolII, after α-amanitin treatment.22 Thus, the proteins and mechanisms mediating interactions between the upstream elements and their promoters are still poorly understood.
This study shows that both enhancer activity and physical looping in the human α-globin cluster predominantly depend on a single cis-acting element (MCS-R2/HS −40) in contrast to the more complex cis-acting interactions observed in gene clusters (including the human and mouse β-globin clusters and the mouse α-globin cluster) involving multiple cis-acting elements. This fact, together with the development of the humanized mouse system, now provides a tractable, in vivo experimental model to understand in detail some of the issues raised in this and other studies concerning the mechanism by which mammalian gene expression is controlled via distal regulatory elements.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are very grateful to Richard Gibbons and David Garrick for critically reading the paper. This work was funded by the Medical Research Council and the National Institute for Health Research (NIHR) Biomedical Research Center Program.
National Institutes of Health
Authorship
Contribution: D.V. designed and performed research, analyzed data, and wrote the first draft of the paper; F.M.-K. designed and performed research and analyzed data; J.A.S., J.A.S.-S., and W.G.W. provided samples and maintained the animal stocks; H.A.C.W. and A.J.H.S. designed and generated the humanized model with ectopic HS −40 reinsertion; and D.R.H. designed research, analyzed data, and wrote the final draft of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Doug R. Higgs, MRC Molecular Hematology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford, OX3 9DS, United Kingdom; e-mail: doug.higgs@imm.ox.ac.uk.