Abstract
Potent graft-versus-leukemia (GVL) effects can be mediated by donor-derived T cells recognizing minor histocompatibility antigens (mHags) in patients treated with donor lymphocyte infusion (DLI) for relapsed hematologic malignancies after HLA-matched allogeneic stem cell transplantation (alloSCT). Donor-derived T cells, however, may not only induce GVL, but also mediate detrimental graft-versus-host disease (GVHD). Because HLA-class II is under noninflammatory conditions predominantly expressed on hematopoietic cells, CD4+ T cells administered late after alloSCT may selectively confer GVL without GVHD. Although a broad range of different HLA-class I–restricted mHags have been identified, the first 2 autosomal HLA-class II–restricted mHags have only recently been characterized. By screening a recombinant bacteria cDNA expression library, we identified 4 new HLA-class II–restricted mHags recognized by CD4+ T cells induced in a patient with relapsed chronic myeloid leukemia who achieved long-term complete remission and experienced only mild GVHD of the skin after DLI. All CD4+ T cells were capable of recognizing the mHags presented by HLA-DR surface molecules on primary hematopoietic cells, but not on skin-derived (cytokine-treated) fibroblasts. The selective recognition of hematopoietic cells as well as the balanced population frequencies and common HLA-DR restriction elements make the novel mHags possible targets for development of immunotherapeutic strategies.
Introduction
Relapsed hematologic malignancies after HLA-matched allogeneic stem cell transplantation (alloSCT) can be efficiently treated with donor lymphocyte infusion (DLI).1-3 This beneficial graft-versus-leukemia (GVL) immunoreactivity is mediated by donor T cells directed against minor histocompatibility antigens (mHags). These mHags are polymorphic peptides that differ between patient and donor, and are able to elicit CD8+ or CD4+ donor T-cell responses in the context of self-HLA.4 A variety of HLA-class I–restricted mHags have been identified,5-17 and the appearance of CD8+ T cells specific for several of these mHags was closely followed by complete remissions of the malignancies,12,14,16,18 indicating the clinical relevance of these T cells.
Because HLA-class I molecules are expressed on all nucleated cells in malignant and nonmalignant tissues of the patient, CD8+ T cells recognizing mHags in the context of HLA-class I molecules may not only mediate GVL, but also induce detrimental graft-versus-host disease (GVHD). GVHD is still the main cause of transplant-related morbidity and mortality,19 and therefore separating GVL and GVHD is a major objective of immunotherapy after alloSCT. One approach is the use of mHags that are encoded by hematopoiesis-restricted genes such as HA-16 and HA-2,5 to specifically stimulate cytotoxicity of donor-originated CD8+ T cells against the malignant hematopoietic cells of the patient.
It is well established that also CD4+ T cells play an important role in antitumor immunity, both as helper cells indispensable for the induction and maintenance of CD8+ T cells20-22 and as effector cells with direct cytolytic activity against HLA-class II+ tumor cells.23-26 Because under noninflammatory or low-inflammatory conditions HLA-class II molecules are predominantly expressed on hematopoietic cells, stimulating CD4+ T-cell immunity late after alloSCT may be another promising strategy to selectively target residual leukemia without inducing severe GVHD.27-29
Although several HLA-class I–restricted mHags have been identified by various methods,5-8,10,11,13-17,30,31 the discovery of HLA-class II–restricted mHags proved to be more difficult and was hampered by technical limitations. We developed a method by which recombinant bacteria cDNA expression libraries were used to identify antigens as targets for CD4+ T cells.32 The feasibility of this method was demonstrated by the recent identification of the first autosomal HLA-class II–restricted mHag LB-PI4K2B-1S.33 Using a combination of genetic linkage analysis and subsequent fine mapping with single nucleotide polymorphism (SNP) markers, another autosomal HLA-DQ–restricted mHag encoded by the B cell–specific CD19 gene was recently identified.34 However, identification of more HLA-class II–restricted mHags is required to further elucidate their role in GVL and GVHD, and to explore their use as targets for CD4+ T cells to treat hematologic malignancies.
Here, we present the molecular identification of 4 new HLA-DR–restricted autosomal mHags recognized by CD4+ T cells induced in a patient who was successfully treated with DLI for relapsed chronic myeloid leukemia (CML). Our data show that CD4+ T cells directed against these mHags recognized primary hematopoietic cells, but not skin-derived (cytokine-treated) fibroblasts. These in vitro findings and the clinical observation that a long-lasting complete remission was induced with only limited GVHD of the skin suggest an important role of the novel HLA-DR–restricted mHags in antitumor reactivity.
Methods
Isolation of hematopoietic cells
Peripheral blood and bone marrow (BM) samples were obtained from patients with hematologic malignancies and healthy persons after approval by the Leiden University Medical Center Institutional Review Board and informed consent according to the Declaration of Helsinki. Mononuclear cells were isolated by Ficoll-Isopaque separation and cryopreserved. Activated CD4+ T cells were isolated from BM of the patient 5 weeks after DLI. BM cells were stained with fluorescein isothiocyanate (FITC)–labeled anti-CD4 (BD Biosciences) and phycoerythrin (PE)–labeled anti–HLA-DR (BD Biosciences) and single cell sorted by flow cytometry. For isolation of patient-derived hematopoietic cells, peripheral blood mononuclear cells (PBMCs) obtained from the patient before alloSCT were stained with FITC-labeled anti-CD14, PE-labeled anti-CD3, and allophycocyanin-labeled anti-CD19 (BD Biosciences), and monocytes (CD14+), T cells (CD3+) and B cells (CD19+) were isolated by flow cytometry. CD34+ CML progenitor cells were isolated from BM cells obtained from the patient during the relapse before DLI by flow cytometric sorting after staining with allophycocyanin-labeled anti-CD34 (BD Biosciences). CD34+ progenitor cells from other patients with CML were isolated by magnetic beads according to the manufacturer's instructions (Miltenyi Biotec). CD33+/CD14− acute myeloid leukemia (AML) cells from 3 patients were isolated by flow cytometric sorting after staining with CD14-FITC and CD33-allophycocyanin (BD Biosciences).
Cell culture
Epstein-Barr virus transformed B-cell lines (EBV-LCLs) were cultured in Iscove modified Dulbecco medium (IMDM; Lonza BioWhittaker) with 10% fetal calf serum (FCS; Cambrex), 1% penicillin/streptomycin (Lonza BioWhittaker), and 1.5% l-glutamine (Lonza BioWhittaker). T-cell clones were cultured in IMDM with 5% human serum, 5% FCS, and 100 IU/mL interleukin-2 (IL-2; Chiron), and restimulated every 10 to 20 days with irradiated allogeneic PBMCs and 0.8 μg/mL phytohemagglutinin (Oxoid) as previously described.14,16 Immature dendritic cells (DCs) were generated by culturing isolated monocytes in medium with 100 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; Novartis) and 500 IU/mL IL-4 (Schering-Plough) for 7 days. During the final 2 days, 100 ng/mL GM-CSF, 10 ng/mL tumor necrosis factor α (TNF-α; Cellgenix), 10 ng/mL IL-1β (Cellgenix), 10 ng/mL IL-6 (Cellgenix), 1 μg/mL prostaglandin E2 (Sigma-Aldrich), and 500 IU/mL interferon-γ (IFN-γ; Boehringer-Ingelheim) were added for maturation. Isolated CD34+ CML progenitor cells were modified to leukemic antigen-presenting cells (APCs) as described previously.35 Fibroblasts were cultured in Dulbecco modified Eagle medium (Lonza, BioWhittaker) with 8% FCS, 1% penicillin/streptomycin, and 1.5% glutamine in the absence or presence of IFN-γ (100 IU/mL) for 5 days.
ELISA
Stimulator cells (3 × 104 cells/well) were coincubated with CD4+ T cells (5 × 103 cells/well) overnight at 37°C in U-bottom 96-well plates. Peptide pulsing was performed by incubating donor EBV-LCLs (106 cells/mL) for 2 hours with synthetic peptides (1 μg/mL) in IMDM containing 2% FCS. Peptide-pulsed donor EBV-LCLs were washed twice and subsequently used as stimulator cells. Cytokine release was measured in 50-μL supernatants by IFN-γ enzyme-linked immunosorbent assay (ELISA; Sanguin) or multi-Th1/Th2/Th17 cytokine ELISA (SABioscience) according to the manufacturer's instructions.
51Cr-release cytotoxicity assay
Target cells were labeled for 1 hour at 37°C with 100 μCi (3.7 MBq) Na251CrO4 (Amersham). After washing, target cells (103 cells/well) were incubated with CD4+ T cells at effector-target ratios of 10:1 for 10 hours. 51Cr was analyzed in 25-μL supernatants. The percentage of specific lysis was calculated as follows: [experimental release (cpm) − spontaneous release (cpm)]/[maximal release (cpm) − spontaneous release (cpm)] × 100%.
Construction of recombinant bacteria cDNA expression library
A recombinant bacterial cDNA expression library was constructed as described previously.33 In brief, RNA of the patient was isolated with TRIzol (Invitrogen) and purified (RNeasy kit; QIAGEN). cDNA was synthesized with oligo-dT and random primers, respectively. Both primers contained an AscI restriction site. After cDNA synthesis, BamHI-EcoRI (Stratagene) adapters were ligated, followed by digestion with AscI and size fragmentation by column chromatography. cDNA fragments were ligated into BamHI and AscI restriction enzyme sites of vector pKE-1,36 which contains an isopropyl β-d-thiogalactoside (IPTG)–inducible tac promoter and resistance genes for ampicillin and kanamycin. Vectors were electroporated into Escherichia coli BL21 (DE3) and recombinant bacteria were selected for ampicillin resistance.
Screening of recombinant bacteria cDNA expression library
Pools of 50 to 100 recombinant bacteria were screened for T-cell recognition, as described previously.33 Bacteria were grown to 600-nm optical density of 0.5 with 50 μg/mL ampicillin (Sigma-Aldrich) and protein expression was induced by 1 mM IPTG (Promega). Subsequently, bacteria were opsonized by adding human serum with 17% (vol/vol) complement (Sigma-Aldrich) and incubated for 1 hour. EBV-LCLs of the fully HLA-matched donor (3 × 104 cells/well) were pulsed with complement-opsonized bacteria in IMDM with 10% FCS and 30 μg/mL gentamycin (Sigma-Aldrich) overnight at 37°C. Mixes of up to 3 CD4+ T-cell clones (3 × 103 cells/well per clone) were coincubated with bacteria-pulsed EBV-LCLs for 20 hours. Supernatants were harvested for IFN-γ ELISA. In a second round, single bacterial clones derived from a positive pool were screened for T-cell recognition. Positive bacterial clones were grown overnight and plasmids were isolated by the plasmid mini prep kit (QIAGEN). Inserted cDNA fragments were sequenced using vector-specific primers.
Isolation and retroviral transduction of HLA-DR alleles
Total RNA of patient-derived EBV-LCLs was obtained using Trizol (Invitrogen) and transcribed into cDNA by reverse transcriptase (Invitrogen) using oligo-dT primers (Roche Diagnostics). HLA-DR genes were amplified with HLA-DRB1– and HLA-DRB3–specific primers. Polymerase chain reaction (PCR) products were cloned into retroviral vector MP71, which contains the marker nerve growth factor receptor (ΔNGF-R).37 PCR products were verified by sequencing. Wild-type φnx A packaging cells were transfected with these vectors as previously described,37 with the exception that the Fugene HD transfection kit (Roche Diagnostics) was used. Viral supernatants were used for transduction of EBV-LCLs on plates coated with recombinant human fibronectin CH 296 (Takara Shuzo) as previously described.37 All EBV-LCLs showed retroviral transduction efficiencies of 3% to 7% based on staining with PE-conjugated anti–ΔNGF-R (BD Biosciences).
SNP genotyping assays
Genomic DNA was isolated from 1400 patient and donor samples (PBMCs or BM cells) by the Gentra Systems PureGene genomic isolation kit (Biocompare). The SNPs rs751019 (PTK2B), rs2236225 (MTHFD1), and rs12692566 (LY75)38 were analyzed using SNP genotyping assay (Applied Biosystems) containing forward and reverse primer for amplification and 2 TaqMan MGB probes labeled with VIC and FAM dyes to detect the different alleles. SNP rs2236410 (MR1) was analyzed using allele-specific primers labeled with VIC and FAM dyes, respectively (KBioScience). Genotyping was performed according to the manufacturer's instructions.
Quantitative RT-PCR
Quantitative real-time reverse-transcriptase (RT)–PCR was performed as described previously.14 Using Primer Express (Applied Biosystems), the following primers were designed: MR1: 5′-AAGGTGGAACTGAAGCGCCT-3′, 5′-GTGGTGCTTCCATCCTCCAG-3′, and probe 5′-(VIC)-CACTACAATCACTCAGGGTCTCACACTTACCAGA-(TAMRA)-3′; PTK2B: 5′-GGAAAGATGGTGAGAAGCGG-3′, 5′-AGCTCTCTGAGAGGTGGGACC-3′, and probe 5′-(VIC)-TGCCCCAGATCCCCATGCTAAACC-(TAMRA)-3′; LY75: 5′-TGCCAGCAATCACAGCTTTC-3′, 5′-TATTGGCCACTCGCTAATTCTTATC-3′, and probe 5′-(VIC)-CTAAAAGCCATCAAAAACAAAATAGCAAATATATCTGGTG-(TAMRA)-3′; MTHFD1: 5′-GGCCTGATGGGAAATACGTG-3′, 5′-GCCCGATTGTAGTTGTGCTTTT-3′, and probe 5′-(VIC)-TGACTGGAATAACTCCAACACCCCTGGG-(TAMRA)-3′. PCR was performed in 25 μL reaction mixture containing 2.5 μL Taqman buffer, 300 nM of each primer, 180 nM probe (LY75: 200 nM), 200 μM deoxynucleotide triphosphate, 3 mM MgCl2 (MTHFD1: 4 mM, PTK2B: 1.5 mM), 0.625 U AmpliTaq Gold polymerase, and 10 and 100 ng cDNA sample. Amplification was started with 10 minutes at 95°C, followed by 50 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 60°C. Expression of the genes was normalized as ratio with the housekeeping gene porphobilinogen deaminase (PBGD).
Results
Isolation and characterization of mHag-specific CD4+ T-cell clones
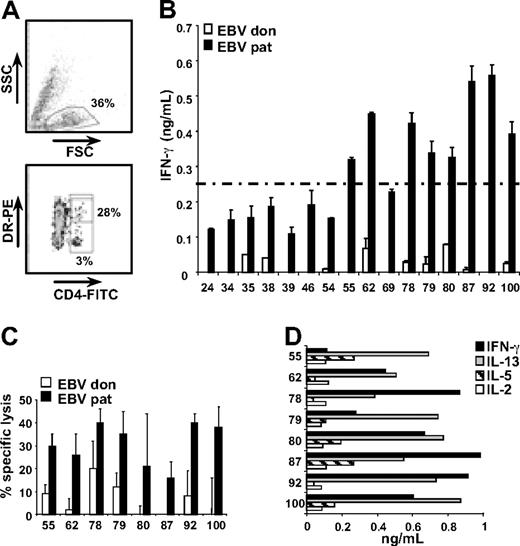
We analyzed the specificity of the CD4+ T-cell response induced in a patient with relapsed CML after HLA-matched alloSCT who responded to DLI with complete donor chimerism and remission of the leukemia, and developed only mild GVHD of the skin. CD4+ T cells were single cell sorted from bone marrow mononuclear cells obtained 5 weeks after DLI based on expression of the activation marker HLA-DR (Figure 1A). A total number of 25 CD4+ T-cell clones were shown to recognize mHags based on differential recognition of patient and donor EBV-LCLs in an overnight IFN-γ ELISA. By pulsing donor EBV-LCLs with a peptide comprising previously identified mHag LB-PI4K2B-1S, 9 of the 25 T-cell clones were shown to be specific for HLA-DQB1*0603–restricted LB-PI4K2B-1S (data not shown). Of the 16 remaining T-cell clones, 8 clones (55, 62, 78, 79, 80, 87, 92, and 100) produced high levels of IFN-γ (> 250 pg/mL) upon incubation with patient EBV-LCLs (Figure 1B), and were selected for further characterization.
Isolation and characterization of mHag-specific CD4+ T-cell clones. (A) Activated (HLA-DR+) CD4+ T cells were single cell sorted from bone marrow cells obtained from the patient 5 weeks after DLI by flow cytometry. Indicated are the percentages of bone marrow cells expressing CD4 (3%) and CD4+ cells expressing HLA-DR (28%). A total number of 106 CD4+ T-cell clones were growing out of 1100 single sorted cells. (B) Sixteen CD4+ T-cell clones were directed against unknown mHags based on differential recognition of patient (pat) and donor (don) Epstein-Barr virus transformed B-cell lines (EBV-LCLs). Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown. (C) All CD4+ T-cell clones recognizing unknown mHags and releasing > 250 pg/mL IFN-γ upon incubation with patient EBV-LCLs were tested for specific lysis of patient and donor EBV-LCLs in a 10-hour 51Cr release assay. Mean specific lysis of triplicate wells at effector-target ratios of 10:1 is shown. (D) CD4+ T-cell clones were tested for cytokine release by multicytokine ELISA. Release of IL-2, IL-5, IL-13, and IFN-γ in 50-μL single supernatants is shown (ng/mL). CD4+ T-cell clones did not produce IL-4, IL-6, IL-10, IL-12, IL-17A, TNF-α, granulocyte CSF, and transforming growth factor β1 (data not shown).
Isolation and characterization of mHag-specific CD4+ T-cell clones. (A) Activated (HLA-DR+) CD4+ T cells were single cell sorted from bone marrow cells obtained from the patient 5 weeks after DLI by flow cytometry. Indicated are the percentages of bone marrow cells expressing CD4 (3%) and CD4+ cells expressing HLA-DR (28%). A total number of 106 CD4+ T-cell clones were growing out of 1100 single sorted cells. (B) Sixteen CD4+ T-cell clones were directed against unknown mHags based on differential recognition of patient (pat) and donor (don) Epstein-Barr virus transformed B-cell lines (EBV-LCLs). Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown. (C) All CD4+ T-cell clones recognizing unknown mHags and releasing > 250 pg/mL IFN-γ upon incubation with patient EBV-LCLs were tested for specific lysis of patient and donor EBV-LCLs in a 10-hour 51Cr release assay. Mean specific lysis of triplicate wells at effector-target ratios of 10:1 is shown. (D) CD4+ T-cell clones were tested for cytokine release by multicytokine ELISA. Release of IL-2, IL-5, IL-13, and IFN-γ in 50-μL single supernatants is shown (ng/mL). CD4+ T-cell clones did not produce IL-4, IL-6, IL-10, IL-12, IL-17A, TNF-α, granulocyte CSF, and transforming growth factor β1 (data not shown).
All 8 T-cell clones showed specific lysis of patient, but not donor, EBV-LCLs in a 10-hour 51chromium release assay (Figure 1C), and produced IFN-γ, IL-13, IL-5, and IL-2 (Figure 1D), but not IL-4, IL-6, IL-10, IL-12, IL-17A, TNF-α, granulocyte CSF, or transforming growth factor β1 as determined by multicytokine ELISA.
To determine the mHag specificity of the 8 T-cell clones, a panel of 22 EBV-LCLs sharing one or more HLA-class II alleles with the patient was tested for recognition. Similar EBV-LCL recognition patterns were observed for T-cell clones 55, 78, and 92 as well as T-cell clones 79 and 100 (data not shown), indicating that the 8 T-cell clones recognized 5 different HLA-class II–restricted mHags. Further characterization and mHag identification were performed for 1 representative clone of each specificity, that is, 62, 78, 80, 87, and 100.
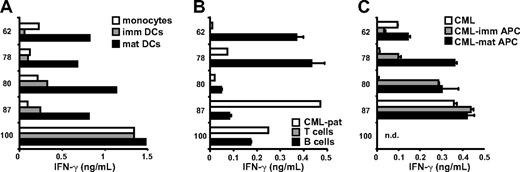
The capacity of the mHag-specific CD4+ T cells to recognize nonmalignant and malignant hematopoietic cells was determined by IFN-γ ELISA. B cells, T cells, and monocytes were purified from PBMCs of the patient before alloSCT, and tested for recognition. In addition, immature and mature dendritic cells (DCs) were generated from isolated monocytes in vitro, and malignant CD34+ CML progenitor cells were purified from BM obtained from the patient during the relapse after alloSCT before treatment with DLI.
Figure 2A shows that all T-cell clones strongly recognized mature DCs, expressing high levels of HLA-class II and costimulatory molecules, whereas monocytes and immature DCs were recognized at variable levels. Clones 62, 78, 80, and 87 weakly recognized monocytes and immature DCs, whereas these cells were strongly recognized by clone 100. All T-cell clones recognized high HLA-class II–expressing B cells, although to a different extent, whereas T cells, lacking expression of HLA-class II, were not recognized (Figure 2B). Purified CD34+ CML progenitor cells from the patient were weakly recognized by clones 62, 78, and 80, whereas strong recognition was observed for clones 87 and 100 (Figure 2B). In addition to purified CD34+ CML progenitor cells of the patient, CD34+ cells were isolated from other patients with CML and differentiated to immature and mature leukemic APCs. The data in Figure 2C show that, upon induction of an APC phenotype in vitro, CML cells were recognized by all T-cell clones.
Recognition of nonmalignant hematopoietic cells and chronic myeloid leukemia. Monocytes (CD14+), B cells (CD19+), and T cells (CD3+) were isolated from PBMCs from the patient obtained before alloSCT by flow cytometry. Isolated monocytes were cultured in vitro to immature and mature DCs. CD34+ chronic myeloid leukemia (CML) progenitor cells were isolated from bone marrow cells obtained from the patient after alloSCT during the relapse before treatment with DLI. In addition, CD34+ cells were isolated from other patients with CML and cultured in vitro to professional APCs. CD4+ T-cell clones were tested against (A) patient-derived monocytes, immature and mature DCs; (B) patient-derived T cells, B cells, and CD34+ CML progenitor cells; and (C) CD34+ CML progenitor cells from other patients with and without in vitro induced APC phenotype. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of single (monocytes, immature DCs, mature DCs, patient-derived CD34+ CML progenitor cells) or duplicate wells is shown. nd indicates not determined
Recognition of nonmalignant hematopoietic cells and chronic myeloid leukemia. Monocytes (CD14+), B cells (CD19+), and T cells (CD3+) were isolated from PBMCs from the patient obtained before alloSCT by flow cytometry. Isolated monocytes were cultured in vitro to immature and mature DCs. CD34+ chronic myeloid leukemia (CML) progenitor cells were isolated from bone marrow cells obtained from the patient after alloSCT during the relapse before treatment with DLI. In addition, CD34+ cells were isolated from other patients with CML and cultured in vitro to professional APCs. CD4+ T-cell clones were tested against (A) patient-derived monocytes, immature and mature DCs; (B) patient-derived T cells, B cells, and CD34+ CML progenitor cells; and (C) CD34+ CML progenitor cells from other patients with and without in vitro induced APC phenotype. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of single (monocytes, immature DCs, mature DCs, patient-derived CD34+ CML progenitor cells) or duplicate wells is shown. nd indicates not determined
In conclusion, these data suggest that the mHag-specific CD4+ T cells have been induced in this patient by malignant CML cells with mature APC phenotype and that the CD4+ T cells then played various roles as effector cells in the direct elimination of the malignant CD34+ CML progenitor cells according to their capability to recognize these cells. Induction by nonmalignant B lymphocytes of the patient is not likely, because before DLI more than 99% of the B cells were of donor origin, whereas 23% of bone marrow cells were bcr-abl–positive leukemic cells (data not shown).
Identification of 4 HLA-class II–restricted mHags as targets for CD4+ T cells
To identify the mHags recognized by CD4+ T-cell clones 62, 78, 80, 87, and 100, recombinant bacterial cDNA expression libraries were constructed from EBV-LCLs of the patient, and screened for T-cell recognition as previously described.33 Two different cDNA libraries were constructed, each containing 4 × 106 random-primed or oligo-dT–primed cDNAs cloned into vector pKE-1 under control of an isopropyl β-d-thiogalactoside (IPTG)–inducible promoter.36 The different cDNAs were size-separated by column chromatography and divided into different fractions. For each fraction, a total number of 960 different pools of approximately 50 different bacteria were screened for T-cell recognition by IFN-γ ELISA. By screening 2 different fractions of the random-primed cDNA library and one fraction of the oligo-dT–primed cDNA, we identified 4 of the 5 HLA-class II–restricted mHags, designated LB-MR1-1R, LB-PTK2B-1T, LB-LY75-1K, and LB-MTHFD1-1Q.
LB-MR1-1R
In a first screening of the fraction containing the largest random-primed cDNA fragments, 1 bacterial pool stimulated IFN-γ release by T-cell clone 62. Subcloning of this pool revealed 2 positive single cDNAs, each comprising a 1556-bp sequence identical to “major histocompatibility complex, class I–related” (MR1) gene (GenBank accession no. NM_001531).39 The MR1 gene contained 5 known missense SNPs, and sequencing of cDNA of the patient and donor EBV-LCLs revealed that the patient was G/A heterozygous at position 121 bp, whereas the donor was A/A homozygous. This A-to-G transition creates a His-to-Arg substitution at amino acid position 39 (H→R39) of the MR1 protein (SNP database ID no. rs2236410).
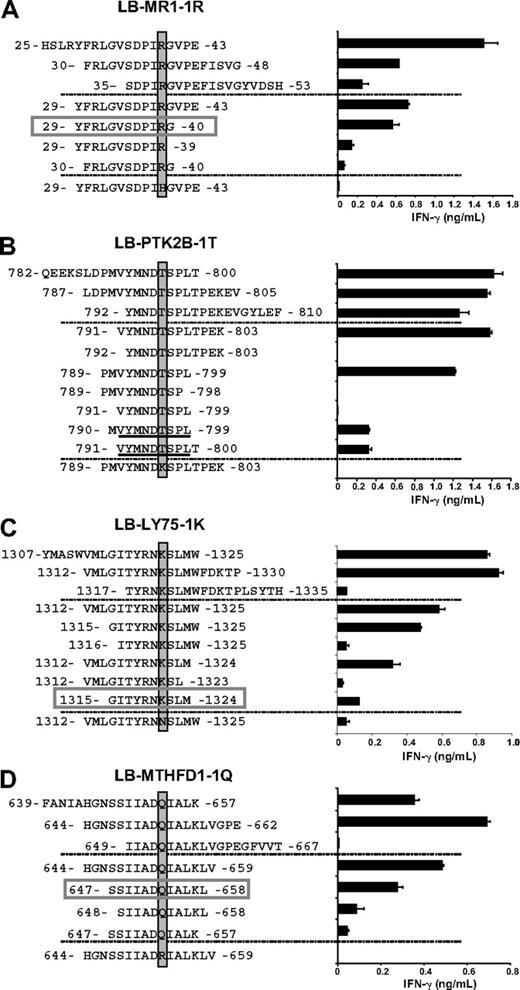
To identify the epitope recognized by clone 62, 3 overlapping 19-mer peptides all comprising the R at position 39 were synthesized and tested for T-cell recognition (Figure 3A). Two of these peptides stimulated IFN-γ release by clone 62. A 15-mer peptide comprising amino acids shared by the 2 19-mer peptides also stimulated IFN-γ release, whereas a 15-mer peptide containing the donor variant H at position 39 was not recognized (Figure 3A). Serial truncations of the 15-mer peptide at the N and C termini led to the identification of a minimal 12-mer YFRLGVSDPIRG peptide (amino acids 29-40), designated LB-MR1-1R (Figure 3A).
Identification of the minimal epitopes of the HLA-class II–restricted mHags. Three partially overlapping 19-mer peptides comprising a region of 14 amino acids upstream and downstream of the identified polymorphisms (top) as well as series of peptides containing N- and C-terminal truncations (middle) and a peptide comprising the polymorphism of the donor (bottom) were synthesized, pulsed onto donor EBV-LCLs, and tested for recognition by the mHag-specific CD4+ T-cell clones in IFN-γ ELISA. Indicated are the variant residue (shaded), the minimal epitope (boxed), and the core region of the minimal epitope (underlined). (A) T-cell clone 62 was tested against peptides comprising LB-MR1-1R. The minimal epitope is the 12-mer YFRLGVSDPIRG. (B) T-cell clone 78 was tested against peptides comprising LB-PTK2B-1T. The core region is the 9-mer VYMNDTSPL. (C) T-cell clone 80 was tested against peptides comprising LB-LY75-1K. The minimal epitope is the 10-mer GITYRNKSLM. (D) T-cell clone 87 was tested against peptides comprising LB-MTHFD1-1Q. The minimal epitope is the 12-mer SSIIADQIALKL. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
Identification of the minimal epitopes of the HLA-class II–restricted mHags. Three partially overlapping 19-mer peptides comprising a region of 14 amino acids upstream and downstream of the identified polymorphisms (top) as well as series of peptides containing N- and C-terminal truncations (middle) and a peptide comprising the polymorphism of the donor (bottom) were synthesized, pulsed onto donor EBV-LCLs, and tested for recognition by the mHag-specific CD4+ T-cell clones in IFN-γ ELISA. Indicated are the variant residue (shaded), the minimal epitope (boxed), and the core region of the minimal epitope (underlined). (A) T-cell clone 62 was tested against peptides comprising LB-MR1-1R. The minimal epitope is the 12-mer YFRLGVSDPIRG. (B) T-cell clone 78 was tested against peptides comprising LB-PTK2B-1T. The core region is the 9-mer VYMNDTSPL. (C) T-cell clone 80 was tested against peptides comprising LB-LY75-1K. The minimal epitope is the 10-mer GITYRNKSLM. (D) T-cell clone 87 was tested against peptides comprising LB-MTHFD1-1Q. The minimal epitope is the 12-mer SSIIADQIALKL. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
LB-PTK2B-1T
Screening the same fraction of the random-primed cDNA library revealed 2 other bacterial pools that stimulated IFN-γ release by clone 78. Subcloning of these 2 pools revealed 2 cDNAs containing 813- to 2558-bp and 2318- to 3414-bp regions of the gene encoding “protein tyrosine kinase 2 beta, variant 4” (PTK2B; GenBank accession no. NM_173175). The 241-bp region that was shared by the 2 cDNAs contained only 1 known missense SNP at position 2542 bp. Sequencing of the donor and the patient confirmed disparity for this SNP, with the patient being C/A heterozygous and the donor A/A homozygous. This A-to-C transition creates a Lys-to-Thr substitution at amino acid position 796 (K→T 796) of the PTK2B protein (SNP database ID no. rs751019).
Three overlapping 19-mer peptides comprising the T at position 796 and subsequently a series of truncated peptides were synthesized and tested for T-cell recognition. The data show that a core region of 9 amino acids (VYMNDTSPL) at positions 791 to 799 is crucial, but not sufficient, for T-cell recognition. T-cell recognition was shown to require addition of at least one additional residue at either N- or C-terminus of the core region. A 15-mer peptide containing the donor variant K at position 796 was not recognized, confirming the identification of LB-PTK2B-1T as the mHag recognized by clone 78 (Figure 3B).
LB-LY75-1K
In another screening containing shorter fragments of the random cDNA library, 3 bacterial pools stimulated IFN-γ release by clone 80. Subcloning of these 3 pools revealed 3 cDNAs containing 2176- to 5739-bp, 2281- to 5739-bp, and 3558- to 4673-bp regions of the gene encoding “lymphocyte antigen 75” (LY75i GenBank accession no. NM_002349). The 1116-bp region shared by the 3 cDNAs contained 5 known missense SNPs. The only observed SNP disparity between patient and donor was at position 4032 bp, with the patient being heterozygous G/T and the donor homozygous T/T. This T-to-G transition creates an Asn-to-Lys substitution at amino acid position 1321 (N→K1321) of the LY75 protein (SNP database ID no. rs12692566).
Following a similar strategy as described for LB-MR1-1R and LB-PTK2B-1T, a minimal epitope of 10 amino acids (GITYRNKSLM) of LB-LY75-1K was shown to be recognized by clone 80 (Figure 3C). A 15-mer peptide containing the donor variant N at position 1321 did not stimulate the release of IFN-γ.
LB-MTHFD1-1Q
Finally, the fraction with the largest fragments of the oligo-dT–primed cDNA library was screened for T-cell recognition. Three bacterial pools stimulated IFN-γ release by clone 87. Subcloning of these 3 pools revealed 2 cDNAs containing 3- to 3116-bp and 1166-to 3116-bp regions of the gene encoding “methylene tetrahydrofolate dehydrogenase (NADP+ dependent) 1” (MTHFD1; GenBank accession no. NM_005956). In the 1951-bp region shared by the 2 cDNAs, 7 missense SNPs were known. Sequencing of the patient and the donor revealed one disparity at position 2011 bp, with the patient being heterozygous G/A and the donor homozygous G/G. This G-to-A transition creates an Arg-to-Gln substitution at amino acid position 653 (R→Q653) of the MTHFD1 protein (SNP database ID no. rs2236225).
Synthesis of 3 overlapping 19-mer peptides and a subsequent series of truncated peptides led to the identification of a 12-mer peptide (SSIIADQIALKL) as the minimal epitope of LB-MTHFD1-1Q. A peptide comprising the donor variant R at position 653 did not stimulate IFN-γ release by T-cell clone 87 (Figure 3D).
Frequencies of HLA-class II–restricted mHag-specific T-cell clones
To determine the frequencies of CD4+ T cells specific for the new mHags, all T-cell clones isolated based on expression of HLA-DR were tested for recognition of donor EBV-LCLs pulsed with the LB-MR1-1R, LB-PTK2B-1T, LB-LY75-1K, and LB-MTHFD1-1Q peptides, respectively. In addition to clone 62, LB-MR1-1R was recognized by T-cell clone 69. Clones 62 and 69 expressed the same T-cell receptor (TCR) beta chain (BV) and complementarity determining region 3 (CDR3) (data not shown). LB-PTK2B-1T was recognized by clones 78, 55, and 92. These 3 clones expressed different TCR BV chains with CDR3 regions variable in length and amino acid composition, demonstrating a polyclonal CD4+ T-cell response against LB-PTK2B-1T (data not shown). LB-LY75-1K was recognized by clones 34 and 80. These 2 clones also expressed different TCR BV chains. LB-MTHFD1-1Q was recognized only by T-cell clone 87.
Population frequencies of HLA-class II–restricted mHags
To determine the population frequencies of the new HLA-class II–restricted mHags, SNP genotyping assays for rs2236410 (LB-MR1-1R), rs751019 (LB-PTK2B-1T), rs2236225 (LB-MTHFD1-1Q), and rs12692566 (LB-LY75-1K) were performed for 1400 patient and donor samples available at our laboratory. The analysis showed balanced population frequencies of 25% (LB-MR1-1R), 33% (LB-LY75-1K), 68% (LB-MTHFD1-1Q), and 70% (LB-PTK2B-1T; Table 1). These population frequencies slightly differ from the white population frequencies in the dbSNP database,38 which were 33%, 40%, 50%, and 60%, respectively.
Population frequencies of HLA-DR–restricted mHags
| . | Immunogenic . | Nonimmunogenic . | ||
|---|---|---|---|---|
| +/+ . | +/− . | −/− . | ||
| LB-MR1-1R | 2 | 23 | 75 | |
| LB-PTK2B-1T | 21 | 49 | 30 | |
| LB-LY75-1K | 3 | 30 | 67 | |
| LB-MTHFD1-1Q | 20 | 48 | 32 | |
| . | Immunogenic . | Nonimmunogenic . | ||
|---|---|---|---|---|
| +/+ . | +/− . | −/− . | ||
| LB-MR1-1R | 2 | 23 | 75 | |
| LB-PTK2B-1T | 21 | 49 | 30 | |
| LB-LY75-1K | 3 | 30 | 67 | |
| LB-MTHFD1-1Q | 20 | 48 | 32 | |
Genotyping was performed for SNP rs2236410 (LB-MR1-1R), rs751019 (LB-PTK2B-1T), rs12692566 (LB-LY75-1K), and rs2236225 (LB-MTHFD1-1Q) in 1400 patient and donor samples. Frequencies (%) of homozygous-positive (+/+), heterozygous (+/−), and homozygous-negative (−/−) samples are shown.
Identification of the HLA-class II restriction elements
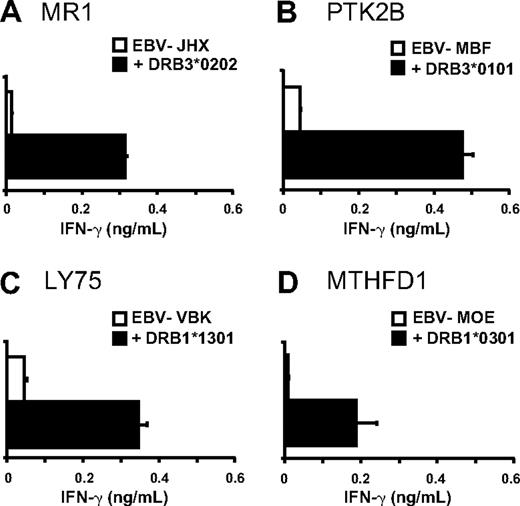
Based on the SNP genotyping results, a panel of mHag+ EBV-LCLs sharing one or more HLA-class II alleles with the patient was composed and tested for T-cell recognition. The data strongly suggested HLA-DRB3*0202, -DRB3*0101, -DRB1*1301, and -DRB1*0301 as restriction elements for LB-MR1-1R, LB-PTK2B-1T, LB-LY75-1K, and LB-MTHFD1-1Q, respectively (data not shown). Restriction by these HLA-class II alleles was confirmed by retroviral transduction of mHag+ EBV-LCLs lacking the appropriate HLA molecule with patient-derived cDNAs encoding the respective HLA-class II molecules (Figure 4).
Identification of the HLA-DR restriction elements. All 4 HLA-DR alleles of the patient (HLA-DRB1*0301, -DRB1*1301, -DRB3*0101, and -DRB3*0202) were isolated and retrovirally transduced into mHag+ EBV-LCLs lacking the appropriate HLA-DR molecules. CD4+ T-cell clones were tested against nontransduced and HLA-DR–transduced EBV-LCLs in IFN-γ ELISA. (A) T-cell clone 62 recognized LB-MR1-1R in the context of HLA-DRB3*0202, (B) T-cell clone 78 recognized LB-PTK2B-1T in the context of HLA-DRB3*0101, (C) T-cell clone 80 recognized LB-LY75-1K in the context of HLA-DRB1*1301, and (D) T-cell clone 87 recognized LB-MTHFD1-1Q in the context of HLA-DRB1*0301. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
Identification of the HLA-DR restriction elements. All 4 HLA-DR alleles of the patient (HLA-DRB1*0301, -DRB1*1301, -DRB3*0101, and -DRB3*0202) were isolated and retrovirally transduced into mHag+ EBV-LCLs lacking the appropriate HLA-DR molecules. CD4+ T-cell clones were tested against nontransduced and HLA-DR–transduced EBV-LCLs in IFN-γ ELISA. (A) T-cell clone 62 recognized LB-MR1-1R in the context of HLA-DRB3*0202, (B) T-cell clone 78 recognized LB-PTK2B-1T in the context of HLA-DRB3*0101, (C) T-cell clone 80 recognized LB-LY75-1K in the context of HLA-DRB1*1301, and (D) T-cell clone 87 recognized LB-MTHFD1-1Q in the context of HLA-DRB1*0301. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
Tissue-specific recognition of HLA-class II–restricted mHags
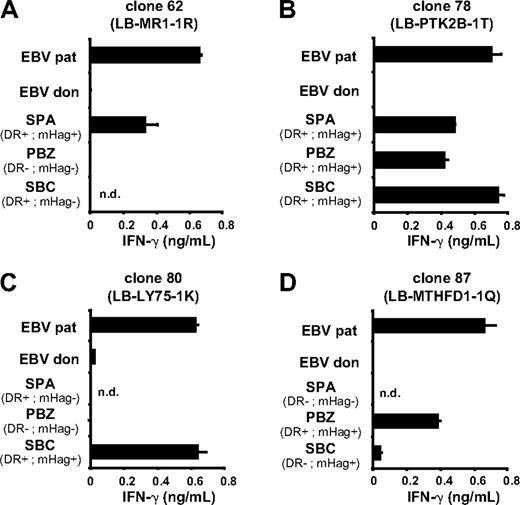
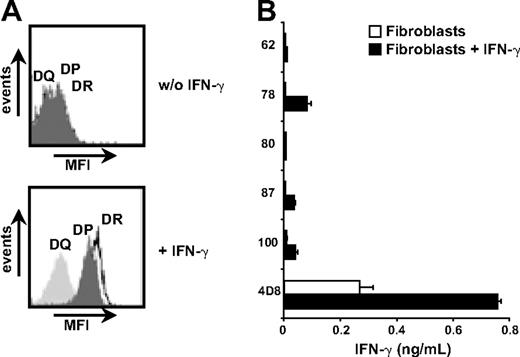
CD33+/CD14− cells from mHag+ patients with AML expressing the appropriate HLA-DR molecules were purified by flow cytometry and tested for T-cell recognition. Figure 5 shows recognition of primary AML cells by all mHag-specific CD4+ T-cell clones. In addition, fibroblasts derived from a skin biopsy of the patient were cultured with and without IFN-γ and tested for recognition. Flow cytometric analysis demonstrated significant up-regulation of HLA-DR surface molecules after treatment with IFN-γ (Figure 6A). Similar results were obtained for skin-derived keratinocytes. None of the CD4+ T-cell clones, however, was capable of recognizing (IFN-γ–treated) fibroblasts (Figure 6B). MHag+ keratinocytes expressing the appropriate HLA-DR molecule were available only for clones 62 and 78. Both clones failed to recognize (IFN-γ–treated) keratinocytes (data not shown), demonstrating that the novel mHags are efficiently presented by HLA-DR surface molecules on various hematopoietic cells, including primary AML cells, but not on nonhematopoietic fibroblasts and keratinocytes.
Recognition of other leukemic cells. Acute myeloid leukemia (AML) cells (CD33+/CD14−) from 3 patients with known HLA and mHag typing were isolated from PBMCs (PBZ, SBC) or bone marrow mononuclear cells (SPA) by flow cytometry. The isolated AML cells were tested for recognition by T-cell clone 62 (A), clone 78 (B), clone 80 (C), and clone 87 (D). EBV-LCLs of donor and patient served as negative and positive control, respectively. The SNP genotyping data (mHag+ or mHag−) and expression of the respective HLA-DR restriction molecule (DR+ or DR−) are indicated for each of the 4 mHags. All 4 T-cell clones recognized mHag+ AML cells expressing the appropriate HLA-DR restriction molecules. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown. nd indicates not determined.
Recognition of other leukemic cells. Acute myeloid leukemia (AML) cells (CD33+/CD14−) from 3 patients with known HLA and mHag typing were isolated from PBMCs (PBZ, SBC) or bone marrow mononuclear cells (SPA) by flow cytometry. The isolated AML cells were tested for recognition by T-cell clone 62 (A), clone 78 (B), clone 80 (C), and clone 87 (D). EBV-LCLs of donor and patient served as negative and positive control, respectively. The SNP genotyping data (mHag+ or mHag−) and expression of the respective HLA-DR restriction molecule (DR+ or DR−) are indicated for each of the 4 mHags. All 4 T-cell clones recognized mHag+ AML cells expressing the appropriate HLA-DR restriction molecules. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown. nd indicates not determined.
Fibroblast recognition by HLA-class II–restricted T cells. Primary fibroblasts of the patient were cultured with and without addition of 100 IU/mL IFN-γ. (A) Up-regulation of HLA-DR, -DQ, and -DP was measured by flow cytometry after 5 days of IFN-γ treatment. (B) T-cell recognition of pretreated and untreated fibroblasts was tested in an IFN-γ ELISA. A CD8+ T-cell clone (4D8) recognizing an unknown mHag in HLA-B8 served as positive control. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
Fibroblast recognition by HLA-class II–restricted T cells. Primary fibroblasts of the patient were cultured with and without addition of 100 IU/mL IFN-γ. (A) Up-regulation of HLA-DR, -DQ, and -DP was measured by flow cytometry after 5 days of IFN-γ treatment. (B) T-cell recognition of pretreated and untreated fibroblasts was tested in an IFN-γ ELISA. A CD8+ T-cell clone (4D8) recognizing an unknown mHag in HLA-B8 served as positive control. Mean release of IFN-γ (ng/mL) in 50-μL supernatants of duplicate wells is shown.
Tissue-specific expression of the mHag-encoding genes
To investigate the tissue distribution of the mHag-encoding genes, nonhematopoietic cells of different origins were cultured (fibroblasts, keratinocytes, biliary epithelial cells, proximal tubular epithelial cells, and gut epithelial cells) or isolated (hepatocytes), and gene expression was analyzed and compared with hematopoietic cells (PBMCs and EBV-LCLs) by quantitative real-time RT-PCR. The data showed overexpression of the LY75 (> 10-fold) and PTK2B (> 4-fold) genes in hematopoietic versus nonhematopoietic cells (Table 2). Expression of the MR1 gene was comparable between hematopoietic cells and fibroblasts, but showed lower expression levels in other nonhematopoietic cells. The MTHFD1 gene was expressed at comparable levels in all analyzed cell types.
mRNA expression of the mHag-encoding genes in hematopoietic and nonhematopoietic cells
| Cell type . | MR1, % . | PTK2B, % . | LY75, % . | MTHFD1, % . |
|---|---|---|---|---|
| EBV-LCLs, n = 2 | 100 | 100 | 100 | 100 |
| PBMCs, n = 2 | 71 | 99 | 34 | 67 |
| Fibroblasts,* n = 2 | 58 | 23 | 0 | 64 |
| Keratinocytes,* n = 2 | 15 | 9 | 2 | 64 |
| Biliary epithelium,† n = 1 | 5 | 3 | 0 | 73 |
| Gut epithelium,* n = 2 | 33 | 7 | 3 | 38 |
| Proximal tubular epithelium,‡ n = 2 | 5 | 26 | 0 | 81 |
| Hepatocytes,§ n = 1 | 3 | 2 | 0 | 49 |
| Cell type . | MR1, % . | PTK2B, % . | LY75, % . | MTHFD1, % . |
|---|---|---|---|---|
| EBV-LCLs, n = 2 | 100 | 100 | 100 | 100 |
| PBMCs, n = 2 | 71 | 99 | 34 | 67 |
| Fibroblasts,* n = 2 | 58 | 23 | 0 | 64 |
| Keratinocytes,* n = 2 | 15 | 9 | 2 | 64 |
| Biliary epithelium,† n = 1 | 5 | 3 | 0 | 73 |
| Gut epithelium,* n = 2 | 33 | 7 | 3 | 38 |
| Proximal tubular epithelium,‡ n = 2 | 5 | 26 | 0 | 81 |
| Hepatocytes,§ n = 1 | 3 | 2 | 0 | 49 |
Expression of the mHag-encoding genes is corrected for expression of the PBGD housekeeping gene and shown relative to the level of gene expression in EBV-LCLs (100%). All measurements have been performed in duplicate with 100 and 10 ng cDNA.
EBV-LCLs indicates Epstein-Barr virus transformed B-cell lines ; PBMCs, peripheral blood mononuclear cells; and n, number of different samples per cell type.
Fibroblasts, keratinocytes, and gut epithelial cells were isolated from biopsies.
Biliary epithelial cells were purchased from ScienCell.
Proximal tubular epithelial cells were kindly provided by Dr C. van Kooten, Department of Nephrology, Leiden University Medical Center (LUMC).
Hepatocytes were kindly provided by Dr A. E. Kremer, Liver Center, Academic Medical Center, University of Amsterdam.
Discussion
In patients with relapsed hematopoietic malignancies after alloSCT, infusions with donor lymphocytes have proven to mediate potent GVL effect resulting in complete remissions.1-3 In this study, we demonstrated that CD4+ T cells directed against 6 different HLA-class II–restricted mHags, including previously identified HLA-DQ–restricted mHag LB-PI4K2B-1S,33 were induced in a patient with relapsed CML after alloSCT who showed long-term complete remission after treatment with DLI.
A broad range of HLA-class I–restricted mHags has been identified using a variety of techniques, including screening of plasmid cDNA libraries,7,10,14,15,17 elution of HLA-bound peptides,5,6,8,11,13,16 genetic linkage analysis,9,12 and recently whole genome association scans.30,31 Most of these techniques did not allow or were difficult to adapt for identification of HLA-class II–restricted mHags. By screening a recombinant bacteria cDNA library,32 we could now identify 4 unknown CD4+ T-cell specificities. This technique is not only extremely efficient for identification of mHags, but may also have broad value for identification of nonpolymorphic antigens in antitumor or autoimmunity. Thus far, most nonpolymorphic HLA-class II–restricted epitopes have been characterized by screening antigens that were known targets for CD8+ T cells or antibodies.40 Although this approach led to the identification of useful targets for T-cell therapy, HLA-class II epitopes from unknown and perhaps clinically more relevant antigens might have been missed.
It is well established that CD4+ T cells are required for efficient induction of in vivo immunity.21,22 In this study, all CD4+ T-cell clones showed not only recognition, but also specific lysis of patient-derived EBV-LCLs, suggesting a role in antitumor immunity as effector cells. Isolated CML progenitor cells, however, were strongly recognized only by T-cell clones 87 and 100, whereas all mHag-specific T-cell clones showed strong recognition of CML cells upon induction of an APC phenotype. Therefore, we hypothesize that all mHag-specific CD4+ T cells were key players as helper cells in the induction of antitumor CD8+ T cells. CD4+ T cells specific for LB-MTHFD1-1Q, represented by clone 87, may have played a more dual role as helper cells as well as effector cells directly eliminating CD34+ CML progenitor cells. In contrast to the variable recognition of isolated CML progenitor cells, all CD4+ T-cell clones strongly recognized AML cells, indicating that all 4 mHags can serve as direct targets for CD4+ effector T cells when presented by the appropriate HLA-DR restriction molecules at the surface of AML cells.
It is unclear why isolated CML progenitor cells, despite significant HLA-DR expression, were not or were hardly recognized by CD4+ T cells specific for LB-MR1-1R, LB-PTK2B-1T, and LB-LY75-1K, whereas CML-APCs were strongly recognized. Possible explanations are overexpression of the mHag-encoding genes or increased surface expression of HLA-DR or costimulatory molecules in CML cells upon induction of an APC phenotype. The latter possibility seems unlikely, because no difference in expression of HLA-DR and various costimulatory (CD40, CD80, CD83, CD86) molecules was observed between CD34+ CML and AML cells (data not shown). A third, and perhaps more likely, explanation is a difference in processing and presentation of endogenous antigens into the HLA-class II pathway in CML-APCs compared with CML progenitor cells. This is supported by recent findings suggesting that maturation of APCs could enhance HLA-class II–restricted antigen presentation of endogenous proteins.41
In several patients treated with alloSCT, high frequencies of mHag-specific CD4+ T cells have been shown to precede and closely correlate with the onset of clinical GVHD.23,42-45 Although constitutive HLA-class II expression is restricted to hematopoietic cells, expression can be induced on nonhematopoietic cells by proinflammatory cytokines. CD4+ T cells recognizing mHags in HLA-class II may therefore contribute to development of GVHD when high levels of proinflammatory cytokines are released as a consequence of conditioning regimens or high pathogenic loads early after transplantation.46
Under noninflammatory conditions, however, mHag-specific CD4+ T cells may exert a selective GVL effect without GVHD. This is supported by one of our recent studies demonstrating high numbers of HLA-DP–specific alloreactive CD4+ T cells in a patient who responded to DLI without GVHD late after HLA-DP–mismatched alloSCT,29 as well as by clinical studies showing that CD8+ T-cell depletion reduces the incidence of GVHD associated with DLI without adversely affecting conversion to donor hematopoiesis.27,28 We here isolated mHag-specific CD4+ T cells from a patient who responded to DLI with only mild GVHD more than 1 year after alloSCT. Our in vitro data demonstrated no recognition of fibroblasts and keratinocytes by the mHag-specific CD4+ T cells, even after up-regulation of HLA-DR by IFN-γ. For LY75 and PTK2B, the lack of recognition can be explained by low gene expression levels in nonhematopoietic cells. For MR1 and MTHFD1, however, gene expression is similar between hematopoietic and nonhematopoietic cells. Our data therefore demonstrate that gene expression is not predictive for T-cell recognition of nonhematopoietic cells, and do not support a role for these mHag-specific CD4+ T cells in development of GVHD. The lack of recognition of nonhematopoietic cells by the mHag-specific CD4+ T cells may be explained by altered protein translation or degradation or inefficient processing and presentation of the novel mHags into the HLA-class II pathway, as postulated for isolated CML progenitor cells.
All 4 novel mHags have potential relevance as targets in antitumor immunity after HLA-matched alloSCT. LB-LY75-1K and LB-PTK2B-1T are attractive based on overexpression of their genes in hematopoietic versus nonhematopoietic cells, whereas LB-MTHFD1-1Q may be useful as direct target for CD4+ T cells against CD34+ CML progenitor cells. Of special interest may be the PTK2B gene, also known as Pyk2/RAFTK/FAK2/CAKβ. The nonreceptor tyrosine kinase has been reported to play a role in dysregulation of various cellular processes in tumor cells, and may be interesting for immunotherapy as potential tumor-associated antigen.47-51 This is supported by the finding that antibodies against PTK2B have been observed in 3 of 19 patients with CML who responded to CD4+ DLI.52 Because PTK2B is an intracellular protein, antibodies against PTK2B are not expected to contribute directly to the elimination of tumor cells in vivo. The presence of antibodies, however, may indicate induction of a coordinated B- and CD4+ T-cell response against PTK2B, emphasizing the potential relevance of LB-PTK2B-1T as target in antitumor immunity after HLA-matched alloSCT.
By screening a recombinant bacteria cDNA expression library, we identified 4 novel HLA-DR–restricted mHags that contributed to GVL reactivity in a patient with complete remission of relapsed CML after DLI. Identification of HLA-class II–restricted mHags is needed to elucidate the role of CD4+ T cells in GVL and GVHD, and is crucial for development of new T-cell–based immunotherapies. The newly identified mHags provide a variety of possible applications. All HLA-DR–restricted mHags have balanced population frequencies and common HLA restrictions, and therefore allow selection of HLA-matched and mHag-mismatched patient-donor pairs for allogeneic stem cell transplantation. The HLA-class II–restricted mHags also open possibilities to specifically induce antileukemic CD4+ T cells, either in vitro for adoptive transfer or in vivo by vaccination. Finally, the identified mHags may be of relevance as helper epitopes to induce alloreactive or autoreactive CD8+ T cells against (HLA-class II−) solid tumors. However, all mHags have been identified as targets for CD4+ T cells isolated from 1 patient, and it therefore remains to be shown whether these mHags are broadly immunogenic in other mHag-disparate patient-donor pairs.
In conclusion, our detailed analysis of the in vivo induced CD4+ T-cell response after DLI provides insight into the role of CD4+ T cells as helper and/or effector cells in therapeutic alloreactivity. The findings establish a basis for fundamental understanding of these phenomena, which is required for optimal use of HLA-class II–restricted mHags in the development of new T cell–based immunotherapies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Harskamp for chimerism analysis and M. van der Hoorn and G. de Roo for technical assistance with flow cytometric isolation. We thank Dr A. E. Kremer, Academic Medical Center Liver Center, University of Amsterdam, and Dr C. van Kooten, Leiden University Medical Center, for kindly providing hepatocytes and renal epithelium, respectively.
This work was supported by grants from the Dutch Cancer Society (UL2008-4111) and the European Union (6th Framework Program Allostem).
Authorship
Contribution: A.N.S. designed and performed research, analyzed data, and wrote the paper; E.D.v.d.M. and C.A.M.v.B. performed research and analyzed data; R.W. wrote the paper; J.H.F.F. designed research, analyzed data, and wrote the paper; and M.G. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marieke Griffioen, Department of Hematology, Leiden University Medical Center, PO Box 9600, 2300 RC, Leiden, The Netherlands; e-mail: m.griffioen@lumc.nl.