Abstract
The sialomucin CD43 is highly expressed on most hematopoietic cells. In this study, we show that the CD43 ectodomain is shed from murine granulocytes, mast cells, and T cells, but not from macrophages. To study the significance of CD43 shedding, we constructed 2 CD43/34 chimeras in which the CD43 membrane-proximal or transmembrane domain was swapped with the corresponding domain from CD34 that is not shed from cells. Viability of cells that normally shed CD43 was negatively affected when forced to express either of the 2 CD43/34 chimeras, but toxicity was reduced when cells coexpressed wild-type CD43. The CD43 cytoplasmic tail (CD43ct) was found to translocate into the nucleus, and inhibition of either its nuclear translocation or its release by γ-secretase was proapoptotic. Involvement of CD43 in regulation of apoptosis is consistent with our findings that CD43ct was modified by small ubiquitin-like modifier-1 and was colocalized with promyelocytic nuclear bodies. CD43-deficient cells exhibited reduced levels of promyelocytic nuclear bodies and had increased sensitivity to apoptosis induced by growth factor withdrawal or T-regulatory cell suppression. Taken together, our data indicate an essential function of CD43 processing and nuclear localization of CD43ct in cell homeostasis and apoptosis.
Introduction
CD43 is a type I transmembrane protein that is heavily expressed on all hematopoietic cells except IgM+ B cells and erythrocytes. Its extracellular domain (224 amino acids) has a rigid rod-like shape that is extensively modified by O-linked carbohydrate chains and abundant terminal sialic acid residues. The 124-amino-acid cytoplasmic tail is highly conserved across species.1
CD43-deficient mice have mild phenotypes, and the functional significance of CD43 continues to be elusive and controversial. CD43 abundance and high cell-surface profile have prompted many studies on CD43 extracellular interactions, and CD43 has been found to have proadhesive as well as antiadhesive properties.2 CD43 can also function as a signaling molecule. Its cytoplasmic tail has conserved threonine and serine residues, which can be phosphorylated,3,4 and ligation with CD43 antibodies leads to T-cell costimulation;5 Fyn/Vav tyrosine phosphorylation through a Src homology 3 binding domain;6,7 protein kinase C activation;8 induction of DNA-binding activity of activator protein-1, nuclear factor of activated T cells, and nuclear factor κB;9 and nuclear translocation of extracellular signal-related kinase 2.7 However, the data in support of a role of CD43 in signaling were obtained by ligating CD43 in experiments in vitro, and it is not yet clear to what degree CD43 deficiency is associated in vivo with corresponding signaling defects.
Several studies firmly established that human CD43 is proteolytically cleaved from the cell surface,10-12 and soluble CD43 has been found in human plasma at approximately 10 μg/mL.13 Human CD43 shedding appears to involve multiple steps and a variety of sheddases, including metalloproteases, serine proteases, and possibly other proteases. However, the physiologic function of CD43 shedding has not yet been determined. In this manuscript, we report that CD43 shedding also occurs in murine hematopoietic cells, and we document that nuclear translocation of CD43 cytoplasmic tail (CD43ct) is a physiologic event and that inhibition of these 2 processes results in cell death, indicating a critical function of CD43ct in regulation of apoptosis. In support of such a function are our observations that CD43ct is modified by small ubiquitin-like modifier 1 (SUMO-1) peptides and is colocalized with PML (promyelocytic) nuclear bodies (NBs), which both are key players in the regulation of apoptosis.
Methods
Mice and cell lines
Mice aged 8 to 14 weeks were used for experiments and were bred at the Biomedical Research Centre. CD43-deficient mice were backcrossed for 10 generations with C57BL/6.14 Bone marrow cells, harvested from the femur and tibia, were suspended in RPMI 1640 medium (GIBCO-BRL), with 10% fetal calf serum, 5 × 10−5 M β-mercaptoethanol, 100 U/mL penicillin, 100 U/mL streptomycin (Stem Cell Technologies), and 2 mM glutamine (Sigma-Aldrich).
To differentiate bone marrow cells into granulocytes, media were supplemented with 2.5 ng/mL interleukin (IL)–3, 10 ng/mL granulocyte colony-stimulating factor (G-CSF), and 5 μM hydrocortisone for 6 to 8 days; for monocyte/macrophage culture, media were supplemented with 2.5 ng/mL stem cell factor (SCF) and 10 ng/mL macrophage colony-stimulating factor (M-CSF) for 7 to 10 days; and for mast cell differentiation, media were supplemented with 2.5 ng/mL IL-3 and 5 ng/mL SCF for 6 to 10 weeks. Differentiation was confirmed by Wright-Giemsa staining for mast cells, and all cells were analyzed by fluorescence-activated cell sorter (FACS) for GR-1, Sca-1, CD34, Mac-1, and/or F4/80. Activated CD8 T cells were derived from concanavalin A (ConA)–activated (4 μg/mL for 48 hours) splenocytes, maintained in 20 ng/mL IL-2 for 10 to 14 days. To maintain stem cell potential bone marrow, cells were obtained from mice injected with 100 μL of 20 mg/mL 5-fluorouracil for 3 days and cultured in 5 ng/mL SCF, 10 ng/mL Flt3, and 100 ng/mL IL-6. Mouse cell lines MC/9 (mast cell), CTLL-2 (cytotoxic T cell), NSF-60 (immature granulocyte), and WEHI274.3 (macrophage) were maintained in media supplemented with either 20 ng/mL IL-2 or 2.5 ng/mL IL-3. The procedures used in this study were reviewed and approved by the University of British Columbia Animal Care Committee.
Antibodies
S11, rat anti–pan-mouse CD43 antibody, was purified in-house from the hybridoma cell line provided by J. Kemp (Department of Pathology, University of Iowa). H18, rabbit polyclonal CD43 antibody, was raised against a peptide corresponding to the C terminus of mouse CD43.15 M2, FLAG antibody, was purchased from Sigma-Aldrich. Antibodies against SUMO-1 and PML were purchased from Santa Cruz Biotechnology, and all secondary antibodies were purchased from Invitrogen and BD Biosciences.
Construction of chimeric proteins, green fluorescent protein, and endoplasmic reticulum fusion proteins
Constructs encoding CD43 and CD43/34 chimeras were subcloned in the murine stem cell virus (MSCV)–based retroviral vector pMX-pie-FLAG16 and contained CD43 5′ untranslated region, CD43 signal peptide, FLAG epitope, 2× glycine spacer, multiple cloning sites, and internal ribosome entry site (IRES)–enhanced green fluorescent protein (GFP; in the order listed). CD43 was amplified with primers 5′-ccatcgatgacagtctgcagaggacgacga and 3′-ccatcgattagagattgaggtgcggcctcatc from cDNA and cloned by ClaI and EcoRI. Chimera membrane-proximal domain (MP) was constructed by swapping the membrane-proximal region of CD43 (residues 180-229) with the membrane-proximal globular region of CD34 (residues 130-252). Chimera trans-membrane domain (TM) was constructed by swapping the transmembrane domain of CD43 (residues 230-252) with the transmembrane domain of CD34 (residues 253-275). Chimeras were amplified by recombinatorial polymerase chain reaction. Vectors encoding GFP-fusion proteins were produced by deleting the stop codon, 3′ untranslated region, and IRES sequences between the CD43 gene and GFP in pMX-pie-FLAG-CD43. Estrogen receptor (ER) fusion was made by combining the ligand binding domain of ER (SAGNMRAADLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSE YDPTRPFSEA SMGLLTNLADR) with the CD43 cytoplasmic domain and by cloning it into pMX-pie.
Retroviral infection
BOSC23 retrovirus packaging cells were seeded onto plates at 2 × 106 cells per 60-mm dish in Dulbecco modified Eagle medium plus 10% fetal bovine serum (FBS) 1 day before transfection. A total of 4 μg of plasmid per 60-mm dish was transfected by Lipofectamine (Invitrogen), according to the manufacturer's protocol. Viral supernatants were harvested after 48 to 72 hours, filtered (0.45-μm membrane), and resuspended with target cell pellets with 5 μg/mL polybrene (Sigma-Aldrich). Twenty-four hours after infection, cells were washed and grown in RMPI plus 10% fetal calf serum and cytokines.
Flow cytometry
Cell sorting was performed using FACSVantage SE high-speed cell sorter (BD Biosciences). Flow cytometric analyses were performed on FACSCalibur and FACScan instruments using CellQuest (BD Biosciences) or FlowJo (TreeStar) software.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis, immunoblotting, and immunoprecipitation
Cells were lysed on ice for 20 minutes (0.5% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 25 mM Tris-HCl (pH 7.5), 450 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA [ethylenediaminetetraacetic acid], and protease inhibitors), sonicated, and clarified at 12 000g for 5 minutes. Proteins, measured by bicinchoninic acid protein assay (Pierce) or spectrophotometry, were transferred after SDS–polyacrylamide gel electrophoresis onto nitrocellulose membranes that were then blocked in Tris-buffered saline skim milk, incubated with a primary antibody in Tris-buffered saline skim milk and 0.1% Tween 20 at room temperature for 2 hours. Bound antibodies were visualized using species-specific secondary antibodies conjugated with alkaline phosphatase or avidin-conjugated alkaline phosphatase by incubating the membranes at room temperature for 1 hour.
For immunoprecipitation, the soluble fraction was incubated with primary antibody (5 μg per 10-mL supernatant or 0.5-mL lysates diluted in 10 ml phosphate-buffered saline [PBS]) at 4°C for 1 hour. Protein G–Sepharose blocked with 2% bovine serum albumin (BSA) was added, and samples were further incubated at 4°C for 90 minutes, then pelleted and washed 3 times with PBS and resuspended in reducing SDS sample buffer.
Confocal microscopy
Cells were cytospun on poly(l-lysine)–coated slides at 500 rpm for 5 minutes, then fixed and permeabilized simultaneously in 4% paraformaldehyde and 0.5% Nonidet P-40 at 4°C for 4 to 16 hours. Slides were washed 3 times in PBS containing 0.1% Nonidet P-40 and 1% BSA, treated with blocking solution (PBS containing 0.1% Nonidet P-40, 1% BSA, and 5% goat or donkey serum) for 30 minutes at 4°C. Target proteins were stained for 1 hour at room temperature with S11, H18, SUMO-1, or PML antibodies. After 2 washes, slides were incubated with a secondary goat anti–rabbit, goat anti–rat, or donkey anti–goat IgG conjugated with Alexa 488 or Alexa 568 for 1 hour at room temperature. After 2 washes, slides were mounted with 10 μL Fluoromount-G (Southern Biotechnology Associates). Images were captured with a confocal microscope (Nikon Eclipse TE2000-U) equipped with Nikon D-Eclipse C1 lasers and analyzed by the Nikon confocal software.
In vitro transcription/translation and sumoylation
CD43ct construct was cloned into pBluescript-KS in T7 orientation and expressed by using TNT T7 wheat germ extract system (Promega), according to the manufacturer's protocol. The expressed CD43ct peptide was sumoylated by adding SUMO-1, SUMO E1-activating enzyme, and SUMO E2-conjugating enzyme (Biomol), according to the manufacturer's protocol.
Cell purification for T regulatory cell suppression and caspase assay
CD4+ T cells were obtained from spleen by negative selection with magnetic cell separation system (MACS; Miltenyi Biotec) using CD8, B220, and CD11b antibodies. CD4+CD25+ (T regulatory [Treg]) cells were positively enriched from the CD4+ T cells by MACS using CD25 antibody, and the remaining fraction yielded CD4+CD25− T responder (Tresp) cell population. Purified Tresp cells were labeled with 3 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) in Hanks balanced salt solution for 5 minutes at 37°C. A total of 100 000 Tresp cells was cultured in 96-well plates (0.2 mL) in RPMI plus 10% FBS with 4 μg/mL ConA at 37°C for 72 hours. Treg or control (CD4+CD25−) cells (30 000) were added to cultures to induce suppression.
Naive T cells were obtained from lymph nodes by negative selection with MACS (Miltenyi Biotec) using B220, GR-1, and CD11b antibodies. Purified T cells (> 90%) were incubated with 5 μM L685,458 (Sigma-Aldrich) in RPMI plus 10% FBS at 37°C for 2 or 16 hours. Caspase-3 activity was determined in cell lysates using the fluorescent substrate Z-DEVD-R110 (Molecular Probes).
Statistical analysis
Statistical analyses were performed using Microsoft Excel. Data are represented as means plus or minus standard deviation. Statistical significance was determined by 2-tailed, unpaired Student t tests. P equals .05 for all tests.
Results
CD43 is selectively shed from murine hematopoietic cells
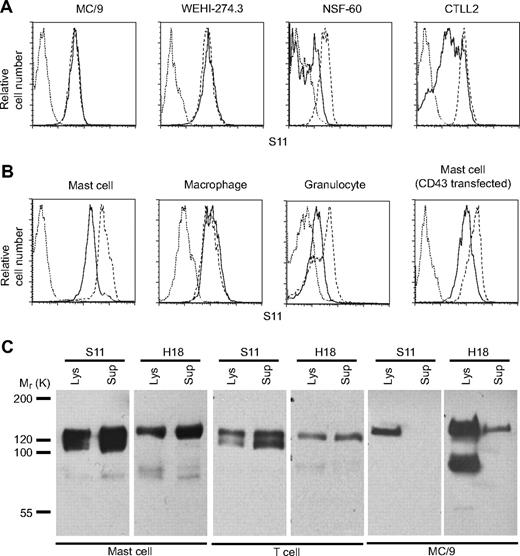
To document shedding of murine CD43, we analyzed activation-induced changes of CD43 expression on several cell lines and primary cells by flow cytometry using S11 mAb that recognizes the CD43 ectodomain in a carbohydrate-independent manner.17 The 12-O-tetradecanoylphorbol-13-acetate (TPA) activation resulted in significant loss of S11 reactivity on CTLL2, NSF-60 cells, bone marrow-derived mast cells, and granulocytes, whereas S11 binding to MC/9, WEHI274.3 cells, and bone marrow-derived macrophages was not affected. N-terminal FLAG-tagged CD43, ectopically expressed in bone marrow-derived mast cells, was also down-regulated after TPA activation (Figure 1A-B). To detect shed CD43 ectodomain, we expressed FLAG-CD43 in bone marrow–derived mast cells and mitogen-activated T cells from CD43-deficient mice and, as a control, in MC/9 cells. FLAG precipitates from culture media and cell lysates were probed with either S11 antibody or H18 antibody that binds to the cytoplasmic tail of CD43. Full-length FLAG-CD43 corresponding to a 135-kDa band was recognized by S11 and H18 antibodies in all 3 target cells (Figure 1C). S11 blots revealed in conditioned media and lysates of mast cells and T cells, but not MC/9 cells, a second 120-kDa band that was not reactive with H18, indicating the presence of shed CD43 ectodomain. Culture supernatant of primary macrophages infected with FLAG-CD43 was also devoid of 120-kDa band reactive with S11 (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article), confirming further that CD43 is not shed from monocytes/macrophages. Full-length FLAG-CD43 was detected in FLAG immunoprecipitates of culture supernatants from all 3 cell types analyzed, indicating that full-length CD43 is also released into culture media, possibly stemming from membrane blebbing.
CD43 is selectively shed from murine cells. (A) Flow cytometry analysis of CD43 cell surface expression on murine cell lines using CD43 antibody S11. Cells were stimulated for 3 hours with 10 nM TPA (solid line), vehicle alone (dashed line), and isotype control (dotted line). (B) Flow cytometric analysis of CD43 cell-surface expression on bone marrow–derived mast cells, macrophages, granulocytes, and CD43-deficient mast cells reconstituted with FLAG-CD43. Cells were stimulated for 3 hours with 10 nM TPA (solid line), vehicle alone (dashed line), and isotype control (dotted line). (C) Western blot analysis of FLAG immunoprecipitates from cell lysates (5 × 106 cells/well) and conditioned media of primary mast cells, activated CD8 T cells (spleen), and MC/9 cells infected with FLAG-CD43. Data are representative of at least 2 repeat experiments.
CD43 is selectively shed from murine cells. (A) Flow cytometry analysis of CD43 cell surface expression on murine cell lines using CD43 antibody S11. Cells were stimulated for 3 hours with 10 nM TPA (solid line), vehicle alone (dashed line), and isotype control (dotted line). (B) Flow cytometric analysis of CD43 cell-surface expression on bone marrow–derived mast cells, macrophages, granulocytes, and CD43-deficient mast cells reconstituted with FLAG-CD43. Cells were stimulated for 3 hours with 10 nM TPA (solid line), vehicle alone (dashed line), and isotype control (dotted line). (C) Western blot analysis of FLAG immunoprecipitates from cell lysates (5 × 106 cells/well) and conditioned media of primary mast cells, activated CD8 T cells (spleen), and MC/9 cells infected with FLAG-CD43. Data are representative of at least 2 repeat experiments.
Ectopic expression of CD43/34 chimeras is associated with toxicity
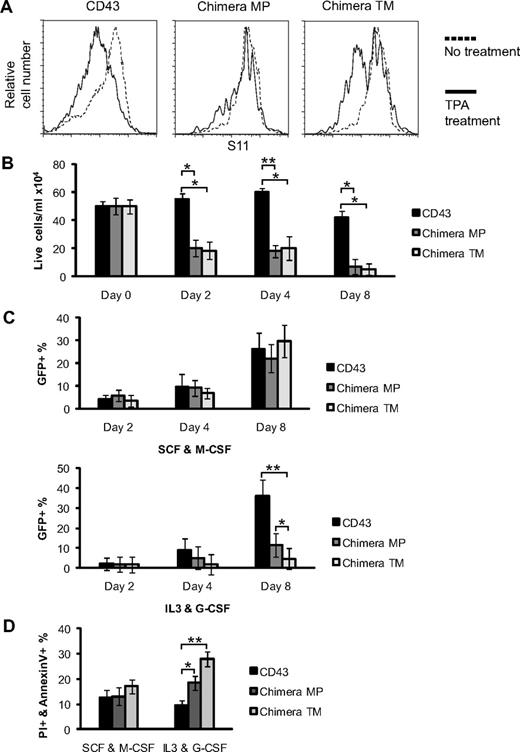
CD34 resembles CD43 in structure, but in the literature there is no indication of CD34 ectodomain release. Indeed, FLAG-CD34 ectopically expressed in bone marrow–derived mast cells is not released after TPA stimulation (supplemental Figure 2A). In an attempt to produce nonsheddable CD43, we constructed 2 retroviral expression vectors encoding CD43/34 chimeras: CD43/34 TM containing the CD34 transmembrane domain and CD43/34 MP containing the globular CD34 membrane-proximal domain (see diagrams in supplemental Figure 2B). The chimeras and control CD43 express FLAG at the N terminus, and are followed by IRES-GFP. CD43 and the CD43/34 chimeras were expressed in CD43-deficient bone marrow cells grown in IL-3/G-CSF. FACS analysis on GR-1+ cells 6 days after infection revealed that TPA activation did not induce CD43 ectodomain shedding from chimera MP, whereas CD43 signal was lost on cells reconstituted with chimera TM or CD43 (Figure 2A), indicating that CD43 membrane-proximal region is required for ectodomain shedding. Immunoblots of FLAG precipitates from CD43 MP- and CD43 TM-infected cells did reveal cleaved CD43 ectodomain in lysates of CD43 TM-infected cells, whereas no cleaved CD43 ectodomain was found in lysates of CD43 MP-infected cells, further confirming that CD43 MP cannot be processed (supplemental Figure 2C). Interestingly, only half of the chimera TM-expressing cells shed CD43. FACS analysis revealed that cells that did not shed CD43/34 TM were positive for the monocyte/macrophage lineage marker F4/80 (supplemental Figure 3), whereas cells that shed CD43/34 TM were negative for F4/80, consistent with our observation that macrophages do not shed CD43.
Ectopic expression of nonsheddable CD43/34 chimeras is toxic. (A) Flow cytometric analysis of CD43 cell surface expression on CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras MP and TM. After retroviral infection, cells were differentiated for 5 days into granulocytes and then stimulated for 3 hours with 10 nM TPA (solid line) or vehicle alone (dashed line). Cells shown were gated on GR-1+, and data are representative of 3 repeat experiments. (B) Cell yield of CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras maintained for up to 8 days in IL-3/G-CSF. Numbers of PI− cells are shown for days 0, 2, 4, and 8. n = 4; *P = .001 and **P = .001 (Student t test). (C) Cell yield of CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras maintained in SCF/M-CSF (top panel) or in IL-3/G-CSF (bottom panel). Percentages of GFP+/PI− cells are shown for days 2, 4, and 8 after infection. *P = .01 and **P = .007 (Student t test). (D) Percentage of annexin V+/PI+ cells is shown at day 8 of CD43-deficient bone marrow cells infected with either wt CD43 or CD43/34 chimeras. During retroviral infection, cells were maintained for 2 days in SCF/Flt3/IL-6, then split and cultured for additional 6 days in either SCF/M-CSF or IL-3/G-CSF. n = 4; *P = .007 and **P = .001 (Student t test). Data are mean ± SD and are representative of at least 3 independent experiments.
Ectopic expression of nonsheddable CD43/34 chimeras is toxic. (A) Flow cytometric analysis of CD43 cell surface expression on CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras MP and TM. After retroviral infection, cells were differentiated for 5 days into granulocytes and then stimulated for 3 hours with 10 nM TPA (solid line) or vehicle alone (dashed line). Cells shown were gated on GR-1+, and data are representative of 3 repeat experiments. (B) Cell yield of CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras maintained for up to 8 days in IL-3/G-CSF. Numbers of PI− cells are shown for days 0, 2, 4, and 8. n = 4; *P = .001 and **P = .001 (Student t test). (C) Cell yield of CD43-deficient bone marrow cells infected with wt CD43 or CD43/34 chimeras maintained in SCF/M-CSF (top panel) or in IL-3/G-CSF (bottom panel). Percentages of GFP+/PI− cells are shown for days 2, 4, and 8 after infection. *P = .01 and **P = .007 (Student t test). (D) Percentage of annexin V+/PI+ cells is shown at day 8 of CD43-deficient bone marrow cells infected with either wt CD43 or CD43/34 chimeras. During retroviral infection, cells were maintained for 2 days in SCF/Flt3/IL-6, then split and cultured for additional 6 days in either SCF/M-CSF or IL-3/G-CSF. n = 4; *P = .007 and **P = .001 (Student t test). Data are mean ± SD and are representative of at least 3 independent experiments.
Surprisingly, bone marrow cells that were infected with either of the 2 CD43/34 chimeras could not be maintained in media supplemented with IL-3/G-CSF, in contrast to cells infected with CD43 (Figure 2B), vector-alone control, or CD34 (data not shown). To investigate whether ectopic expression of CD43/34 chimeras also affected cell viability of primary macrophages that do not shed CD43 and whether there was a possible connection between CD43 sheddability and cell viability, we infected CD43-deficient bone marrow cells with either CD43 or the CD43/34 chimeras and cultured the cells in either M-CSF/SCF or G-CSF/IL-3. Retroviral infection efficiency and yield of GFP+ cells were similar for each of the CD43 forms in cultures maintained in M-CSF/SCF, with 22% to 30% cells being GFP+ 8 days after infection, documenting that the retroviral vectors had comparable transfection efficiencies and that ectopic expression of CD43/34 chimeras is not toxic in macrophage/monocytic cells (Figure 2C). In contrast, cells maintained in IL-3/G-CSF yielded significantly less GFP+ cells in CD43/34 chimera-infected cultures (11% and 4% GFP+) than in CD43-infected cultures (35% GFP+). To exclude a potential influence of the different growth factors used during infection, we infected bone marrow cells while maintaining them under stem cell/progenitor-enriching conditions. Cultures were then split and maintained for an additional 6 days in either IL-3/G-CSF or M-CSF/SCF. CD43/34 transfectants grown in IL-3/G-CSF had higher apoptotic and necrotic cells compared with corresponding CD43 transfectants, whereas transfectants maintained in M-CSF/SCF did not differ (Figure 2D), indicating that both CD43/34 chimeras exert toxic effects when they are expressed in cells in which CD43 shedding normally occurs.
Endogenous CD43 reduces toxicity of CD43/34 chimeras
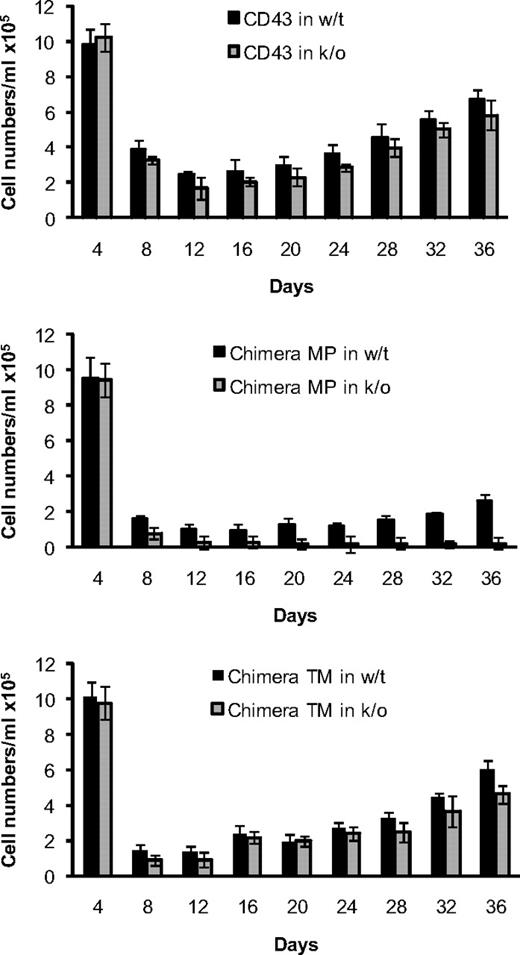
To determine whether endogenous CD43 can modulate the CD43/34 chimera-associated toxicity and to examine the chimera-associated toxicity for a more extended period, we established mast cell cultures from wild-type (wt) and CD43-deficient bone marrow cells that were infected with the different CD43 constructs. C57BL/6 wt and CD43-deficient bone marrow cells that were infected with wt-CD43 followed the same typical growth curve of mast cell differentiation (Figure 3). The number of GFP+ cells was lowest by day 12 (∼ 2 × 104 GFP+ cells/mL), and GFP+ cells increased steadily thereafter. In contrast, cultures infected with either of the CD43/34 chimeras contained approximately 50% less GFP+ cells at day 12. Cells expressing CD43/34 chimera MP showed a retarded cell growth that was especially apparent in CD43-deficient cells, in which GFP+ cells were steadily lost over time and cell numbers remained very low. CD43+ cells tolerated expression of chimera MP to some degree, and by day 36 there was a 50-fold higher number of GFP+ cells than in CD43-deficient cells, indicating that endogenous CD43 can rescue the chimera MP-associated toxicity to some extent. Chimera TM-infected cells showed a relative good recovery after day 12 that was again somewhat better for CD43+ than CD43-deficient cells. Control experiments confirmed that measurement of GFP was a reliable indicator for CD43 cell surface expression (supplemental Figure 4). In summary, our data show that CD43 membrane-proximal and CD43 transmembrane domains are both of functional importance. Their replacement with a corresponding CD34 segment may thus affect a common downstream event. We hypothesize that this common downstream event may constitute inhibition of processing by γ-secretase in that chimera MP may be resistant to γ-secretase because the cleavage would be dependent on a prior cleavage by α-secretase and chimera TM may not be cleaved by γ-secretase because its CD34 transmembrane domain is not a substrate.
Endogenous expressed CD43 reduces toxicity of CD43/34 chimeras. Mast cell differentiation of wt and CD43-deficient bone marrow cells after retroviral infection with wt CD43 (top panel), CD43/34 chimera MP (middle panel), or chimera TM (bottom panel). Yields of GFP+/PI− cells are shown in 4-day intervals up to day 36 after retroviral infection. Data are mean ± SD (n = 3) and are representative of 2 independent experiments.
Endogenous expressed CD43 reduces toxicity of CD43/34 chimeras. Mast cell differentiation of wt and CD43-deficient bone marrow cells after retroviral infection with wt CD43 (top panel), CD43/34 chimera MP (middle panel), or chimera TM (bottom panel). Yields of GFP+/PI− cells are shown in 4-day intervals up to day 36 after retroviral infection. Data are mean ± SD (n = 3) and are representative of 2 independent experiments.
Nuclear translocation of CD43ct is of physiologic importance
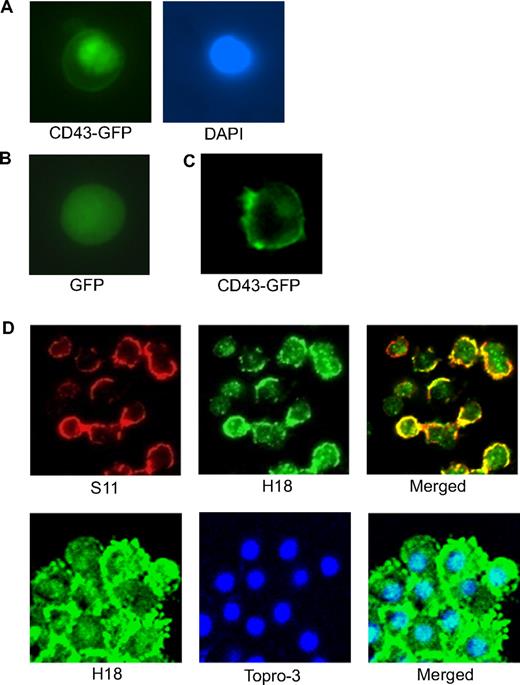
Many γ-secretase products are transported into the nucleus, and indeed, the membrane-proximal CD43 cytoplasmic domain contains 2 stretches of basic amino acids, which resemble a classical nuclear localization signal motif (RQRQKRRTGALTLSGGGKR; residues 272-291). It has furthermore been shown that ectopically expressed human CD43ct can be detected in the nucleus, suggesting that CD43ct might be released from membrane-anchored full-length CD43 and translocate into the nucleus.18 Using 2 different approaches, we show that CD43ct translocates to the nucleus in murine cells. Transfection of cells with CD43 fused with GFP at its C terminus resulted in GFP fluorescence signals in the nucleus (Figure 4A), and confocal microscopy of bone marrow cells showed that S11 signals were consistently located only in the membrane areas, whereas H18 signals could be found associated with membrane and nuclear regions (Figure 4D). Costaining of activated T cells with H18 and the nuclear stain Topro-3 further confirmed the nuclear localization of CD43ct (Figure 4D). When CD43-GFP fusion protein was expressed in primary macrophages, fluorescent signals were exclusively associated with the cell membrane and no nuclear GFP signal was observed (Figure 4C), suggesting that CD43 processing is required for CD43ct release and its nuclear translocation.
CD43ct is localized in the nucleus. (A) Fluorescent microscopy of CD43-deficient bone marrow cells infected with CD43-GFP. DAPI (4,6 diamidino-2-phenylindole) staining shows the location of the nucleus. (B) Fluorescent microscopy of CD43-deficient bone marrow cells infected with GFP alone as a control. (C) Fluorescent microscopy of CD43-deficient macrophages infected with CD43-GFP. (D) Confocal microscopy of total bone marrow cells stained with S11 and H18 antibodies (top panel) and confocal microscopy of mitogen-activated T cells stained with H18 antibody and Topro-3 nuclear stain (bottom panel).
CD43ct is localized in the nucleus. (A) Fluorescent microscopy of CD43-deficient bone marrow cells infected with CD43-GFP. DAPI (4,6 diamidino-2-phenylindole) staining shows the location of the nucleus. (B) Fluorescent microscopy of CD43-deficient bone marrow cells infected with GFP alone as a control. (C) Fluorescent microscopy of CD43-deficient macrophages infected with CD43-GFP. (D) Confocal microscopy of total bone marrow cells stained with S11 and H18 antibodies (top panel) and confocal microscopy of mitogen-activated T cells stained with H18 antibody and Topro-3 nuclear stain (bottom panel).
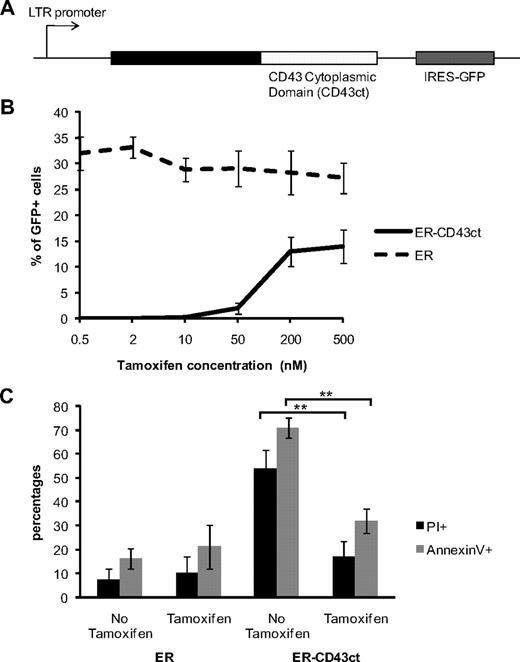
To determine whether nuclear translocation of CD43ct was of physiologic importance, we used an ER-CD43ct fusion protein,19 which allowed us to control the distribution of CD43ct between cytoplasm and nucleus. We infected CD43-deficient bone marrow cells grown in IL-3/G-CSF with a retroviral vector encoding an ER-CD43ct fusion protein, followed by IRES-GFP (Figure 5A). Successful infection of cells with this vector required a continuous supplement of high concentrations of the estrogen analog tamoxifen that induces nuclear translocation of the ER. Infection efficiency was generally low, but survival of infected cells was tamoxifen dependent (Figure 5B). Withdrawal of tamoxifen resulted in a significant increase of propidium iodide (PI) and annexin V–positive cells (Figure 5C), indicating that inhibition of nuclear localization of CD43ct negatively affects cell viability and confirming our hypothesis that impaired nuclear translocation of CD43ct is the underlying cause of cell toxicity. Control cells infected with ER alone did not show an increase in PI+ and annexin V+. Collectively, our data indicate that CD43ct nuclear translocation is necessary for cell homeostasis.
Inhibition of nuclear translocation of CD43ct induces apoptosis. (A) Diagram of the plasmid encoding the ER-CD43ct fusion protein. (B) CD43-deficient bone marrow cells infected with ER-CD43ct or ER were grown for 5 days in IL-3/G-CSF. Percentages of GFP+/PI− cells are shown in cultures supplemented with increasing concentrations of tamoxifen. (C) Aliquots of cultures kept in 100 nM tamoxifen were deprived of tamoxifen at day 5 for 1 day. Percentages of PI+ and percentages of annexin V+ cells are shown at day 6 in cultures maintained without tamoxifen and cultures with tamoxifen. **P = .001. Data are mean ± SD (n = 3) and are representative of 2 independent experiments.
Inhibition of nuclear translocation of CD43ct induces apoptosis. (A) Diagram of the plasmid encoding the ER-CD43ct fusion protein. (B) CD43-deficient bone marrow cells infected with ER-CD43ct or ER were grown for 5 days in IL-3/G-CSF. Percentages of GFP+/PI− cells are shown in cultures supplemented with increasing concentrations of tamoxifen. (C) Aliquots of cultures kept in 100 nM tamoxifen were deprived of tamoxifen at day 5 for 1 day. Percentages of PI+ and percentages of annexin V+ cells are shown at day 6 in cultures maintained without tamoxifen and cultures with tamoxifen. **P = .001. Data are mean ± SD (n = 3) and are representative of 2 independent experiments.
CD43 protects cells from apoptosis
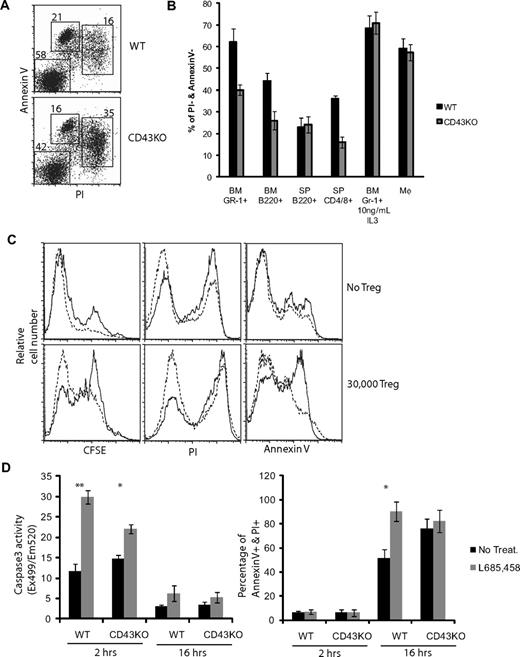
Apoptosis, associated with retention of CD43ct in the membrane and/or cytoplasm caused by inhibition of CD43ct nuclear translocation or inhibition of CD43 processing, could be indicative of a role of CD43 in cell homeostasis. To elucidate whether lack of CD43 affected apoptotic responses in hematopoietic cells, we compared the extent of apoptosis induction, measured by PI and annexin V staining, in bone marrow cells, lymphocytes, and splenocytes from wt and CD43-deficient mice. Of the apoptotic inducers analyzed, which included treatment with γ-irradiation, interferon-γ, Fas antibody, cycloheximide, and tumor necrosis factor-α, only growth factor withdrawal and Treg suppression showed significant differences between wt and CD43-deficient cells, indicating that CD43 may be involved in specific apoptotic signaling pathways. CD43-deficient bone marrow cultures maintained 36 hours in media alone contained more than 34% of apoptotic (PI/annexin V double-positive) and only 41.1% live (PI/annexin V–negative) GR-1+ cells, whereas in corresponding cultures less than 16% of the GR-1+ cells were apoptotic and more than 57% were negative for PI/annexin V (Figure 6A). Similarly, wt B220+ bone marrow cells and CD4+/CD8+ splenocytes (Figure 6B) showed better survival in growth factor–deficient media than corresponding CD43-deficient cells, whereas spleen-derived B220+ cells, which do not express CD43,1 and macrophages, which do not shed CD43, did not differ in PI/annexin staining. These observations show that CD43 expression can convey a survival advantage and suggest that CD43 can affect apoptotic responses.
CD43 has an antiapoptotic function. (A) PI and annexin V staining of Gr-1+ cells purified from bone marrow cells of wt and CD43-deficient mice after 36-hour culture in RPMI without growth factors. (B) Data show percentage of live and nonapoptotic cells (PI−/annexin V−) in cultures of wt (■) and CD43-deficient (▩) cells; Gr-1+ cells purified from bone marrow (BM), B220+ cells purified from bone marrow and from spleen (SP), CD4+/CD8+ T cells purified from spleen, and macrophages (Mφ) cultured from bone marrow cells. Data for Gr-1+ and Mφ cells are shown after a 36-hour time point; B220+ and CD4/CD8+ cells are shown after a 16-hour time point, n = 3. (C) Purified CD4+ T cells from wt (dashed line) and CD43-deficient (solid line) mice were stained with CFSE and stimulated with ConA. FACS profiles show CFSE dilution, PI, and annexin V profiles of cultures without Treg (top panels) and with 30 000 Treg added (bottom panels) 3 days after mitogen stimulation. (D) Naive T cells maintained in RPMI without growth factors were treated with γ-secretase inhibitor (▩; L685,458) or vehicle control (■; No treatment). Induction of apoptosis was assessed by determination of caspase-3 activity or annexin V and PI staining 2 hours and 16 hours after initiation of culture conditions, respectively. n = 3; *P < .05 and **P < .01 (Student t test). Data are mean ± SD and are representative of at least 3 independent experiments.
CD43 has an antiapoptotic function. (A) PI and annexin V staining of Gr-1+ cells purified from bone marrow cells of wt and CD43-deficient mice after 36-hour culture in RPMI without growth factors. (B) Data show percentage of live and nonapoptotic cells (PI−/annexin V−) in cultures of wt (■) and CD43-deficient (▩) cells; Gr-1+ cells purified from bone marrow (BM), B220+ cells purified from bone marrow and from spleen (SP), CD4+/CD8+ T cells purified from spleen, and macrophages (Mφ) cultured from bone marrow cells. Data for Gr-1+ and Mφ cells are shown after a 36-hour time point; B220+ and CD4/CD8+ cells are shown after a 16-hour time point, n = 3. (C) Purified CD4+ T cells from wt (dashed line) and CD43-deficient (solid line) mice were stained with CFSE and stimulated with ConA. FACS profiles show CFSE dilution, PI, and annexin V profiles of cultures without Treg (top panels) and with 30 000 Treg added (bottom panels) 3 days after mitogen stimulation. (D) Naive T cells maintained in RPMI without growth factors were treated with γ-secretase inhibitor (▩; L685,458) or vehicle control (■; No treatment). Induction of apoptosis was assessed by determination of caspase-3 activity or annexin V and PI staining 2 hours and 16 hours after initiation of culture conditions, respectively. n = 3; *P < .05 and **P < .01 (Student t test). Data are mean ± SD and are representative of at least 3 independent experiments.
To explore in a more physiologic setting CD43 involvement in regulation of apoptosis, we tested whether CD43 can modulate the sensitivity of activated CD4 T cells to Treg-induced killing. To exclude influences of intercell culture differences, CD4 T cells from wt congenic Ly5.1 mice were mixed at a 1:1 ratio with CD43-deficient CD4 T cells (Ly5.2). Proliferation of mitogen-activated T cells was measured by CFSE dilution, and induction of apoptosis was assessed by PI and annexin V staining in cultures with or without Treg cells.20 ConA-induced cell division was not affected by CD43 status in absence of Treg cells (Figure 6C). Addition of Treg cells was, as expected, associated with an inhibition of cell proliferation that was more pronounced for CD43-deficient than wt CD4 T cells. Similarly, CD43-deficient T cells had a more pronounced increase of PI+ and annexin V+ cells than CD43+ T cells, consistent with the notion that CD43 has an antiapoptotic function.
To directly investigate whether CD43 processing by γ-secretase is involved in apoptosis regulation, we maintained naive T cells from wt and CD43-deficient mice in media without growth factors and treated the cells with the γ-secretase inhibitor L685,458. Measurement of the early marker of apoptosis induction, caspase 3, showed that after 2 hours, wt T cells had an approximately 3-fold increase in caspase-3 activity compared with vehicle-treated cells (Figure 6D). CD43-deficient cells in contrast had only a modest (approximately 0.5-fold) increase in caspase 3 activity over vehicle-treated cells, indicating that CD43+ cells are more sensitive to γ-secretase inhibition. Analysis of induction of the late apoptosis markers annexin V and PI after 16 hours of treatment showed that the percentage of annexin V/PI double-positive cells increased in wt T-cell cultures from 50% in vehicle control cultures to 90% in L685,458-treated cultures, whereas γ-secretase inhibition did not significantly increase annexin V/PI double-positive cells in CD43-deficient T cells. Collectively, these observations support the concept that processing of CD43 by γ-secretase is associated with regulation of apoptosis.
CD43ct is sumoylated and colocalized with PML NB
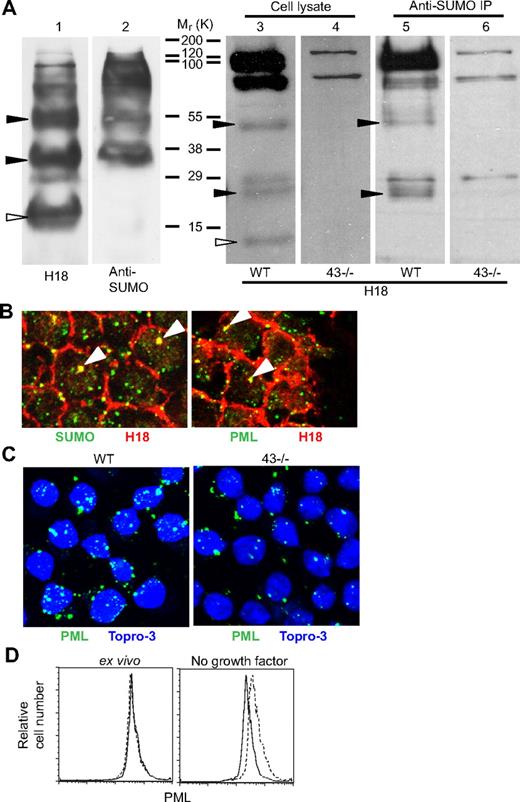
The observation that CD43ct translocates to the nucleus and may be involved in regulation of apoptosis prompted us to search for protein domain motifs indicative of a nuclear role of CD43ct. The search revealed 3 potential SUMO modification consensus sequence sites (ψKXE) located close to the C terminus (LKPG, residues 353-356; LKGE, residues 361-364; AKDE, residues 386-389). Using an in vitro sumoylation assay, we found that CD43ct can be modified by SUMO-1. H18 blots of in vitro sumoylated CD43ct revealed 3 bands with molecular mass 20 kDa, 36 kDa, and 55 kDa (Figure 7A), whereas corresponding SUMO-1 antibody blots revealed 2 bands with molecular mass 36 kDa and 55 kDa, suggesting that the 20-kDa (open triangle) band corresponds to CD43ct and the other 2 bands represent CD43ct modified with 1 and 3 SUMO-1, respectively. The somewhat higher than expected molecular mass estimates (13.5 kDa, 26 kDa, and 52 kDa for CD43ct, CD43ct + 1 SUMO, and CD43ct + 3 SUMO, respectively) are most likely due to posttranslational modifications associated with the in vitro wheat germ protein expression system used.
CD43ct is modified by SUMO-1 and localized in PML NB. (A) In vitro transcribed/translated CD43ct was sumoylated with SUMO-1 in vitro, and the products were then separated by SDS–polyacrylamide gel electrophoresis and probed with H18 antibody (lane 1) and SUMO-1 antibody (lane 2). Western blot analysis of total bone marrow cell lysates from wt (lanes 3 and 5) and CD43-deficient (lanes 4 and 6) mice is shown. Lanes 3 and 4 are whole cell lysates probed with H18 antibody. Lanes 5 and 6 are SUMO-1 immunoprecipitates probed with H18 antibody. Filled arrows indicate bands representing CD43ct modified with 1 and 3 SUMO-1. Open arrows indicate nonsumoylated CD43ct. (B) Confocal microscopy of total bone marrow cells from wt mice stained with H18 and SUMO-1 antibodies (left panel) and H18 and PML antibodies (right panel). (C) Confocal microscopy of wt (left panel) and CD43-deficient (right panel) total bone marrow cells stained with PML antibody after 24 hours in media alone. Data are representative of at least 3 repeat experiments. (D) FACS analysis of wt (blue line) and CD43-deficient (red line) total bone marrow cells stained for intracellular PML; ex vivo (left panel) and after 24 hours in media alone (right panel).
CD43ct is modified by SUMO-1 and localized in PML NB. (A) In vitro transcribed/translated CD43ct was sumoylated with SUMO-1 in vitro, and the products were then separated by SDS–polyacrylamide gel electrophoresis and probed with H18 antibody (lane 1) and SUMO-1 antibody (lane 2). Western blot analysis of total bone marrow cell lysates from wt (lanes 3 and 5) and CD43-deficient (lanes 4 and 6) mice is shown. Lanes 3 and 4 are whole cell lysates probed with H18 antibody. Lanes 5 and 6 are SUMO-1 immunoprecipitates probed with H18 antibody. Filled arrows indicate bands representing CD43ct modified with 1 and 3 SUMO-1. Open arrows indicate nonsumoylated CD43ct. (B) Confocal microscopy of total bone marrow cells from wt mice stained with H18 and SUMO-1 antibodies (left panel) and H18 and PML antibodies (right panel). (C) Confocal microscopy of wt (left panel) and CD43-deficient (right panel) total bone marrow cells stained with PML antibody after 24 hours in media alone. Data are representative of at least 3 repeat experiments. (D) FACS analysis of wt (blue line) and CD43-deficient (red line) total bone marrow cells stained for intracellular PML; ex vivo (left panel) and after 24 hours in media alone (right panel).
Western blot analysis of bone marrow cell lysates from wt and CD43-deficient mice with H18 antibody revealed bands with molecular mass 13 kDa, 26 kDa, and 52 kDa, which were absent in lysates from CD43-deficient cells (Figure 7A). H18 blots of SUMO-1 immunoprecipitates revealed bands at 26 kDa and 52 kDa that were absent in corresponding blots from CD43-deficient immunoprecipitates, indicating that in vivo CD43ct sumoylation is identical to the pattern observed using the in vitro approach. Thus, all the 3 SUMO consensus sequences on CD43ct can be recognized and modified by SUMO-conjugating enzymes, but interestingly, CD43ct is modified with either 1 or 3, but not 2 SUMO peptides only.
The majority of SUMO-modified proteins have been shown to be localized in a subnuclear structure called the PML NB, which are made up of PML protein. Confocal microscopy of bone marrow cells with PML and H18 antibodies confirmed that CD43ct is also colocalized with PML in NB (Figure 7B). To determine whether CD43 affects PML expression quantitatively, we used FACS and confocal microscopy and found that under apoptotic conditions, induced by growth factor withdrawal, PML expression was much reduced in CD43-deficient cells (Figure 7C-D), indicating that CD43ct directly or indirectly influences the status of PML, and thereby may affect apoptosis.
Discussion
Physiologic cleavage of CD43 in human cells causes the release of the entire CD43 ectodomain that was originally isolated from human plasma as Galgp.13 However, a physiologic function of CD43 shedding has not yet been established. Our work confirms that CD43 shedding also occurs on murine cells and that it is similarly regulated in that cell activation by TPA (this report) or immobilized CD43 antibody (our unpublished observations) can induce release of CD43 ectodomain. Western blotting combined with CD43/34 chimera expression confirmed that murine CD43 is also cleaved at a juxta-membrane site. Whereas shedding of human CD43 has been studied mainly in neutrophils, our survey of murine hemopoietic cells revealed that CD43 shedding occurred on most hemopoietic cell types. It was, however, not shed from primary macrophages, indicating that CD43 function and/or processing may differ in these cells. It will be of interest to determine whether human macrophages also lack CD43 shedding activity and whether macrophages in general use different strategies to process CD43.
The unexpected cell toxicity associated with ectopic expression of the CD43/34 chimeras was suggestive of important physiologic functions associated with cleavage of both transmembrane and membrane-proximal domains of CD43. The observation that after 5 days of culture in IL-3/G-CSF, half of the CD43/34 TM-transfected bone marrow cells differentiated into F4/80+ monocytes/macrophages suggests that chimera TM could influence cell lineage differentiation, but it could also simply be reflective of the fact that cells to survive were forced to differentiate into monocyte/macrophage. Interestingly, the CD43/34 MP-expressing bone marrow cells did not differentiate into F4/80+ monocytes/macrophages in the 5-day time window of cell culture, indicating that chimeras MP and TM vary in their impact on differentiating cells. The fact that the CD43/34 chimeras were not toxic in monocytes/macrophages shows furthermore that these cells can process CD43 independent of ectodomain shedding and that the toxicity associated with ectopic expression of the CD43/34 chimeras in other hemopoietic cells is not due to nonspecific effects such as a protein misfolding response.
Processing of human CD43 by α- and γ-secretases has been demonstrated in several carcinoma cell lines18,21 and primary human neutrophils,22 raising the possibility that CD43 might be functionally involved in regulated intramembrane proteolysis–dependent signaling similarly to Notch-1.23 CD43 processing by regulated intramembrane proteolysis is consistent with our hypothesis that the common downstream event associated with ectopic expression of CD43/34 chimeras is blockage of γ-secretase cleavage, and is coherent with our findings that CD43ct translocates to the nucleus and inhibition of CD43 processing by γ-secretase induces apoptosis.
A nuclear function of CD43ct is further supported by our discovery that it is a substrate of SUMO-1. Sumoylation is a cellular process in which a 12.5-kDa SUMO peptide is covalently linked to lysine residues on target proteins. Unlike their cousin, ubiquitin, SUMOs do not mark proteins for degradation, but affect the stability, structure, activity, and localization of modified proteins (for reviews, see Seeler and Dejean,24 Muller et al,25 and Hay26 ). SUMO substrates are functionally involved in many biologic functions, and deficiency in the SUMO-1 E2-conjugating enzyme Ubc9 is embryonic lethal.27 The SUMO conjugation machinery is localized in and around the nucleus, indicating sumoylation occurs en route to the nucleus.28 However, modification of transmembrane- and organelle-associated proteins has also been reported,29,30 consistent with our observation that full-length CD43 may also be sumoylated.
Many SUMO-modified proteins are recruited into a subnuclear structure, known as PML leukemia NB. Whereas regulation of apoptosis is believed to be a major function of PML NB and the proteins localized within this structure,31 a variety of other nuclear functions, including genome stability32 and transcriptional regulation,33 has also been proposed, and the exact function of PML NB remains enigmatic (for reviews, see Hofmann and Will34 and Bernardi and Pandolfi35 ). CD43ct localization in PML NB and its involvement in regulation of apoptosis are thus consistent with the proposed function of PML NB-localized proteins. It is of interest that of the many different apoptosis inducers we tested, only growth factor withdrawal and regulatory T-cell inhibition were found to be affected by CD43, suggesting a specific role for CD43 signaling in some, but not all apoptotic pathways.
The observation that CD43-deficient T cells are more susceptible to Treg-cell inhibition is in agreement with our previous data showing that mice lacking CD43 have an elevated Mycobacterium tuberculosis burden in the chronic phase of infection.36 It has been shown that Treg cells can suppress protection from M tuberculosis infection.37,38 Increased parasite burden in CD43-null mice may thus be due to the increased susceptibility of CD43-deficient T cells to regulatory T-cell suppression.
The list of SUMO-modified proteins and PML NB-associated proteins is constantly growing. Addition of CD43 to this list is even more noteworthy in that CD43 is the only SUMO-1 substrate and PML NB-localized protein that is exclusively expressed in hematopoietic cells. Many functions of CD43 have been proposed and established in different settings.2 Our discovery that CD43 has an antiapoptotic effect on leukocytes further expands to functional range of this molecule. Recognition that the antiapoptotic effect of CD43 is a normal physiologic function might explain why some carcinomas, especially colon carcinomas, express CD43 and support the concept that aberrantly expressed CD43 may promote cell survival in oncogenesis.39,40
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Kelly M. McNagny and Douglas A. Carlow for helpful discussions and for critically reading the manuscript. Contribution by Kelly M. McNagny to the nonsheddable CD43 and ER approaches is also acknowledged.
This work was supported by grants MOP-77552 and MOP-82867 from the Canadian Institutes for Health Research.
Authorship
Contribution: W.S. designed, performed, and analyzed the experiments and cowrote the manuscript; and H.J.Z. supervised the experiments and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hermann J. Ziltener, Biomedical Research Centre, University of British Columbia, 2222 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; e-mail: hermann@brc.ubc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal