Abstract
Gene transfer into hematopoietic stem cells by γ-retroviral vectors (RVs) is an effective treatment for inherited blood disorders, although potentially limited by the risk of insertional mutagenesis. We evaluated the genomic impact of RV integration in T lymphocytes from adenosine deaminase-deficient severe combined immunodeficiency (ADA-SCID) patients 10 to 30 months after infusion of autologous, genetically corrected CD34+ cells. Expression profiling on ex vivo T-cell bulk population revealed no difference with respect to healthy controls. To assess the effect of vector integration on gene expression at the single-cell level, primary T-cell clones were isolated from 2 patients. T-cell clones harbored either 1 (89.8%) or 2 (10.2%) vector copies per cell and displayed partial to full correction of ADA expression, purine metabolism, and T-cell receptor-driven functions. Analysis of RV integration sites indicated a high diversity in T-cell origin, consistently with the polyclonal T-cell receptor-Vβ repertoire. Quantitative transcript analysis of 120 genes within a 200-kb window around RV integration sites showed modest (2.8- to 5.2-fold) dysregulation of 5.8% genes in 18.6% of the T-cell clones compared with controls. Nonetheless, affected clones maintained a stable phenotype and normal in vitro functions. These results confirm that RV-mediated gene transfer for ADA-SCID is safe, and provide crucial information for the development of future gene therapy protocols. The trials described herein have been registered at http://www.clinicaltrials.gov as #NCT00598481 and #NCT00599781.
Introduction
Gene transfer into hematopoietic stem/progenitor cells (HSCs) by γ-retroviral vectors (RVs) has been demonstrated as an effective treatment for inherited blood disorders, including X-linked severe combined immunodeficiency (SCID-X1),1,2 adenosine deaminase-deficient SCID (ADA-SCID),3 and X-linked chronic granulomatous disease (X-CGD).4 However, after the occurrence of leukemic proliferation in 5 of 19 SCID-X1 patients5,6 and of clonal expansion in 2 CGD patients,7 the safety of the use of retroviral vectors turned out to be a primary concern. Because the viral RNA genome is reverse transcribed into a DNA copy that becomes integrated into the host cell genome, RV may induce cell transformation by insertional mutagenesis leading to proto-oncogene trans-activation, in combination with transgene- and disease-specific cofactors.
The genomic features driving the preferential integration of RVs have been described in several large-scale survey of mapping studies in mice, nonhuman primates, and humans enrolled in clinical trials.8-14 These studies have shown that RVs integrate nonrandomly in the host genome favoring transcription start sites (TSSs) and expressed genes, which makes productive interactions between the vector and host transcriptional machinery more probable. As a result of this bias, integrations that alter the expression of flanking genes involved in cell cycle, transcriptional activity, and signal transduction might influence the biologic fate of the affected cell clone, thereby conferring a growth or survival advantage.4,15,16 However, to gain relevant information on the influence of vector integrations on transcriptional activity of nearby genes, studies should be carried out in relevant primary cells at the level of a clonal population containing defined insertion sites. Previous work conducted in the context of gene therapy with retrovirally transduced peripheral blood (PB) T cells revealed that RV insertion induced overexpression of a proximal gene at a frequency of almost 20%, although without measurable consequences on the biology and function of infused cells.17
We showed previously that promoters and transcriptionally active genes are preferential sites of RV integration also in bone marrow-derived, transduced CD34+ progenitor cells, and in their progeny isolated ex vivo from ADA-SCID patients. We also found that, in an ADA-deficient context, selective growth advantage of ADA+ lymphocytes is associated with a significant bias toward integrations in genomic regions, allowing a sustained transgene expression. Nevertheless, infusion of ADA-transduced HSCs is safe long-term and does not result in in vivo clonal expansion or malignant transformation up to 8 years after treatment.11,18
Here we report a detailed analysis aimed at fully assessing the potential transcriptional interference caused by vector integration, in both bulk populations and single clones of transduced T cells. Our data show that RV integration induces minor changes in the transcriptional activity of nearby endogenous genes at the clonal level, which, however, does not impact on the biology of the T-cell clones. In addition, they provide crucial information for the development of future gene therapy protocols.
Methods
Patients
The ADA-SCID gene therapy (GT) clinical trial was approved by the San Raffaele Scientific Institute and Hadassah University Ethical Committees and the respective national regulatory authorities. The patients' characteristics and treatment have been described previously.3,11 Patients received a single infusion of autologous CD34+ cells transduced with the GIADAl vector. Blood samples from ADA-SCID patients and healthy donors were obtained after informed consent in accordance with the Declaration of Helsinki, following standard ethical procedures and with approval of the Ethical Committee.
T-cell cloning and manipulation
Mononuclear cells from PB of patient 2 and patient 318 were isolated by Ficoll-Hypaque gradient separation and enriched for T lymphocytes by immunomagnetic-based depletion of monocytes (CD14+), and B (CD19+) and natural killer (CD56+) cells. Individual T-cell clones were obtained by plating cells in 96-well plates at a concentration of 0.3, 1, and 3 cells/well, respectively. Cells were stimulated with 1 μg/mL phytohemagglutinin (Roche Diagnostics), 100 IU/mL recombinant human interleukin-2 (Chiron) in the presence of an allogeneic feeder mixture of 105/well peripheral blood mononuclear cells (X-ray irradiated at 60 Gy) and 104/well JY cells19 (X-ray irradiated at 100 Gy). The cultures were performed in Iscove modified Dulbecco medium (Cambrex Bio Science) supplemented with 10% YSSEL medium (Dyaclone), 5% Hyclone (Cambrex Bio Science), and 100 U/mL penicillin/streptomycin (Bristol-Myers Squibb). Under these conditions, cloning efficiencies were 1.2% for patient 2, 1.9% for patient 3, and 5.1% for a healthy donor. T-cell clones were expanded by adding fresh medium with 600 IU/mL recombinant human interleukin-2 and restimulated as described every 14 days. The generation of T-cell clones at sufficient numbers for performing experiments required 4 cycles of restimulation. Phenotypic analysis, quantitative reverse-transcribed polymerase chain reaction (RT-PCR), biochemical, and functional assays were performed in resting T-cell clones sampled after 4 to 6 cycles at day 14 after restimulation.
Analysis of T-cell phenotype and function
Anti-CD4, -CD8, –T-cell receptor (TCR) αβ, –TCRγδ monoclonal antibodies (mAbs) directly coupled with fluorescein isothiocyanate or phycoerythrin were from BD Biosciences. Expression of ADA protein was assessed by intracellular staining using an anti–human ADA mAb, as described.19 To assess TCR β-chain usage, T cells (2 × 105) were stained with a mix of directly conjugated TCR Vβ Abs corresponding to 24 different specificities, according to the manufacturer's instructions (IOTest Beta Mark kit; Immunotech). Samples were acquired using FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (TreeStar). Intracellular ADA and S-adenosylhomocysteine hydrolase (SAHH) enzyme activities were analyzed in cell lysate (0.5 × 106 cells) as described.19,20
Resting T-cell clones were seeded at 105 cells/well in a 96-well plate round-bottom precoated with the indicated dose of anti-CD3 mAb (OKT3, Janssen Cilag Ltd) with or without soluble anti-CD28 mAb (10 μg/mL; BD Biosciences), in a final volume of 200 μL. Proliferation was assessed after 48 hours by [3H]thymidine incorporation. All experiments were performed in duplicate or triplicate wells. The secretion of cytokines was measured in the culture supernatants by the luminex technique according to the manufacturer's instructions (Bio-Rad).
Analysis of RV integration sites
Genomic DNA was extracted from T-cell clones using the QIAamp DNA Blood Mini kit (QIAGEN). Inverse PCR was performed as previously described.11 The resultant nested PCR products were either separated on 2% agarose excised and directly sequenced. Integration was mapped onto human genome by BLAST analysis using the ENSEMBL search engine (November 2004 freeze).
Quantitative tracking of specific integrants
Quantitative PCR analysis was performed on 50 ng of genomic DNA, directly isolated from sorted CD3+ T cells, using the specific genomic flanking primers in combination with common long terminal repeat (LTR) primer and probe (Primm). The primer sequence were as follows: 3′LTR primer, 5′-GTTTGCATCCGAATCGTGGT-3′; 3′LTR probe, 5′-6-FAM-TCTCCTCTGAGTGATTGACTACCCACGACG-TAMRA-3′; 5′ LTR primer, 5′-CCTGACCTTGATCTGAACTTCTCT-3′; ID-04 primer, 5′-TTGCCAGTCTAAATGGGTGTGCG-3′; 5′ LTR probe, 5′-6-FAM-CACCTGTAGGTTTGGCAAGCTAGCTT-TAMRA-3′; ID-13 primer, 5′-CAGAGGTAGACAGAGAAGGCAT-3′; ID-16 primer, 5′-TGCACAGACTGATCAAAAGTCTCTAA-3′; ID-20 primer, 5′-CGGCTCGGGTGTCCTG-3′. The frequency of each individual clone, expressed as the frequency of CD3+ T cells containing the specific vector-genome junction sequence, was calculated on the basis of a standard curve of the relative integrants diluted in untransduced cells.
Gene expression analysis
Transcriptional profiling was carried out in CD4+ and CD8+ T cells purified with immunomagnetic beads (Miltenyi Biotec) from the PB lymphocytes of ADA-SCID patients (patients 1, 3, and 4) 10 to 30 months after GT and age-matched healthy controls (n = 4). The study was not performed in patient 2 because of limited availability of cells. RNA was isolated from 106 cells using Eurozol reagent (Euroclone SpA). The transcribed biotinylated cRNA was fragmented and hybridized to Affymetrix HG-U133A GeneChip arrays for 16 hours (Affymetrix). Scanned images were processed by the Affymetrix MAS 5.0 suite. Data analysis was performed with the GeneSpring GX, Version 7.3.1 (Agilent Technologies). Custom 384-well TaqMan Low Density Arrays (LDAs; Applied Biosystems) were used to assess potential transcriptional interference at the site of RV integration. LDA format was customized online with 4 replicates per target gene. A total of 135 probes (assays) corresponding to 120 single genes were included in this study. Samples were analyzed using the 7900HT system with a TaqMan LDA Upgrade (Applied Biosystems) and SDS2.2 software, according to the manufacturer's instructions. Data were normalized to the housekeeping gene GAPDH. In the case of genes analyzed with 2 probes, the average of corresponding ΔCt was considered. Relative quantification was calculated using the ΔΔCt methods as relative to the average of ΔCt in all samples as calibrator. The amount of target relative to the calibrator was 2 −ΔΔCt, and is indicated in Table 2 as fold changing.
Statistical analysis
In the gene expression study on ex vivo bulk populations, transcript levels have been compared by means of a t type statistic (Welch statistic for not equal variances assumptions). P values have been computed based on distribution-free permutation procedures and corrected for multiplicity by Benjamini and Hochberg false discovery rate. The permutation procedure21 was considered for each single retroviral integration site (RIS) to detect significant difference between the expression of gene(s) hit by RV in the target cell clone(s) and that of control clones. The computation of P values by this resampling nonparametric procedure is not affected by the sparseness of observations and the different size of the 2 groups of interest, which, in standard parametric methods (like t test) could lead to strong biases in estimating the false positives because of large variances that would artificially spread up the t values. In functional assays, a 2-tailed Mann-Whitney U test was used for statistical comparison of patients versus healthy controls. Responses within the same group were compared by the (2-tailed) Wilcoxon signed-rank test for paired data.
Results
Normal gene expression profile in T lymphocytes purified ex vivo from GT-treated patients
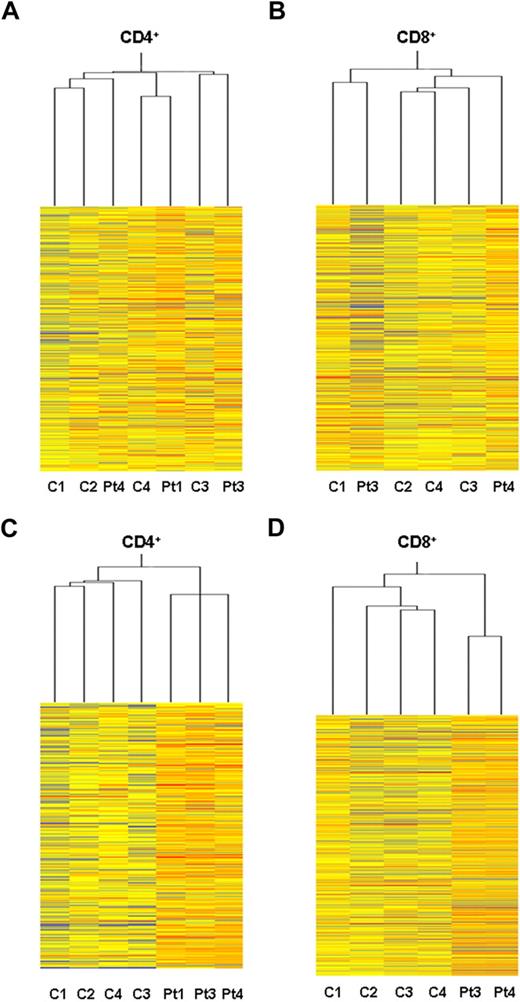
Global gene expression profiling analysis of ex vivo PBT lymphocytes from ADA-SCID patients was performed to assess potential alterations in the transcriptional activity after GT. CD4+ and CD8+ T cells were isolated from 3 patients (patient 1, patient 3, and patient 4) 10 to 30 months after autologous transplantation with genetically corrected CD34+ cells. At the indicated time points, the percentage of vector-positive T cells by quantitative PCR was more than 75%. Transcriptional profiles were determined on Affymetrix HG-U133A microarray and compared with those of age-matched healthy controls. The microarray data have been deposited into the GEO public database under accession no. GSE17354. Heat maps originating from CD4+ and CD8+ datasets are depicted in Figure 1. An unsupervised cluster analysis identified 2 main branches corresponding to the CD4+ and CD8+ cell types (not shown). Within each T-cell subset, samples from GT-treated patients did not form distinct clusters compared with healthy controls, indicating substantial overlap between the gene expression profiles of the 2 groups (Figure 1A-B).
Gene expression profile in purified T-cell subset from GT-treated patients. Heat maps representing the unsupervised Affymetrix gene expression analysis of PB CD4+ (A) and CD8+ (B) T cells isolated from 3 GT-treated patients (patient 1, +30 months; patient 3, +26 months; patient 4, +10 months) and 4 age-matched healthy controls (C). Expression levels are represented by a color key in which bright red represents the highest levels (up-regulated genes) and bright blue represents the lowest levels (down-regulated genes); less saturated shades represent intermediate levels of expression. Heat maps resulting from supervised hierarchical clustering of CD4+ (C) and CD8+ (D) T-cell samples according to the type of treatment (ie, GT vs normal controls). P value cutoff = .05; multiple testing correction, none. This restriction tested 9364 genes. Approximately 4682 genes would be expected to pass the restriction by chance.
Gene expression profile in purified T-cell subset from GT-treated patients. Heat maps representing the unsupervised Affymetrix gene expression analysis of PB CD4+ (A) and CD8+ (B) T cells isolated from 3 GT-treated patients (patient 1, +30 months; patient 3, +26 months; patient 4, +10 months) and 4 age-matched healthy controls (C). Expression levels are represented by a color key in which bright red represents the highest levels (up-regulated genes) and bright blue represents the lowest levels (down-regulated genes); less saturated shades represent intermediate levels of expression. Heat maps resulting from supervised hierarchical clustering of CD4+ (C) and CD8+ (D) T-cell samples according to the type of treatment (ie, GT vs normal controls). P value cutoff = .05; multiple testing correction, none. This restriction tested 9364 genes. Approximately 4682 genes would be expected to pass the restriction by chance.
Using a supervised hierarchical ordering approach, one-way analysis of variance was performed to search for genes that varied significantly between patients' and control samples. We identified as few as 235 (2.5%) genes, among the CD4+ dataset, and 479 (5.1%) genes, among the CD8+ dataset, differentially expressed in the cluster of patients compared with that of controls (Figure 1C-D). However, when we applied a multiple testing corrections with the least stringent Benjamini and Hochberg false discovery rate procedure, no significant differences were observed in the gene expression patterns between the 2 groups (not shown). Such findings are in keeping with the lack of sample segregation according to the treatment, as observed by the clustering analysis.
These results indicate that our GT protocol causes no major alterations in the gene expression program of T lymphocytes developed from genetically corrected CD34+ cells.
Analysis of RV integrations and diversity of T-cell clones
To study the effect of vector integration on gene expression and cell function at clonal level, primary T-cell clones were isolated ex vivo by limiting dilution from PB lymphocytes of 2 ADA-SCID patients 18 months after GT11 (patient 2 [X], 26 clones; patient 3 [Y], 27 clones). Among the clones generated from patient 2, 4 (15.4%) did not contain an integrated provirus, in agreement with the lower percentage (70%) of gene marking in CD3+ cells at the time of cell cloning. In all the vector-marked clones, vector-genome junctions were retrieved by inverse PCR, sequenced, and mapped onto the human genome by BLAST analysis. Overall, 41 RISs were unambiguously assigned to a chromosomal position, 26 of which were from patient 3 and 15 from patient 2 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The large majority of clones contained a single provirus per cell (n = 44, 89.8%), with few clones carrying 2 proviruses (n = 5, 10.2%), as confirmed by quantitative PCR.
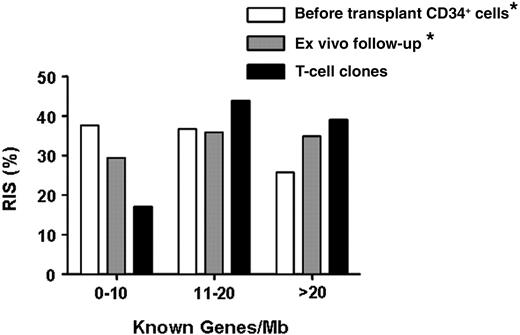
The distribution of RIS with respect to the closest RefSeq genes is shown in Table 1, in comparison with the insertion profile described for pretransplant CD34+ cells and for cells obtained ex vivo during the patients' follow-up.11 Intergenic and promoter-proximal (± 5 kb from transcription start sites [TSSs]) RISs were overrepresented in T-cell clones with respect to pre-GT and post-GT bulk samples (70.7% vs 49.1% and 58.7%, and 36.6% vs 23.6% and 28.8%, respectively). The characteristic cluster of RIS in gene-dense chromosomal regions was confirmed in T-cell clones (83% RIS in > 10 genes/Mb regions; Figure 2).
RIS distribution in T-cell clones from ADA-SCID patients
| . | Intragenic, % . | Intergenic, % . | Less than 10 kb upstream, % . | Within 5 kb from TSS, % . |
|---|---|---|---|---|
| T-cell clones | 29.3 | 70.7 | 29.3 | 36.6 |
| Ex vivo follow-up | 41.3 | 58.7 | 19.6 | 28.8 |
| Pretransplant CD34+ cells | 50.9 | 49.1 | 12.3 | 23.6 |
| . | Intragenic, % . | Intergenic, % . | Less than 10 kb upstream, % . | Within 5 kb from TSS, % . |
|---|---|---|---|---|
| T-cell clones | 29.3 | 70.7 | 29.3 | 36.6 |
| Ex vivo follow-up | 41.3 | 58.7 | 19.6 | 28.8 |
| Pretransplant CD34+ cells | 50.9 | 49.1 | 12.3 | 23.6 |
Distribution of RV integration sites mapped in T-cell clones isolated ex vivo from the peripheral blood of patients 2 and 3 at 18 months after gene therapy, compared with RV integrations in pre-GT and post-GT bulk samples from a previously published collection.11 Integrations were distributed as inside (intragenic) or outside (intergenic) RefSeq genes, and at a distance of less than 10 kb upstream or plus or minus 5 kb from TSSs.
Distribution of vector integration site in ex vivo T-cell clones. Distribution of integration sites, retrieved from T-cell clones isolated ex vivo from patient 2 (X, n = 22) and patient 3 (Y, n = 27), relative to gene density within a 1-Mbp window, compared with insertion sites retrieved from ex vivo and in vitro cell samples, as reported in Aiuti et al11 (*). Each bar represents the percentage of integration sites within the corresponding gene density region.
Distribution of vector integration site in ex vivo T-cell clones. Distribution of integration sites, retrieved from T-cell clones isolated ex vivo from patient 2 (X, n = 22) and patient 3 (Y, n = 27), relative to gene density within a 1-Mbp window, compared with insertion sites retrieved from ex vivo and in vitro cell samples, as reported in Aiuti et al11 (*). Each bar represents the percentage of integration sites within the corresponding gene density region.
Phenotypically, all clones were CD4+ T cells and displayed a high diversity in the TCR β chain usage (Figure 3A), mirroring the complexity of the immune reconstitution occurred in patients. To track the in vivo origin of T-cell clones, we combined the RIS mapping with TCR Vβ analysis. In 5 cases, we could identify T-cell clones harboring distinct Vβ TCR that shared the same vector insertion, indicating that they derived from the same prethymic progenitor (Figure 3B, supplemental Table 1). Interestingly, RISs identified in multiple cell clones (ie, ID-04 and ID-16 found in 5 and 3 clones, respectively) are present at relatively higher frequency within the ex vivo isolated CD3+ population, thus demonstrating that RISs sampled in the T-cell clones are directly representative of the in vivo clonal composition at the time of cloning. Moreover, some of these T-cell clones shared the same RIS previously identified in CD15+ granulocytes and CD19+ B lymphocytes in the same patients,11 confirming their common origin from transduced multipotent HSCs (Figure 3D).
Analysis of TCR diversity and hematopoietic origin of T-cell clones. (A) Immune repertoire of T-cell clones isolated from the PB of patient 2 and patient 3. Each bar represents the percentage of clones displaying the specific TCR Vβ chain rearrangement, as analyzed by flow cytometry. (B) Analysis of Vβ repertoire in 3 clones from patient 3 sharing a common RIS. (C) Quantification of specific RIS-containing clones (ID-04 and ID-13 from patient 2 identified in 5 and 2 independent T-cell clones; ID-16 and ID-30 from patient 3 identified in 3 and one T-cell clone, respectively) within the total T-cell population. The frequency of insertions was measured by sequence-specific real-time PCR in CD3+ T cells purified from PB at different time points (months) during the follow-up after GT. Rectangle represents the time point at which the T-cell cloning was performed. (D) T-cell clones from patient 3 share RIS with distinct hematopoietic cell subsets. Integration analysis was performed, by random cloning and sequence-specific PCR, on cells purified ex vivo from PB or bone marrow (BM) at different time points (months [mo]) during the follow-up.
Analysis of TCR diversity and hematopoietic origin of T-cell clones. (A) Immune repertoire of T-cell clones isolated from the PB of patient 2 and patient 3. Each bar represents the percentage of clones displaying the specific TCR Vβ chain rearrangement, as analyzed by flow cytometry. (B) Analysis of Vβ repertoire in 3 clones from patient 3 sharing a common RIS. (C) Quantification of specific RIS-containing clones (ID-04 and ID-13 from patient 2 identified in 5 and 2 independent T-cell clones; ID-16 and ID-30 from patient 3 identified in 3 and one T-cell clone, respectively) within the total T-cell population. The frequency of insertions was measured by sequence-specific real-time PCR in CD3+ T cells purified from PB at different time points (months) during the follow-up after GT. Rectangle represents the time point at which the T-cell cloning was performed. (D) T-cell clones from patient 3 share RIS with distinct hematopoietic cell subsets. Integration analysis was performed, by random cloning and sequence-specific PCR, on cells purified ex vivo from PB or bone marrow (BM) at different time points (months [mo]) during the follow-up.
Quantitative transcript analysis of genes targeted by RVs
We next investigated any potential transcriptional perturbation of the genes surrounding RIS in prospectively isolated ex vivo T-cell clones. Adequate RNA material was available for 43 clones, corresponding to 35 different RV insertion sites. We studied the expression level of all genes contained in an interval of 200 kb centered on the site of RV integration. This broad interval was chosen because previous studies have suggested that insertional gene activation is not merely a function of the distance of the RV from the TSSs.16,17 Overall, we analyzed a total of 120 different genes (T-cell clone gene set). The transcriptional profile of the T-cell clone gene set was preliminarily evaluated within the Affymetrix datasets of purified T cells from healthy donors, to assess their relative expression with respect to the global average transcription levels. The proportion of expressed genes in the array did not increase significantly when the analysis was restricted to genes landing within 100 kb from RIS (43.3% vs 44.9%; supplemental Figure 1A), indicating that no bias toward expressed genes was introduced with the chosen cutoff. Functional classification indicated that the RV hit genes belonged to major networks involved in cellular process and metabolism groups, and without any particular skewing with respect to the expected distribution (supplemental Figure 1B). Three genes (2.5%) are known proto-oncogenes, of which one (BLM) was identified as common insertion site in our previously published collection.11
Using custom TaqMan LDAs, the expression of the genes hit by the vector was evaluated in the clone(s) in which the integration was identified and compared with the level of expression found in the other clones carrying integrations in different loci. Untransduced (UT) clones from patient 2 (n = 4) as well as clones from healthy donors (n = 3) were used as additional controls. Normalized expression levels of genes neighboring RV insertion sites are represented in Figure 4A-B. Because normal distribution of data was doubtful and normal approximations to obtain standard errors and confidence intervals cannot be correctly used, we used a nonparametric permutation test based on resampling, which shifts the assignment of sample within each single RIS and recomputes the t statistic each time.
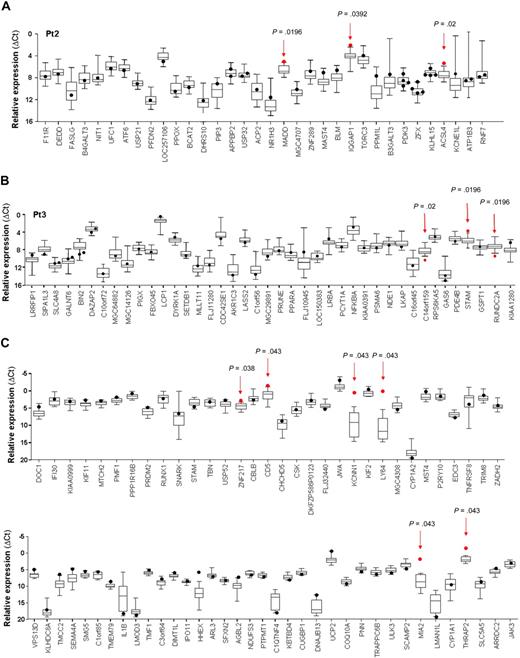
Expression analysis of genes targeted by RIS in T-cell clones. Quantitative transcript analysis of genes landed in a 200-kb window around the RV integration site in individual T-cell clones isolated ex vivo from patient 2 (A) and patient 3 (B), as performed by real-time RT-PCR on low-density arrays. Only genes found expressed by the majority of clones (≥ 95%) are depicted. (C) Quantitative PCR analysis of the expression of genes hit by RV integration in randomly selected T-cell clones transduced in vitro. Expression level (y-axis) of each gene (x-axis) hit by the nearby RIS, measured as relative mRNA quantity after normalization for the level of the housekeeping gene GAPDH (ΔCt), is presented as a dot for the target clone(s) and by box-and-whisker plots for all control clones. Box-and-whisker plots illustrate the median, interquartile range, and range that contains the central 95% of the observation and the maximum and the minimum values. Red arrows and dots represent the expression of the dysregulated gene in the T-cell clone containing the RIS and its relative P value.
Expression analysis of genes targeted by RIS in T-cell clones. Quantitative transcript analysis of genes landed in a 200-kb window around the RV integration site in individual T-cell clones isolated ex vivo from patient 2 (A) and patient 3 (B), as performed by real-time RT-PCR on low-density arrays. Only genes found expressed by the majority of clones (≥ 95%) are depicted. (C) Quantitative PCR analysis of the expression of genes hit by RV integration in randomly selected T-cell clones transduced in vitro. Expression level (y-axis) of each gene (x-axis) hit by the nearby RIS, measured as relative mRNA quantity after normalization for the level of the housekeeping gene GAPDH (ΔCt), is presented as a dot for the target clone(s) and by box-and-whisker plots for all control clones. Box-and-whisker plots illustrate the median, interquartile range, and range that contains the central 95% of the observation and the maximum and the minimum values. Red arrows and dots represent the expression of the dysregulated gene in the T-cell clone containing the RIS and its relative P value.
This analysis revealed that 7 of the 120 genes assayed (5.8%) had a significantly perturbed expression in the clones carrying a RV integration at a distance of less than 100 kb (Table 2, supplemental Table 2). ACSL4, MADD, IQGAP1, STAM, and LOC57228 genes were up-regulated (frequency, 11.4%; 2.8-4.5-fold increase), whereas C14orf159 and RUNDC2A appeared to be down-regulated (frequency, 5.7%; 4.7-5.2-fold decrease; a description of deregulating RIS genomic position is available in supplemental Table 1).
Genes with deregulated expression induced by the nearby RIS
| Clone . | Gene . | Description . | Vector orientation . | Insertion type . | Distance from TSS, kb . | Relative expression . | Fold change . |
|---|---|---|---|---|---|---|---|
| X24 | ACSL4 | Long-chain acyl-CoA synthetase 4 | Antisense | Intergenic | −0.45 | 4.5 | 3.8 |
| X22 | MADD | MAP-kinase activating death domain protein | Sense | Intergenic | −53.8 | 5.3 | 2.8 |
| X12 | IQGAP1 | Ras GTPase-activating-like protein | Antisense | Intergenic | 213.1 | 2.4 | 3.9 |
| Y7 | STAM | Signal transducing adapter molecule 1 | Antisense | Intergenic | 99.9 | 3.7 | 4.5 |
| Y20/Y26 | LOC57228 | Unknown | Antisense | Intergenic | −54.4 | 13.9/14.6 | NA |
| Y3 | C14orf159 | Mitochondrial precursor | Antisense | Intergenic | −0.68 | 10.4 | −4.7 |
| Y16 | RUNDC2A | RUN domain-containing protein 2A | Antisense | Intragenic | 34.4 | 9.4 | −5.2 |
| Clone . | Gene . | Description . | Vector orientation . | Insertion type . | Distance from TSS, kb . | Relative expression . | Fold change . |
|---|---|---|---|---|---|---|---|
| X24 | ACSL4 | Long-chain acyl-CoA synthetase 4 | Antisense | Intergenic | −0.45 | 4.5 | 3.8 |
| X22 | MADD | MAP-kinase activating death domain protein | Sense | Intergenic | −53.8 | 5.3 | 2.8 |
| X12 | IQGAP1 | Ras GTPase-activating-like protein | Antisense | Intergenic | 213.1 | 2.4 | 3.9 |
| Y7 | STAM | Signal transducing adapter molecule 1 | Antisense | Intergenic | 99.9 | 3.7 | 4.5 |
| Y20/Y26 | LOC57228 | Unknown | Antisense | Intergenic | −54.4 | 13.9/14.6 | NA |
| Y3 | C14orf159 | Mitochondrial precursor | Antisense | Intergenic | −0.68 | 10.4 | −4.7 |
| Y16 | RUNDC2A | RUN domain-containing protein 2A | Antisense | Intragenic | 34.4 | 9.4 | −5.2 |
List of genes displaying altered mRNA level in the ex vivo isolated T-cell clone(s) harboring RIS. T-cell clones retrieved from patient 2 are indicated with X; clones from patient 3 with Y. Vector orientation is relative to targeted gene transcription. Relative gene expression level is indicated as normalized for the housekeeping gene GAPDH in each hit clone, and as fold change relative to the calibrator, as described in “Gene expression analysis.”
NA indicates not applicable.
All the up-regulated genes are functionally involved in metabolic and signaling pathways and are expressed at low (ACSL4) or intermediate levels (MADD, IQGAP1, STAM) in T cells, as assayed by Affymetrix profiling (supplemental Figure 1 and data not shown).
To compare the genotoxic potential of RV vector insertion in T cells in the absence of in vivo selection, we used the same quantitative assay and analysis criteria to reanalyze perturbation of gene expression around RV insertion site in T-cell clones from one of our previous studies.17 By this analysis, 6 of 29 (20.7%) randomly selected clones showed significantly higher expression in 6 of 68 (8.8%) genes close to an integrated provirus. In nontransplanted T cells, activating RIS increased mRNA transcript level at a similar frequency but in a more prominent fashion, ranging from 3.2-fold (ZNF217) to more than 1900-fold (LY64) the average value in the control clones (Figure 4C, supplemental Table 3). Deregulation of gene expression occurred preferentially when a provirus was integrated in close proximity of a TSS (≤ 50 kb; supplemental Figure 2) and in opposite transcriptional orientation, in both ex vivo and in vitro clones.
RIS-associated altered gene expression has no impact on phenotype and functional behavior of T-cell clones
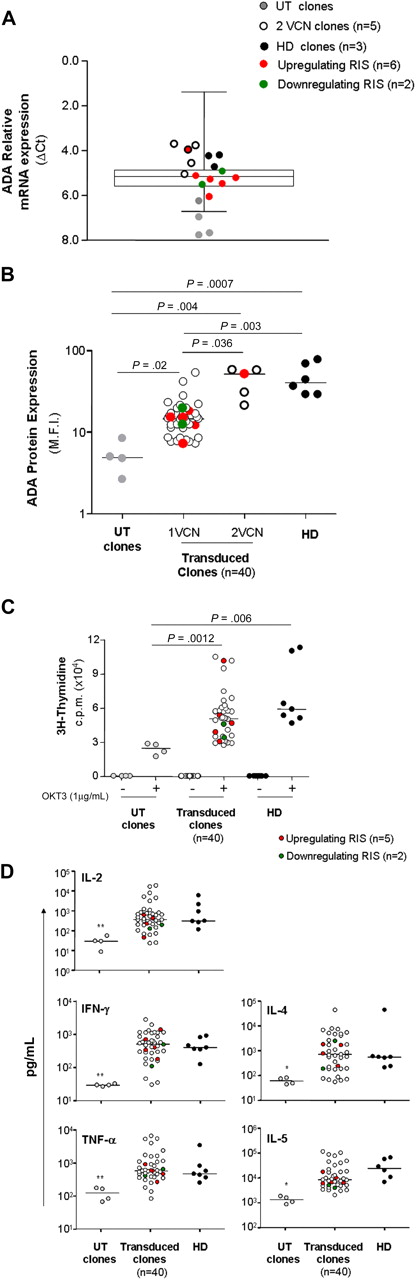
To evaluate the consequences of vector integration on the biology of T-cell clones, we first asked whether perturbation in neighboring gene transcription could influence the ADA transgene expression. ADA transcript analysis was measured with LDA in (1) clones carrying deregulating RIS (n = 6), (2) other transduced clones (n = 37), (3) untransduced clones from ADA-SCID patients (n = 4), and (4) control clones from healthy subjects (n = 3; Figure 5A). Relative ADA mRNA level was significantly lower in untransduced clones compared with transduced and normal clones (P = .023), whereas no difference in ADA expression was observed between the last 2 groups. Interestingly, clones with 2 proviruses generally showed higher ADA mRNA levels with respect to clones carrying one provirus, and closer to normal levels. Remarkably, deregulating RIS had no discernible effect on transgene expression in all the hit clones, with the exception of clone Y7, which harbored 2 integration events (Figure 5A). Similar results were achieved when ADA expression was evaluated at the protein level by intracellular staining (Figure 5B). Nonetheless, a more robust correlation was observed between the protein expression and the vector copy number because only clones carrying 2 proviruses restored ADA to normal levels (P = .036, Figure 5B). A significant tendency to select for integrations in genomic regions potentially more favorable to vector-derived ADA expression has been previously reported for ex vivo purified bulk T cells.11 This tendency was further enhanced in in vitro expanded clones (Table 1) and confirmed by the higher protein expression detected in T-cell cultures (bulk T-cell line mean fluorescence intensity [MFI], 20.6; average MFI in T-cell clones, 24.2) compared with freshly isolated T cells from patient 3 (MFI, 12.2; supplemental Figure 3A). Possible position effects were additionally evaluated in T-cell clones, after grouping according to the type of integration and genomic location harboring RIS. No difference in ADA expression was observed among clones carrying intragenic versus intergenic RIS, or RIS landing in high dense gene regions (not shown). However, when the analysis was restricted to clones with TSS-proximal (< 5 kb) intergenic RIS, ADA protein levels showed a tendency to increase, although not statistically significant (supplemental Figure 3B). Interestingly, hit genes were transcriptionally active and enriched for the intermediate-high level of expression (not shown). Overall, these data confirm that productive interaction between the vector and host transcriptional machinery is one of the crucial parameters determining the selective advantage of T-cell clones expressing adequate ADA levels.
Phenotypic and functional characterization of T-cell clones. (A) Relative mRNA expression of ADA in T-cell clones. Expression was measured with the microfluidic card in UT clones from patient 2 (gray dots), clones from healthy donors (black dots), clones carrying up-regulating (red dots) or down-regulating (green dots) RIS, clones with 2 vector copy/cell (white dots) and 1 vector copy/cell (box and whiskers). (B) ADA protein expression. Expression was determined by intracytoplasmic staining as described in “Analysis of T-cell phenotype and function.” M.F.I. indicates mean fluorescence intensity. (C) Proliferative responses and cytokine production. T-cell clones generated from patients and age-matched controls were stimulated with immobilized anti-CD3 monoclonal antibody (OKT3, 1 μg/mL), and 3H-thymidine incorporation was assessed after 48 hours. In parallel, cytokine secretion was evaluated in the culture supernatants collected at different time points (18 hours for IL-2 and 48 hours for the other cytokines; D). *P < .05 vs transduced clones. **P < .01 versus transduced clones.
Phenotypic and functional characterization of T-cell clones. (A) Relative mRNA expression of ADA in T-cell clones. Expression was measured with the microfluidic card in UT clones from patient 2 (gray dots), clones from healthy donors (black dots), clones carrying up-regulating (red dots) or down-regulating (green dots) RIS, clones with 2 vector copy/cell (white dots) and 1 vector copy/cell (box and whiskers). (B) ADA protein expression. Expression was determined by intracytoplasmic staining as described in “Analysis of T-cell phenotype and function.” M.F.I. indicates mean fluorescence intensity. (C) Proliferative responses and cytokine production. T-cell clones generated from patients and age-matched controls were stimulated with immobilized anti-CD3 monoclonal antibody (OKT3, 1 μg/mL), and 3H-thymidine incorporation was assessed after 48 hours. In parallel, cytokine secretion was evaluated in the culture supernatants collected at different time points (18 hours for IL-2 and 48 hours for the other cytokines; D). *P < .05 vs transduced clones. **P < .01 versus transduced clones.
We then asked whether insertional dysregulation of genes flanking the retroviral insertion could confer preferential proliferative advantage to the targeted clones. None of the cell clones here analyzed showed a cytokine-independent in vitro growth during the relatively long-term culture period. The functional behavior of cell clones was further analyzed on TCR stimulation with plate-bound anti-CD3 mAb (OKT3). Proliferative responses of gene-corrected T clones were significantly increased with respect to those observed for untransduced ADA-deficient T-cell clones (P > .005) and reached the range of normality (Figure 5C). Interestingly, 3 of 4 clones carrying 2 vector copy numbers displayed the highest range of proliferation, further confirming the strong selective advantage conferred by ADA expression to T-cell function.19 The ability to produce both Th1- and Th2-like cytokines was also restored to normal level (Figure 5D). Finally, it is worth noting that cell clones displaying significantly dysregulated gene expression did not show increased basal proliferation or aberrant functional responses on mitogen stimulation.
ADA transgene expression correlates with normalization of purine metabolism
We took advantage of the availability of untransduced, gene-corrected, and normal T-cell clones to study at single-cell level the effect of restored ADA expression on other enzymes involved in purine metabolism (Figure 6A). Indeed, in the absence of ADA, increased amounts of adenosine (Ado) and deoxyadenosine (dAdo) may “spill over” into additional intracellular metabolic pathways normally only minimally used,22-24 thus contributing to the pathogenetic mechanisms of the disease.
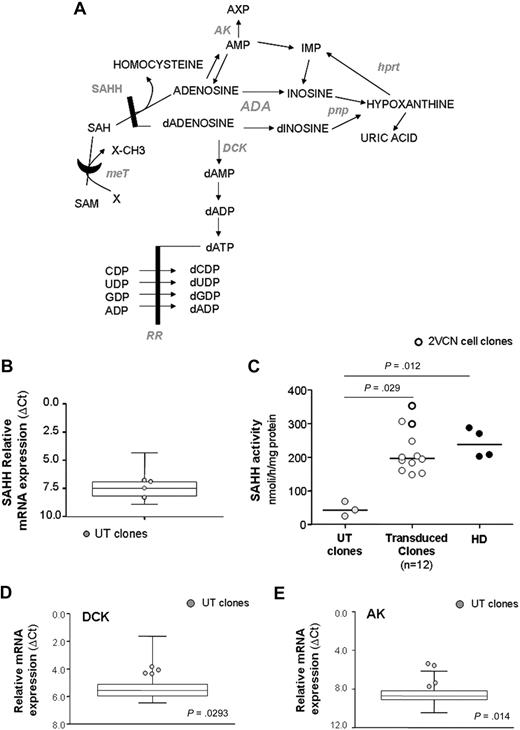
Expression analysis of purine metabolic enzymes. (A) Illustration of purine pathway. HPRT indicates hypoxanthine phosphorybosil transferase; PNP, purine nucleoside phosphorylase; MeT, methyl transferase; AK, adenosine kinase; DCK, deoxycytidine kinase; RR, ribonucleotide reductase. (B) Relative mRNA expression of SAHH, as measured with quantitative RT-PCR, in UT clones from patient 2 (n = 4; gray dots) and controls (transduced and healthy clones, n = 46; box-and-whiskers). (C) SAHH enzymatic activity. Relative mRNA expression of DCK (D) and AK (E) in UT clones and controls. P values refer to gene expression of UT clones versus controls.
Expression analysis of purine metabolic enzymes. (A) Illustration of purine pathway. HPRT indicates hypoxanthine phosphorybosil transferase; PNP, purine nucleoside phosphorylase; MeT, methyl transferase; AK, adenosine kinase; DCK, deoxycytidine kinase; RR, ribonucleotide reductase. (B) Relative mRNA expression of SAHH, as measured with quantitative RT-PCR, in UT clones from patient 2 (n = 4; gray dots) and controls (transduced and healthy clones, n = 46; box-and-whiskers). (C) SAHH enzymatic activity. Relative mRNA expression of DCK (D) and AK (E) in UT clones and controls. P values refer to gene expression of UT clones versus controls.
First, we focused on the SAHH enzyme, as reduction of its activity has been extensively documented in cells from ADA-SCID patients and considered a hallmark of the disease.19,25 Whereas no difference in the relative mRNA expression was detected between untransduced and gene-corrected or normal clones (Figure 6B), a marked block in SAHH activity was observed in untransduced clones (P = .029 vs gene-corrected clones, P = .012 vs normal clones; Figure 6C). In contrast, normalization of SAHH activity was detected in gene-corrected cells (Figure 6C). Of note, T-cell clones carrying 2 proviruses showed the best recovery of enzyme function, in keeping with the highest expression of ADA protein (Figure 6C).
Although dAdo is a weak substrate for adenosine kinase (AK) and deoxycytidine kinase (DCK), in the absence of ADA these enzymes can phosphorylate dAdo. In turn, the resulting dATP pool expansion may interfere with several metabolic pathways critically required for cell viability.26 Similarly, Ado phosphorylation-induced pyrimidine depletion has been one of the first mechanisms proposed as the cause of lymphopenia in ADA deficiency.27
Remarkably, expression analysis in our cohort of T-cell clones revealed that mRNA transcript levels of both kinases were significantly up-regulated (2- and 2.6-fold increase for DCK and AK, respectively; Figure 6D-E) in untransduced compared with gene-corrected and normal clones (P > .05 for DCK; P = .014 for AK), strongly supporting the cooperative role of DCK- and AK-mediated nucleoside phosphorylation in the intracellular pathogenetic mechanisms of the disease.
Discussion
The great therapeutic potential of retroviral-mediated gene transfer is accompanied by the inherent risk of insertional mutagenesis through deregulation of host gene transcription. Indeed, GT has been shown to be a beneficial treatment for patients affected by SCID-X1 and CGD but seriously limited by the emergence of leukemic proliferation (5 of 19 treated SCID-X1 patients)5,6 and clonal expansion of myeloid cells (2 CGD patients)4 associated with RV insertions near cellular proto-oncogenes. In the context of ADA-SCID GT, the cumulative experience of our study of 10 patients18 as well as of other clinical trials28-30 did not unveil such complications, indicating that retroviral-mediated gene transfer for ADA-SCID retains a favorable safety profile. Consistently, our previous extensive sequencing analysis of RIS in multiple hematopoietic lineages revealed the typical genomic preferences of γ-RVs but the lack of in vivo skewing for sensitive genes.11 Here we have performed a comprehensive study of the influence of vector integration on gene expression in primary bulk populations and in single T-cell clones obtained from ADA-SCID patients transplanted with autologous, transduced CD34+ HSCs.
A global gene expression profiling was performed on CD4+ and CD8+ T-cell subsets purified ex vivo from 3 ADA-SCID patients at different times after GT. The microarray analysis showed a substantial overlap with the expression patterns of T cells from controls, indicating the absence of gross abnormalities in the development and function of T cells deriving from genetically corrected HSCs.
However, gene expression studies in bulk cell populations may underscore vector-induced gene deregulation occurring only in a small fraction of transduced cells, unless the altered cell clones contribute to a relevant amount of the analyzed cell population. To overcome the limited resolution power of the microarray technique and to provide a direct estimate of the transcriptional interference caused by vector integration at the single-cell level, we carried out a quantitative transcript analysis of genes close to integrated proviruses in mature T-lymphocyte clones isolated from 2 of the patients. T-cell clones expanded ex vivo from single cells represent a unique and powerful experimental tool to directly establish a relationship between integration sites and host gene expression profile. A potential bias of this approach is that circulating T cells represent only a limited fraction of the whole-body T-cell pool. In addition, we cannot exclude that certain clones may be less prone to ex vivo stimulation and long-term culture and thus potentially negatively selected. The large majority of the clones contained a unique integration, demonstrating the heterogeneity of the T-cell population. Indeed, the few cell clones carrying the same vector integrant showed distinct patterns of TCR rearrangement, indicating the major contribution of polyclonal thymopoiesis in immune reconstitution rather than peripheral expansion. Moreover, the occurrence of shared integrants between lymphoid and myeloid cells further confirmed the engraftment of transduced HSC with multilineage potential.
The analysis of the RIS genomic distribution in T-cell clones reflected the known preferences of γ-RVs.9,10 Although not significant, an additional tendency to favor integrations in intergenic regions, and particularly in the proximity of TSSs, was observed in T-cell clones with respect to RIS retrieved from uncultured T-cell populations collected during the follow-up.11 The increased expression of ADA protein detected, particularly, in clones with RIS closer (≤ 5 kb) to TSS of active genes, suggests that such vector insertion profiles could be biased by a selection in vitro for clones carrying RIS in genome sites more favorable for a high level of vector-derived expression, without altering the transcription of the target gene(s) in the majority of cases. Beyond the position effects, it appears that a complete normalization of ADA activity in peripheral T cells was mainly dependent on 2 copies of ADA transgene inserted into HSCs, leading to full correction of TCR-driven effector function.
In the absence of ADA, Ado and dAdo may be metabolized differently and give rise to alternative pathogenetic pathways, ultimately causing the intracellular poisoning.31 Our data indicate that untransduced T-cell clones had significantly higher expression of the kinases in charge of the purine nucleoside phosphorylation, compared with gene-corrected and healthy clones. Therefore, the increased conversion of ADA metabolites to nucleotide derivatives is probably responsible for the intracellular toxicity associated with SAHH inhibition, which persisted in vitro in ADA-deficient but not in corrected clones.19
The quantitative transcript analysis in single cells revealed an incidence of transcriptional perturbation in 18.6% of the interrogated ex vivo clones, with 5.8% of genes assayed showing altered expression as a consequence of a nearby vector insertion (7 of 35, frequency 20%). Enhanced gene expression, resulting from the transcriptional read-through from the nearby LTR or the activity of viral enhancer present in the LTR, was observed in 4 of the 120 genes analyzed (3.3%), whereas one gene (0.83%) showed actual insertional activation in 2 independently isolated clones sharing the same RIS. Interestingly, in 2 cases (1.6%), the integrated vector caused down-regulation of gene expression. Reduced transcript accumulation might be the consequence of aberrant splicing and/or premature transcript termination resulting from viral polyadenylation signals in intronic integrations or transcriptional interference in the case of intergenic insertions.
The extent of dysregulation in the expressed genes ranged from 2.8- to 5.2-fold the average value detected in control clones, whereas in case of putative RIS-mediated insertional activation the expression level of target gene was at the limit of detection in quantitative PCR for both the cellular clones. The Affymetrix analysis performed on the polyclonal population of patient 3, purified 8 months after the cell cloning, revealed a remarkable transcriptional identity with the expression profile detected in other GT-treated patients and healthy controls, with none of the genes deregulated by RIS at the clonal level showing significant difference. Hence, minor alterations of gene expression at the single-cell level have no discernible effect on the bulk population. Interestingly, the extent of altered gene expression detected in our cohort of ex vivo derived clones was on average 100-fold lower compared with that induced by RIS in T cells cloned transduced in vitro and not subjected to in vivo selection. It is also worth noting that an up-regulation of more than 10-fold was detected for LMO2 and CCND2 in leukemic cells (with respect to control T cells or thymocytes) from patients of the SCID-X1 trial5,6 and for MDS-EVI1 (36- to 74-fold increase) in CGD patients.4
Overall, our results suggest that in the ADA-SCID context dangerous insertional activations, if any, result in a competitive disadvantage, leading to extinction rather than selection of the affected clones. An alternative and nonmutually exclusive hypothesis for the lack of dominant/malignant clones in ADA-SCID disease settings is that the selective advantage conferred by the restoration of ADA expression is strong and acts on a wide number of cell types. Thus, any possible additional proliferative advantage conferred by vector integrations near oncogenes will not be relevant in an environment in which virtually all transduced cells compete equally in repopulating the host.32 Previous studies on mice transplanted with RV-transduced bone marrow have proposed a causal link between insertional deregulation and enhancement of cell “fitness.” Accordingly, dominant clones were consistently found to have increased expression of a gene, within the region of the integrated RV, which could be implicated in potentially causing survival or proliferative advantage, even in the lack of malignant transformation.15,16 The murine system, however, might be biased by the strong selection process determined by serial transplantation and repopulation experiments, which could cause per se the emergence of clones displaying altered expression of sensitive genes. In the case of the X-SCID and CGD cells, the nature of the therapeutic transgene or of the particularly strong LTR enhancer could have played a role in the selection of malignant or premalignant cell clones carrying integrations into proto-oncogenes.33-35 In our study, the majority of the genes showing vector-induced transcriptional perturbation belonged to “housekeeping” pathways in T cells and, remarkably, none of them is a known proto-oncogene. Based thereon, overexpression of such genes induced by the nearby vector insertion may theoretically confer a growth advantage to the affected clones. Although we were unable to directly assess the effect of increased gene transcription at the protein level because of the reduced availability and limited life span of T-cell clones, the stable phenotype and the normal functional behavior observed in the target clones ruled out the possibility that any transformation event has occurred. Accordingly, the polyclonal pattern of vector integrations and T-cell repertoire and the inability to retrieve these insertions by random cloning in the ex vivo and in vitro-expanded bulk populations at later time points11 (and B.C. and A.A., unpublished observations, January 2009) suggest that these clones do not represent a major fraction of the cell populations tested.
In conclusion, besides the actual risk for RV-mediated perturbation in the host cellular transcriptional activity, these results indicate that GT for ADA-SCID retains a low-risk profile and that disease-specific cofactors need to be accounted for in future design of clinical trials.33,36-39 In this regard, the construction of so-called self-inactivating vectors should allow further reduction of the risk of insertional gene activation.40-42 Different approaches would be to incorporate suicide genes in therapeutic vector,43 or insulators.44 However, the impact of such modifications on vector safety remains speculative, and it has to be demonstrated in appropriate preclinical models.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Filippo Carlucci and Dr Antonella Tabucchi from the University of Siena for biochemical assays and Dr Samantha Scaramuzza for technical assistance and helpful discussions.
This work was supported by grants from the Italian Telethon Foundation (HSR-TIGET grant; GGP06101), Association Française contre les Myopathies-Telethon (GAT0205), the independent drug research program of the Italian Medicines Agency (FARM5JRXRM), and the European Commission (Concerted Safety and Efficiency Evaluation of Retroviral Transgenesis in Gene Therapy of Inherited Diseases; LSBH-CT-2004-005242 and Clinigene LSHB-CT2006-018933).
Authorship
Contribution: B.C. designed and performed research experiments, analyzed data, and wrote the paper; E.M. contributed to research design, acquisition, and data analysis, and revised the paper; G.M. contributed to research experiment and data analysis; A. Ambrosi and C.D.S. performed statistical analysis; M.M. and S.S. performed research experiments; E.B., I.F., and V.H.-T. provided clinical care of the patients; M.G.R. and L.N. supervised the research and revised the paper; F.M. contributed to study design and revised the paper; A. Aiuti designed and supervised the research and wrote the paper; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current address for B.C. is Fondazione Humanitas per la Ricerca, Rozzano (MI), Italy.
Correspondence: Alessandro Aiuti, San Raffaele Telethon Institute for Gene Therapy, via Olgettina 58, 20132 Milan, Italy; e-mail: a.aiuti@hsr.it.
References
Author notes
B.C. and E.M. contributed equally to this study.

![Figure 3. Analysis of TCR diversity and hematopoietic origin of T-cell clones. (A) Immune repertoire of T-cell clones isolated from the PB of patient 2 and patient 3. Each bar represents the percentage of clones displaying the specific TCR Vβ chain rearrangement, as analyzed by flow cytometry. (B) Analysis of Vβ repertoire in 3 clones from patient 3 sharing a common RIS. (C) Quantification of specific RIS-containing clones (ID-04 and ID-13 from patient 2 identified in 5 and 2 independent T-cell clones; ID-16 and ID-30 from patient 3 identified in 3 and one T-cell clone, respectively) within the total T-cell population. The frequency of insertions was measured by sequence-specific real-time PCR in CD3+ T cells purified from PB at different time points (months) during the follow-up after GT. Rectangle represents the time point at which the T-cell cloning was performed. (D) T-cell clones from patient 3 share RIS with distinct hematopoietic cell subsets. Integration analysis was performed, by random cloning and sequence-specific PCR, on cells purified ex vivo from PB or bone marrow (BM) at different time points (months [mo]) during the follow-up.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/17/10.1182_blood-2009-02-202085/4/m_zh89990942470003.jpeg?Expires=1769108279&Signature=HF5dexhcZX5o8r58vKVTNLd6rPmIg~CNz8rCDH--WkLU0bvPvkpB~7rhg9-M71tY2KdrgX42FMeDLYyLIFjIEUJZ1BfDyFusq4qC53JuPzV7NI6FbQ8mPBkK2865fPkZHmoVareWuwvM7ppQ-56jcY~XTdokl6yO9MUDdY9VUZYQGTJWAAmaZgq6ivnMbM5IktC3Q61DTrFK28D9ZRNQj0S0KU46A-qfoUhSISXtY-Vzyj6B5pq0ujfG5ihxA3YE8x2JLOU2rWvFsD0mcHJl46tPpKL68bY3xIgEVSN8NTQ1~37TU5Y1oEbKz~o2ognqzwRnwZbT8~xPB63y8Lg1JQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)