In essential thrombocythemia (ET), the JAK2-V617F mutation is usually restricted to a subpopulation of neutrophils and platelets, and production of JAK2 wild-type (WT) platelets is not suppressed. Nonmutated precursor cells may, therefore, be susceptible to the acquisition of further JAK2 mutations. We used a common single nucleotide polymorphism (SNP) in the JAK2 coding sequence to genotype V617F alleles obtained either by allele-specific restriction enzyme digestion (RED) or by cloning. Both SNP alleles were detected in JAK2 mutant–positive alleles from neutrophils of 10 of 11 ET patients studied using RED compared with 0 of 5 with polycythemia vera. These results were confirmed in cloned products from 5 ET patients and indicate the occurrence of at least 2 separate JAK2 mutation events in the majority of ET patients investigated. In a further ET patient, JAK2 mutant–positive erythroid colonies with either X-allele inactivated were detected, demonstrating they could not have arisen from a common clonal precursor. These results indicate that at least 2 independent JAK2-V617F events occur commonly in ET patients, and they may arise on a polyclonal background. The presence of a JAK2 mutation in ET patients should not, therefore, be equated with a malignant disease.
Introduction
Essential thrombocythemia (ET) is a chronic myeloproliferative disorder (MPD) characterized by the persistent overproduction of platelets in the absence of a recognizable cause of thrombocytosis such as chronic inflammation, a concurrent nonhematologic malignancy, or another MPD (eg, chronic myeloid leukemia, polycythemia vera [PV], or primary myelofibrosis). It is usually an indolent disorder with a low incidence of transformation to acute leukemia unless leukemogenic therapeutic agents are used. The mortality from the disease is also low and when a causally related death occurs, it is usually due to a thrombotic event.1
Studies using X-chromosome inactivation patterns (XCIPs) have demonstrated that ET is a heterogeneous condition.2,,–5 In some cases, all the blood and bone marrow myeloid cells are monoclonal, clearly indicating that the disease has arisen from a common myeloid stem cell. In other cases, the XCIP is polyclonal and this typically remains stable over many years.6,7 The presence of a nonclonal XCIP does not, however, exclude the presence of a small, or sometimes medium-sized, clone in a polyclonal background, although why the mutated clone should enlarge to a certain size and then remain stable is not fully clear.7
A Val617Phe mutation in the JAK2 gene (JAK2-V617F) has been identified in the majority of cases of PV.8,,–11 The mutation leads to constitutive activation of JAK2 and increased sensitivity of erythroid progenitors to erythropoietin (Epo).12 As the red cell mass rises, oxygen sensors switch off Epo production by the kidney and, under the low Epo conditions, only the JAK2-mutated cells can proliferate. Consequently they rapidly outgrow the nonmutated cells. The JAK2-V617F mutation is also found in approximately half the cases of ET.8,,–11 However, although the mutation is sometimes present in all the myeloid cells in ET and the XCIP pattern is fully clonal (analogous to PV), more often the JAK2-V617F mutation is present in only a subpopulation of neutrophils and platelets, and the XCIP in these mutant-positive patients is frequently nonclonal.7,13,14
Furthermore, the JAK2-V617F mutant level can remain stable over many years.7,15 We have recently suggested16 that JAK2 wild-type (WT) platelet production is not suppressed by the presence of JAK2 mutant–positive platelets in ET because, although their regulator thrombopoietin is largely controlled by end organ consumption, the thrombopoietin level remains normal or even raised in ET despite the increased platelet mass.17,–19 With the JAK2 WT cell population unsuppressed by the presence of a JAK2 mutant clone, there would be the opportunity for further JAK2 mutations to arise in these nonmutated cells, and this might be particularly likely if some individuals were genetically predisposed to develop ET as suggested by several studies.20,–22 The presence of more than one mutant JAK2 clone would not readily be apparent as the JAK2 sequence in both clones would be identical. We have, therefore, taken advantage of a frequent single nucleotide polymorphism (SNP) in exon 19 of the JAK2 gene to ascertain whether the JAK2-V617F mutation can be found on both chromosome copies in the same ET patient, indicating the presence of at least 2 separate JAK2 mutation events. The results indicate that this is a frequent occurrence, indicative of a new paradigm for this disease.
Methods
Patients and samples
In total, 111 patients with ET, 30 JAK2-V617F–positive PV patients, and 114 hematologically normal controls were investigated. Peripheral blood neutrophils, T cells (CD3+), and platelets were purified as previously described.4 Bone marrow CD34+ cells were purified using the CD34 Microbead Kit (Miltenyi Biotec) per the manufacturer's instructions. DNA and RNA were prepared as previously described.4 Complementary DNA (cDNA) was prepared from approximately 1 μg RNA using SuperScript III First-Strand Synthesis Supermix (Invitrogen) according to the manufacturer's instructions. Approval for these studies was provided by the London Multicenter Research Ethics Committee and patient consent was obtained in accordance with the Declaration of Helsinki.
JAK2 mutation and SNP analysis
JAK2-V617F screening and quantification of mutant level.
DNA was screened for presence of the G-to-T mutation in exon 14 of the JAK2 gene as previously described using the polymerase chain reaction (PCR) with primers here called 14(MM)F and 14R, followed by digestion with AflIII (New England Biolabs) to discriminate between WT (G) alleles that are cut by AflIII and mutant (T) alleles that remain undigested.7 Primer sequences are provided in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Quantification of JAK2-V617F mutant level was performed as previously described.7
JAK2 SNP genotyping.
DNA samples from T cells of ET patients were screened for the synonymous A/G SNP rs2230724 in exon 19 of the JAK2 gene using PCR with primers in the flanking introns (19F and 19R) followed by BstNI digestion (New England Biolabs; Figure 1A). For A alleles, the product remained undigested (214 bp), whereas G alleles were reduced to 2 fragments of 121 and 93 bp. For cDNA analysis, exons 18 to 20 were amplified using primers 18F and 20R and digested with BstNI. Fragment size for A alleles was 235 bp (uncut) and for G alleles, 127 + 108 bp.
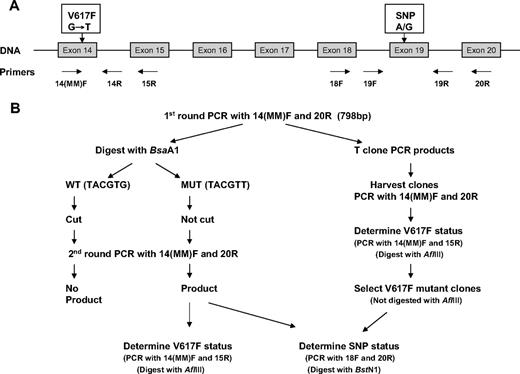
Schematic representation of technique used to determine JAK2 exon 19 SNP genotype of V617F-positive mutant alleles. (A) Genomic structure of JAK2 gene, showing primers used. (B) Procedure to determine the JAK2 exon 19 SNP genotype of JAK2 V617F mutant–positive alleles using either total cDNA (lefthand side) or individual clones (righthand side).
Schematic representation of technique used to determine JAK2 exon 19 SNP genotype of V617F-positive mutant alleles. (A) Genomic structure of JAK2 gene, showing primers used. (B) Procedure to determine the JAK2 exon 19 SNP genotype of JAK2 V617F mutant–positive alleles using either total cDNA (lefthand side) or individual clones (righthand side).
Mutant allele-specific SNP genotyping
The procedure to determine the genotype of JAK2-V617F mutant alleles in mutant-positive patients who were heterozygous for the SNP is outlined in Figure 1B and method details are given in the supplemental material. Briefly, a 798-bp product from exons 14 to 20 covering both the exon 14 mutation and the exon 19 SNP was first amplified from cDNA using primers 14(MM)F and 20R. Products were digested with BsaA1, which would cut once in WT but not mutant alleles. AflIII digestion could not be used here as it cut at another site within the full-length product. The digests were then used as template for a second round of exons 14 to 20 PCR as before and the products were screened for JAK2-V617F mutant status (PCR with primers 14(MM)F and 15R plus AflIII digestion) and exon 19 SNP genotype (PCR with primers 18F and 20R plus BstNI digestion) as appropriate.
In selected cases, the products obtained from the first round of exons 14 to 20 PCR were gel purified then cloned (TOPO TA cloning kit; Invitrogen). JAK2-V617F mutant status of individual clones was determined by PCR with primers 14(MM)F and 15R followed by AflIII digestion. For mutant-positive clones, the SNP genotype was determined as before.
Genotyping of peripheral blood erythroid burst-forming units
Mononuclear cells were purified from peripheral blood by Ficoll-Hypaque density centrifugation and cultured at a density of 1 to 2 × 105/mL in MACS HSC-CFU media complete with Epo (Miltenyi Biotec) at 37°C in 5% CO2. Individual erythroid burst-forming units (BFU-Es) were harvested at day 14, and DNA was extracted (supplemental materials and methods) and screened for the presence of the JAK2-V617F mutation by PCR with exon 14(MM)F and 14R primers followed by AflIII digestion. Mutant-positive colonies were then analyzed using human androgen receptor analysis (HUMARA) as previously described, except that only 24 cycles of amplification were used.7
Results
SNP genotyping of JAK2-V617F mutant alleles
ET patients.
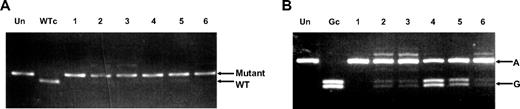
Of the 111 ET patients studied, 39 (35%) were SNP genotype AA, 55 (50%) AG, and 17 (15%) GG. These results are comparable with the SNP genotypes of 114 hematologically normal controls (29% AA, 50% AG, 21% GG). A JAK2-V617F mutation was detected in 43 (39%) of the ET patients, and of these, 16 (37%) were SNP genotype AA, 23 (53%) AG, and 4 (9%) GG. Neutrophil RNA was available from 11 of the 23 ET patients who were both JAK2-V617F mutant positive and SNP heterozygotes. The V617F mutant level, as measured in neutrophil DNA, varied between 11% and 100%. The SNP genotype of V617F mutant-carrying alleles in cDNA from these 11 patients was determined by a PCR–restriction enzyme digestion–PCR method in which PCR products from WT alleles were cut by digestion with BsaAI and were, therefore, not reamplified in a second round of PCR. In all patients, reamplification of only mutant-positive, not WT, alleles was confirmed by exon 14 PCR and AflIII digestion (Figure 2A). SNP genotyping of the second-round PCR products from 10 of the 11 patients showed a mixture of A and G alleles (Figure 2B) with only 1 patient (patient 1, lane 3) showing only A alleles; this patient had a relative mutant level of 100%, consistent with loss of heterozygosity on chromosome 9 (9pLOH). The relative A:G allele proportions varied in the 10 patients (Table 1). This suggested that JAK2 mutations were present on both chromosomes in most of the patients studied.
JAK2 exon 19 SNP genotype analysis of V617F mutant alleles. (A) Gel showing AflIII restriction enzyme digest of 14(MM)F + 15R PCR products amplified from 14(MM)F + 20R second-round PCR products that were obtained from 6 ET patients. V617F mutant–positive products remained undigested. Un indicates no restriction enzyme added; WTc, wild-type control. (B) Gel showing BstNI restriction enzyme digest of 18F + 20R PCR products amplified from the same 14(MM)F + 20R second-round PCR products. Exon 19 SNP allele G, but not A, products were digested. Un indicates no restriction enzyme added; Gc, allele G control.
JAK2 exon 19 SNP genotype analysis of V617F mutant alleles. (A) Gel showing AflIII restriction enzyme digest of 14(MM)F + 15R PCR products amplified from 14(MM)F + 20R second-round PCR products that were obtained from 6 ET patients. V617F mutant–positive products remained undigested. Un indicates no restriction enzyme added; WTc, wild-type control. (B) Gel showing BstNI restriction enzyme digest of 18F + 20R PCR products amplified from the same 14(MM)F + 20R second-round PCR products. Exon 19 SNP allele G, but not A, products were digested. Un indicates no restriction enzyme added; Gc, allele G control.
JAK2 exon 19 SNP analysis of PCR products from 11 patients with JAK2-V617F mutant–positive ET
| Patient . | % Mutant JAK2, neutrophil DNA . | SNP allele A:G ratio in V617F PCR products,* neutrophil DNA . | % SNP allele A V617F+ clones† (no. clones examined) . |
|---|---|---|---|
| 1 | 100 | A | N/A |
| 2 | 13 | A>G | N/A |
| 3 | 40 | A>G | N/A |
| 4 | 25 | A<G | N/A |
| 5 | 24 | A=G | N/A |
| 6 | 42 | A>G | N/A |
| 7a (Oct. 2000) | 20 | A>G | 82% (33) |
| 7b (Dec. 2004) | 33 | A>G | Platelets 78% (27) |
| 8 | 26 | A<G | 41% (31) |
| 9 | 14 | N/A | 76% (29) |
| 10a (July 2000) | 27 | A>G | 77% (33) |
| Platelets 72% (29) | |||
| 10b (June 2002) | 20 | A>G | 87% (31) |
| CD34+ 78% (32) | |||
| 11 | 11 | A<G | 23% (31) |
| Patient . | % Mutant JAK2, neutrophil DNA . | SNP allele A:G ratio in V617F PCR products,* neutrophil DNA . | % SNP allele A V617F+ clones† (no. clones examined) . |
|---|---|---|---|
| 1 | 100 | A | N/A |
| 2 | 13 | A>G | N/A |
| 3 | 40 | A>G | N/A |
| 4 | 25 | A<G | N/A |
| 5 | 24 | A=G | N/A |
| 6 | 42 | A>G | N/A |
| 7a (Oct. 2000) | 20 | A>G | 82% (33) |
| 7b (Dec. 2004) | 33 | A>G | Platelets 78% (27) |
| 8 | 26 | A<G | 41% (31) |
| 9 | 14 | N/A | 76% (29) |
| 10a (July 2000) | 27 | A>G | 77% (33) |
| Platelets 72% (29) | |||
| 10b (June 2002) | 20 | A>G | 87% (31) |
| CD34+ 78% (32) | |||
| 11 | 11 | A<G | 23% (31) |
ET indicates essential thrombocythemia; PCR, polymerase chain reaction; and N/A, not assessed.
Visual estimate of alleles from gel.
Neutrophil samples unless otherwise indicated.
To ensure that these results were not due to incomplete BsaAI digestion of the WT PCR product, which would mimic the mutant product, full-length PCR products covering the V617F mutation and the SNP (exon 14-20) were cloned using neutrophil cDNA from 5 of these patients. Clones were harvested and first screened for presence of the V617F mutation using exon 14 PCR (primers 14(MM)F and 15R) plus AflIII digestion (Figure 1B). Clones showing just V617F mutant–positive products, which excluded possible contamination with WT clones, were then SNP genotyped. Between 29 and 33 V617F mutant–positive clones were genotyped from each sample. In each case, both allele A and allele G clones were detected (Table 1, patients 7-11). In 3 cases, the majority of clones had allele A (82%, 76%, and 77% of clones, respectively, in patients 7, 9, and 10). In one case, A and G clones were approximately equal (patient 8, 41% A clones), and in the remaining case there were more G clones (patient 11, 23% A clones). Platelet samples from 2 patients gave comparable results to the neutrophil samples, 82% and 78% allele A clones, respectively, in neutrophils and platelets from patient 7, and 77% and 72%, respectively, from patient 10. Neutrophil samples taken from the same patient 23 months apart (patient 10) were analyzed and gave comparable results with 77% and 87% allele A clones, respectively. RNA from a purified bone marrow CD34+ sample was also available from this patient and yielded similar results to the neutrophil sample (78% allele A clones).
X-allele analysis of JAK2 mutant–positive BFU-Es
Fresh peripheral blood samples were available in a further V617F mutant–positive ET patient who was not informative for the exon 19 SNP. The patient had a polyclonal XCIP by HUMARA (31%:69% for her alleles of 278 bp and 281 bp, respectively, in both neutrophils and T cells) and a mutant JAK2 level of 16% in neutrophil DNA. Mononuclear cells purified from this patient were cultured under conditions that supported erythroid colony formation. In total, 58 individual BFU-Es were harvested and good PCR products obtained from 47 of them, of which 13 (28%) were V617F mutant positive. All colonies were heterozygous for presence of the mutation. This result is consistent with the observed allele mutant level of 16%, which would be expected to give rise to a relative proportion of 32% mutant-positive cells in the total population. HUMARA of the 13 V617F mutant–positive colonies gave a 278-bp PCR product in 5 (38%) and a 281-bp product in 8 (62%), confirming that the JAK2 mutation had arisen in more than one cell of this patient.
PV patients.
Neutrophil DNA from 30 JAK2-V617F–positive PV patients was SNP genotyped. In 4 cases the JAK2-V617F level was greater than 80%, consistent with 9pLOH in the majority of cells, and therefore these were excluded. Of the remaining 26 cases, 10 were SNP heterozygotes. Neutrophil RNA was available for 5 of these and was investigated using the PCR–BsaAI digestion–PCR technique. Only 1 SNP allele was observed after digestion in all 5 cases.
Discussion
The discovery of the JAK2-V617F mutation in nearly all cases of PV led to the assumption that the mutation was the primary pathogenic event in this disorder. In a murine model, transplantation of JAK2-V617F–transfected bone marrow cells into lethally irradiated mice was sufficient to give rise to an elevated hematocrit,8 and transgenic mice expressing JAK2-V617F developed thrombocytosis, erythrocytosis, or a mixed phenotype, depending on the expression level of the mutant gene relative to the WT gene.23 There have been some data, however, that have challenged this concept. First, in many cases of JAK2-V617F–positive ET that transform to acute myeloid leukemia, the leukemic blasts are JAK2-V617F negative, suggesting that they arose from a pre–JAK2-V617F ancestor.24,25 Second, several groups have reported that in some patients with JAK2-V617F–positive PV, a proportion of the Epo-independent erythroid colonies have WT JAK2, implying that the JAK2 mutation is not the initiating event in at least these patients.26,27 Third, the description of families that appear to have a propensity to develop MPDs and in which only a proportion of affected individuals harbor the JAK2-V617F mutation raises the possibility of a heritable factor that predisposes to the acquisition of this mutation, but that also can cause an MPD in the absence of the mutation.20,–22 It has therefore been suggested that the JAK2-V617F mutation is the second genetic event involved in the pathogenesis of MPD, and there is some evidence to support this. For example, some PV and ET patients have been reported to have a JAK2-V617F–positive population that is smaller than the clonal myeloid population as determined by XCIP analysis or by quantification of the proportion of granulocytic cells carrying the chromosome del20q abnormality.28 In addition, the recent identification in some JAK2-V617F–positive patients of TET2 mutations in JAK2-V617F–negative myeloid precursors suggests that acquisition of TET2 mutations may predate acquisition of JAK2-V617F in at least some MPD patients and, therefore, be the initiating mutation.29
In the current study, SNP genotyping of the JAK2-V617F mutant alleles suggested that there were at least 2 JAK2-V617F mutational events in 10 of 11 ET cases studied. As this assay is dependent on full digestion of the WT PCR product, and could lead to misinterpretation if digestion was incomplete, the finding was confirmed by genotyping individual V617F-positive clones from 5 patients. It is not possible to state accurately how many mutational events had occurred. It must be more than one, and the fact that the incidence of monoallelic mutations in our cohort is very low suggests from Poisson statistics that there is likely to be more than 2 independent mutations in most cases. Previous studies investigating haplotypes or the presence of more than one marker in individual patients have provided evidence for multiple independent clones in a limited number of cases. Olcaydu et al demonstrated that in 3 (2.8%) of 109 unspecified MPD patients investigated, there was evidence for JAK2-V617F being acquired on more than one occasion.30 Similarly, analysis of large cohorts of unselected ET patients indicates that coexistence of JAK2-V617F and another specific cytogenetic or molecular mutation such as del20q or a MPL mutation occurs rarely, accounting for no more than 1% of patients.31,32 Study of in vitro cultures of hematopoietic colonies from such double mutant–positive patients has shown that these events can occur separately, with some clones positive for only one or the other abnormality, although in some cases both abnormalities occurred in the same clone.33,34 In addition, X-allele analysis of a single double mutant–positive ET patient demonstrated that the BFU-E populations carrying either JAK2-V617F or a MPL mutation expressed different X alleles, indicative of separate events.33
Our data go further and suggest that in most ET patients there is more than one, possibly multiple, independent JAK2-V617F event(s). It is possible that in such patients the disease arises from a single stem cell, possibly after a mutation in another gene such as TET2, and that subsequent JAK2-V617F mutations arise in the transformed clone. As the JAK2-V617F–mutated megakaryocytic clone does not suppress the nonmutated cells, these JAK2-WT cells continue to divide and are vulnerable to further JAK2-V617F mutations, which are then also selectively expanded. However, our data are also compatible with the notion that V617F mutations in the JAK2 gene might be arising on a polyclonal background without a preceding transforming event, and this model is supported by our previous studies where we have shown by XCIP analysis that, in patients in whom only a subpopulation of myeloid cells carry a JAK2-V617F–positive allele, the large majority of remaining cells are polyclonal.7 The XCIP analysis of individual erythroid BFU-Es cultured from a patient in the current study is particularly informative in this regard. The observation that the JAK2-V617F mutation is found in cells that have different X chromosomes inactivated not only further confirms the presence of more than one JAK2-V617F mutational event, but also shows that in this case the mutations are not arising in a single pretransformed clone where all the transformed cells would have the same XCIP. The 2 models proposed are not mutually exclusive in that a different sequence of events could occur in different patients.
Regardless of whether the multiple JAK2-V617F mutations arise in a pretransformed cell or on a polyclonal background, the presence of a recurrent mutation in the same gene raises the possibility that there is an increased susceptibility to such mutations in some individuals. This susceptibility may relate to the rapid proliferation of myeloid progenitors in patients with ET. For example, there is a good precedent in mucosa-associated lymphomas for cells that are constantly proliferating in response to an external stimulus to undergo clonal cytogenetic change.35 There are also data from MPD patients that indicate that there may be a genetic predisposition to the development of the disease. Family studies have implied the presence of a heritable tendency to acquire JAK2 mutations in patients with familial MPD,20,22 and recent work by 3 independent groups suggests a strong association between the development of sporadic JAK2-V617F–positive MPDs and a specific JAK2 haplotype.30,36,37 Alternatively, JAK2-V617F mutations may arise at a similar frequency in the hematopoietic progenitors of healthy individuals but confer no survival advantage compared with nonmutated cells, so that the JAK2-V617F–positive population remains small and rapidly fades. In patients with an MPD, the presence of an altered bone marrow environment, whether due to an inherited or acquired abnormality, might confer a proliferative advantage to cells carrying this mutation, thereby prolonging their survival.
Our findings in ET are in contrast to the results obtained in 5 V617F-positive PV patients, where we could find only a single SNP allele associated with the V617F mutation. Although this does not exclude the presence of multiple V617F mutations in some cases of PV, the difference from ET is highly significant (P < .001, Fisher exact test).
The situation we describe of more than one, possibly multiple, independent mutational events occurring in a single gene in ET is similar to the pathogenesis of paroxysmal nocturnal hemoglobinuria, which is considered a benign clonal stem cell disorder. Multiple clones, each with a different mutation in the phosphatidylinositol glycan anchor gene, are seen in some patients, and the extent to which the clones expand varies widely among patients.38 The presence of a JAK2-V617F mutation should not, therefore, be equated with a malignant disease. This is in accord with the long-term stability of the size of the total JAK2 mutant–positive pool that is observed in patients where this pool coexists with JAK2 WT platelets,7 and with the low incidence of transformation to acute myeloid leukemia.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the UK Leukemia Research Fund and the sample bank by the UK Medical Research Council. The work was undertaken at UCL Hospitals (UCLH)/UCL, which receive a proportion of funding from the Department of Health's National Institute for Health Research (NIHR) Biomedical Research Centres' funding scheme.
Authorship
Contribution: J.R.L. performed experimental work, analyzed data, and wrote the paper; T.E. performed sample preparation and experimental work; D.C.L. designed the study, analyzed data, and wrote the paper; and R.E.G. designed the study, performed experimental work, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, UCL Cancer Institute, Paul O'Gorman Bldg, 72 Huntley St, London WC1E 6DD, United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal