Abstract
Chronic myeloid leukemia (CML) has been regarded as the paradigmatic example of a malignancy defined by a unique molecular event, the BCR-ABL1 oncogene. Decades of research zeroing in on the role of BCR-ABL1 kinase in the pathogenesis of CML have culminated in the development of highly efficacious therapeutics that, like imatinib mesylate, target the oncogenic kinase activity of BCR-ABL1. In recent years, most research efforts in CML have been devoted to developing novel tyrosine kinase inhibitors (TKIs) as well as to elucidating the mechanisms of resistance to imatinib and other TKIs. Nonetheless, primordial aspects of the pathogenesis of CML, such as the mechanisms responsible for the transition from chronic phase to blast crisis, the causes of genomic instability and faulty DNA repair, the phenomenon of stem cell quiescence, the role of tumor suppressors in TKI resistance and CML progression, or the cross-talk between BCR-ABL1 and other oncogenic signaling pathways, still remain poorly understood. Herein, we synthesize the most relevant and current knowledge on such areas of the pathogenesis of CML.
Introduction
Chronic myeloid leukemia (CML) is characterized by the Philadelphia (Ph) chromosome, which results from the t(9;22)(q34;q11) balanced reciprocal translocation.1 The molecular consequence of this translocation is the generation of the BCR-ABL1 oncogene that encodes the chimeric BCR-ABL1 protein with constitutive kinase activity.1 Structural studies have facilitated the rational design of therapeutics targeting the tyrosine kinase activity of BCR-ABL1. The impressive results obtained with the first agent of this kind, imatinib mesylate, spurred the development of targeted therapies in cancer medicine. However, these initial results were tempered by the fact that BCR-ABL1 transcripts are readily detectable in most patients receiving imatinib and that responses in the accelerated (AP) or the blastic phase (BP) of the disease, when they occur, are generally short-lived.2 These findings fueled the interest in elucidating the mechanisms of resistance to tyrosine kinase inhibitor (TKI) therapy and in developing novel agents to override these limitations. This article synthesizes recent information generated by researchers on critical aspects of the molecular biology of CML.
The BCR-ABL1 oncogene
Several experimental models, such as BCR-ABL1–expressing CD34+/− cells in culture3,4 or retrovirally transduced BCR-ABL1-positive mouse cells,5,6 have established a causal relationship between BCR-ABL1 and CML. Mice expressing a BCR-ABL1 isoform with a lysine-to-arginine substitution at residue 1176 (K1176R) in the ATP-binding pocket of ABL1, which inactivates its kinase activity, do not develop leukemia even when the mutant is expressed in hematopoietic stem cells (HSCs).7 The critical role of BCR-ABL1 in CML was further demonstrated in transgenic mice in which the tetracycline-responsive element (tet-O) was used to inducibly drive BCR-ABL1 expression in HSCs. When the BCR-ABL1 tet-O mice were crossed with mice expressing the tetracycline transactivator (tTA) under the control of the murine stem cell leukemia (SCL) gene 3′ enhancer, the resultant SCL-tTA/BCR-ABL-tetO mice developed a MPD that mimicked human CML upon withdrawal of tetracycline treatment.8
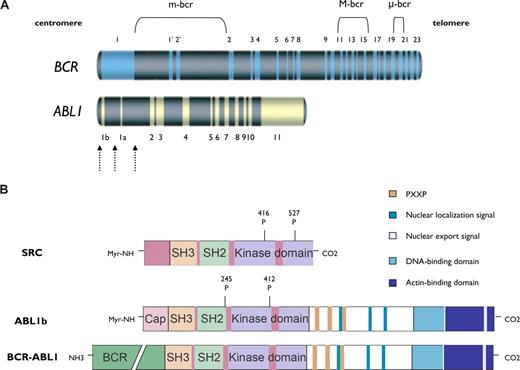
The breakpoints within the ABL1 take place either upstream of exon Ib, downstream of exon Ia, or, more frequently, between exons Ib and Ia (Figure 1A).9 In most patients with CML and in one-third of those with Ph-positive B-cell acute lymphoblastic leukemia (Ph+ B-ALL) the breakpoints within BCR map to a 5.8-kilobase (kb) area spanning exons e12-e16 (formerly called b1-b5), referred to as the major breakpoint cluster region (M-bcr). Alternative splicing gives rise to fusion transcripts with either b2a2 or b3a2 junctions that generate a 210-kDa protein (p210BCR-ABL1).10 In two-thirds of patients with Ph+ B-ALL and in rare cases of CML, the BCR breakpoint localizes to a 54.4-kb area between exons e2′ and e2 (minor breakpoint cluster region or m-bcr), which generates an e1a2 transcript that translates into p190BCR-ABL1. A third breakpoint cluster region (μ-bcr) giving rise to a 230-kDa fusion protein (p230BCR-ABL1), is found in chronic neutrophilic leukemia.11
Schematic representation of the ABL1 and BCR genes and the BCR-ABL1 kinase. (A) BCR contains 23 exons. Exons 1′ and 2′ of BCR are alternative exons within the first intron. The 3 main breakpoint cluster regions (m-bcr, M-bcr, and μ-bcr) in BCR are presented. ABL1 contains 2 alternative first exons (1b and 1a). The dashed arrows represent the breakpoints within ABL1. The combination of breakpoints within BCR and ABL1 genes generates different fusion transcripts encoding proteins with distinct molecular weights. (B) The structural modularity of SRC, ABL1b, and BCR-ABL1 kinases is shown. SRC and ABL1 kinases share a common central core (42% overall homology) composed of a tyrosine kinase domain, an SRC-homology-2 (SH2) domain, and an SH3 domain. The domains upstream of the SH3 domain and downstream of the kinase domain differ significantly between SRC and ABL1 kinases. The NH2 terminus in ABL1 and BCR-ABL1 kinases is the “Cap” region. Two isoforms of ABL1 (human types 1a and 1b) are generated by alternative splicing of the first ABL1 exon. ABL1b contains a myristate site (Myr-NH) at the extreme end of the amino-terminal segment, which binds to the kinase domain and keeps the SH2-SH3 autoinhibitory structure in place (ie, in the “off state”). The homology region in SRC family kinase is the N-terminal membrane-localization domain (also referred to as the SH4 domain). Tyrosine phosphorylation sites are shown.
Schematic representation of the ABL1 and BCR genes and the BCR-ABL1 kinase. (A) BCR contains 23 exons. Exons 1′ and 2′ of BCR are alternative exons within the first intron. The 3 main breakpoint cluster regions (m-bcr, M-bcr, and μ-bcr) in BCR are presented. ABL1 contains 2 alternative first exons (1b and 1a). The dashed arrows represent the breakpoints within ABL1. The combination of breakpoints within BCR and ABL1 genes generates different fusion transcripts encoding proteins with distinct molecular weights. (B) The structural modularity of SRC, ABL1b, and BCR-ABL1 kinases is shown. SRC and ABL1 kinases share a common central core (42% overall homology) composed of a tyrosine kinase domain, an SRC-homology-2 (SH2) domain, and an SH3 domain. The domains upstream of the SH3 domain and downstream of the kinase domain differ significantly between SRC and ABL1 kinases. The NH2 terminus in ABL1 and BCR-ABL1 kinases is the “Cap” region. Two isoforms of ABL1 (human types 1a and 1b) are generated by alternative splicing of the first ABL1 exon. ABL1b contains a myristate site (Myr-NH) at the extreme end of the amino-terminal segment, which binds to the kinase domain and keeps the SH2-SH3 autoinhibitory structure in place (ie, in the “off state”). The homology region in SRC family kinase is the N-terminal membrane-localization domain (also referred to as the SH4 domain). Tyrosine phosphorylation sites are shown.
Anatomy and autoregulation of the BCR-ABL1 protein
BCR-ABL1 kinase contains a series of functionally distinct domains (Figure 1B). The N terminus of BCR-ABL1 consists of the “Cap” region, which is present in 2 different isoforms generated by alternative splicing of the first exon, termed 1a and 1b. ABL1b contains a C14 myristoyl moiety covalently linked to the N terminus and is expressed at higher levels than type 1a, which is not myristoylated. ABL1 also contains a tyrosine kinase domain preceded by highly conserved Src-homology-2 (SH2) and SH3 domains.12 The last exon region contains 4 proline-rich SH3 motifs that function as binding sites for the SH3 domains of adaptor proteins such as Crk, GRB2 (growth-factor-receptor-bound 2), and Nck,13,14 a DNA-binding domain, an actin-binding domain, 3 nuclear localization signals, and 1 nuclear export signal, which determines ABL1 subcellular localization in response to environmental stimuli (Figure 1B).
BCR also exhibits a complex spatial modularity that includes a coiled-coil oligomerization domain, a serine/threonine kinase domain, a Dbl/CDC24 guanine-nucleotide exchange factor homology domain, a pleckstrin homology domain, a putative calcium-dependent lipid binding site, and a Ras-related C3 botulinum toxin substrate (RAC) guanosine triphosphatase–activating protein domain. Tyr177 at BCR serves as docking site for GRB2, GRB10, 14-3-3, and the ABL1 proteins through its SH2 domain.15
The myristoyl modification at the extreme end of the N-terminal segment of ABL1b engages the C-terminal lobe of the ABL1 catalytic domain, facilitating the docking of the SH2 and SH3 domains onto the kinase domain,16,17 in a manner that resembles the inactive conformation of SRC.12,18,19 Forms of ABL1b lacking the myristoyl group exhibit constitutive tyrosine kinase activity.12 The structural elements in ABL1a that replace the function of the myristoyl group of ABL1b are currently unknown. In the inactive conformation of the kinase domain, a prominent α-helix (helix αC) in the N-terminal lobe of the kinase is inwardly displaced toward the active site, forming an ion pair between the conserved Lys271 and Glu286 residues,20 whereas the SH2 domain remains tightly bound to the C-lobe of the kinase. In this conformation, the rotation of the helix αC results in the displacement of important catalytic residues out of the active site, which impairs the access of ATP and the peptide substrate to the active site.12 However, the last exon region of ABL1 may be dispensable for SH3-domain–dependent autoinhibition in vitro.21 X-ray crystal structural analysis of the oligomerization domain of BCR-ABL1 (residues 1-72 or BCR1-72), which includes the coiled-coil domain (BCR30-65), has shown that 2 monomers associate in an antiparallel dimer that stacks to form a tetramer.22 The kinase activity, phosphorylation of tyrosine residues in the activation loop of the catalytic domain, and the leukemogenic potential of BCR-ABL1 are remarkably impaired by mutations of the coiled-coil domain secondary to impairment of oligomerization.23
Signaling pathways downstream of BCR-ABL1 kinase
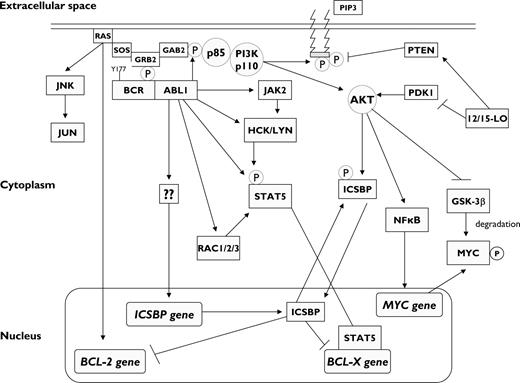
BCR-ABL1 activates numerous BCR-ABL1 downstream signaling pathways (Figure 2). Phosphorylation of BCR Tyr177 is essential for BCR-ABL1–mediated leukemogenesis.24 Mutation of Tyr177 to phenylalanine (Tyr177Phe) largely abolishes GRB2 binding and diminishes BCR-ABL1–induced RAS activation.25 Thus, BCR-ABL1-Tyr177Phe impairs the transformation of primary bone marrow cultures despite preservation of the kinase activity of ABL1.25 Transfection of the mutant BCR-ABL1-Tyr177Phe limits remarkably the induction of a myeloproliferative disorder (MPD) in a murine stem cell transplantation (SCT) model of CML.25 Tyr177 functions as a high-affinity docking site for the SH2 domain of GRB2, which in turn recruits SOS (son of sevenless; a guanine-nucleotide exchanger of RAS), resulting in activation of RAS15 and the scaffold adapter GRB2-associated binding protein 2 (GAB2).15 Therefore, BCR-ABL1–induced GAB2 phosphorylation is mediated by the GRB2/GAB2 complex,26 which causes constitutive activation of the phosphatidylinositol 3-kinase (PI3K)/AKT27 and extracellular signal-regulated kinase (ERK) in primary CML cells. BCR-ABL1 fails to transform primary myeloid cells from GAB2−/− mice.26 Interestingly, MEK-ERK activation is cytokine-dependent in chronic phase (CP) but becomes constitutively activated in BP CML and is readily detectable in CD34+ progenitors.28
Molecular signaling in BCR-ABL1–positive myeloid progenitors. The phosphorylated Tyr177 residue of BCR serves as a docking site for growth factor receptor-bound protein 2 (GRB2), which binds GRB2-associated binding protein 2 (GAB2), as well as SOS (a guanine-nucleotide exchanger of RAS), resulting in RAS-MAPK activation, which in turn results in BCL-2 gene transcription. Upon phosphorylation by BCR-ABL1, GAB2 recruits phosphatidylinositol 3-kinase (PI3K), which activates AKT. AKT activation results in increased transcription of MYC gene and stabilization of MYC protein via inhibition of its degradation by GSK-3β. BCR-ABL1 also activates STAT5, both directly and indirectly through activation of JAK2 and the SRC kinases HCK and LYN. The end result is the activation of BCL-X gene transcription. By contrast, BCR-ABL1 abrogates the transcription of interferon consensus sequence binding protein (ICSBP) through an unknown mechanism, which releases the ISCBP-mediated inhibition of BCL-2 and BCL-X gene transcription and results in increased survival of myeloid progenitors. The same effect is attained via 12/15-lipoxygenase (12/15-LO), which may either inhibit PDK1 or activate PTEN (both regulators of AKT), thus resulting in increased phosphorylation of ICSBP. The net effect of BCR-ABL1 kinase activation is the promotion of cell proliferation and survival. Pointed arrows indicate direct interactions and/or activation. Blunt-ended arrows indicate inhibitory effects. GSK-3β, glycogen synthase kinase-3β; PIP3, phosphatidylinositol-3,4,5-triphosphate.
Molecular signaling in BCR-ABL1–positive myeloid progenitors. The phosphorylated Tyr177 residue of BCR serves as a docking site for growth factor receptor-bound protein 2 (GRB2), which binds GRB2-associated binding protein 2 (GAB2), as well as SOS (a guanine-nucleotide exchanger of RAS), resulting in RAS-MAPK activation, which in turn results in BCL-2 gene transcription. Upon phosphorylation by BCR-ABL1, GAB2 recruits phosphatidylinositol 3-kinase (PI3K), which activates AKT. AKT activation results in increased transcription of MYC gene and stabilization of MYC protein via inhibition of its degradation by GSK-3β. BCR-ABL1 also activates STAT5, both directly and indirectly through activation of JAK2 and the SRC kinases HCK and LYN. The end result is the activation of BCL-X gene transcription. By contrast, BCR-ABL1 abrogates the transcription of interferon consensus sequence binding protein (ICSBP) through an unknown mechanism, which releases the ISCBP-mediated inhibition of BCL-2 and BCL-X gene transcription and results in increased survival of myeloid progenitors. The same effect is attained via 12/15-lipoxygenase (12/15-LO), which may either inhibit PDK1 or activate PTEN (both regulators of AKT), thus resulting in increased phosphorylation of ICSBP. The net effect of BCR-ABL1 kinase activation is the promotion of cell proliferation and survival. Pointed arrows indicate direct interactions and/or activation. Blunt-ended arrows indicate inhibitory effects. GSK-3β, glycogen synthase kinase-3β; PIP3, phosphatidylinositol-3,4,5-triphosphate.
BCR-ABL1 also phosphorylates the SRC family kinases (SFKs) HCK, LYN, and FGR. Phosphorylated HCK recruits the transcription factor signal transducer and activation of transcription 5 (STAT5),15,29 which modulates gene transcription and up-regulates cyclin D1, leading to cell-cycle progression from G1 to S phase.30 However, the role of STAT5 in BCR-ABL1–mediated leukemogenesis remains controversial. Retroviral transduction of BCR-ABL1 into bone marrow induces a CML-like MPD in STAT5a−/−STAT5b−/− mice.31 Inactivation of STAT5 with siRNA in primary CML samples impairs Ph+ myeloid colony formation. Fetal liver hematopoietic progenitors from STAT5a−/−STAT5b−/− mice failed to induce leukemia in recipient mice after retroviral transduction with BCR-ABL1.32 Furthermore, the antiapoptotic protein BCL-X, which is repressed by the transcription factor interferon consensus sequence binding protein (ICSBP),33 is transcriptionally activated by STAT5.34 Although the individual contribution of some BCR-ABL1 downstream pathways may appear negligible when evaluated individually, a cooperative interplay may be necessary for the full realization of the leukemogenic potential of BCR-ABL1. For instance, when BCR-ABL1–positive K562 cells were induced to express dominant-negative forms of RAS, PI3K, or STAT5, marked apoptosis was observed in cells coexpressing 2 of the 3 dominant-negative mutants in any combination.35
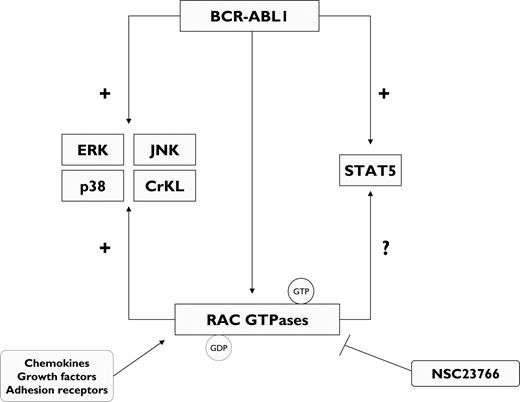
The RAC subfamily of guanosine triphosphatases (GTPases) encompasses RAC1, RAC2, and RAC3 and modulates multiple cell functions.36,37 RAC GTPases are activated in primary CML cells.37 In a murine model of p210BCR-ABL1-induced MPD, targeting of RAC1 and RAC2 genes delayed remarkably the development of myeloproliferation and abrogated the phosphorylation of the BCR-ABL1 downstream signaling molecules CrKL, ERK, c-Jun N-terminal kinase (JNK), and p38,37 suggesting that the BCR-ABL1 signaling network is highly dependent on RAC GTPases (Figure 3). The specific RAC1/RAC2 inhibitor NSC2376636 markedly reduced the growth of primary bone marrow BP CML cells as well as Ba/F3 cells ectopically expressing BCR-ABL1 T315I.37
BCR-ABL1 kinase regulates the activation of RAC guanosine triphosphatases (RAC GTPases). In CML progenitors, RAC GTPases include RAC1, RAC2, and RAC3, which integrate signal chemokines, growth factors, and adhesion receptors to coordinate cell responses. BCR-ABL1 activates a variety of effectors to promote cell proliferation, including RAC GTPases. In turn, RAC GTPases may mediate STAT5 phosphorylation. Inhibition of RAC GTPases by the specific RAC inhibitor NSC23766 abrogates BCR-ABL1–induced cell proliferation. GDP, guanosine diphosphate; GTP, guanosine triphosphate.
BCR-ABL1 kinase regulates the activation of RAC guanosine triphosphatases (RAC GTPases). In CML progenitors, RAC GTPases include RAC1, RAC2, and RAC3, which integrate signal chemokines, growth factors, and adhesion receptors to coordinate cell responses. BCR-ABL1 activates a variety of effectors to promote cell proliferation, including RAC GTPases. In turn, RAC GTPases may mediate STAT5 phosphorylation. Inhibition of RAC GTPases by the specific RAC inhibitor NSC23766 abrogates BCR-ABL1–induced cell proliferation. GDP, guanosine diphosphate; GTP, guanosine triphosphate.
Mice lacking the enzyme 12/15-lipoxygenase (12/15-LO) develop an MPD with 100% penetrance that progresses to transplantable leukemia independent from ABL1 dysregulation.38 Cells isolated from chronic-stage 12/15-LO–deficient mice (Alox15) exhibit increased PI3K/AKT activation as well as interferon (IFN) consensus sequence binding protein (ICSBP), which results in decreased direct DNA binding, limiting its ability to repress BCL-2 gene transcription, thus promoting leukemic cell survival (Figure 2).38 ICSBP is a negative regulator of granulocyte differentiation. ICSBP−/− and ICSBP+/− mice exhibit deregulated hematopoiesis manifested as a CML-like MPD.39 Forced expression of ICSBP inhibited BCR-ABL1–induced CML-like disease in vivo.40 All the effects observed in Alox15 mice were reversed upon treatment with a PI3K inhibitor, which suggests that 12/15-LO is an important suppressor of MPD by virtue of its PI3K-dependent regulation of ICSBP and their downstream target genes in vivo.38
JUNB is a component of the activator protein 1 family of transcription factors that functions as a tumor suppressor in myeloid cells,41 antagonizing the RAS downstream target JUN, thus inhibiting cell proliferation and survival. Transgenic mice specifically lacking JUNB in the myeloid lineage (JunB−/−Ubi-JunB mice) developed an MPD that closely resembled human CML, including progression to BP.42,43 Furthermore, only JUNB-deficient long-term self-renewing HSCs from diseased mice were able to induce CML-like disease in recipient mice after transplantation.43
Transformation to blast phase
The mechanisms responsible for CML progression and transformation are complex and only partially understood. This is due in part to the lack of adequate animal models of BCR-ABL1–induced leukemia with a long “chronic” myeloproliferative phase necessary for an adequate assessment of the mechanisms of disease progression. Various factors have been identified as critical in the transition from CP to BP CML.
BCR-ABL1 expression
The continuous unrestrained expression and activity of BCR-ABL1 kinase is paramount not only in CML maintenance but also in progression. In fact, BCR-ABL1 mRNA and protein levels are higher in BP than in CP.44,45 BCR-ABL1 transcript levels may be up to 200-fold greater in the most primitive CML CD34+ progenitors relative to their more differentiated counterparts.46 In vitro, BCR-ABL1 directly regulates growth factor dependence,46 clonogenicity,46,47 protection against apoptosis,47 and motility46 in a dose-dependent fashion. BCR-ABL1 dosage is also a critical mediator of disease latency when BCR-ABL1–positive cells are injected subcutaneously into syngeneic mice.46 Although the reason for the increment of BCR-ABL1 transcript levels is not well understood, this phenomenon may result from selective pressure favoring the expansion of highly proliferative/poorly differentiated leukemic clones. Supporting this contention is the fact that elevated BCR-ABL1 transcript levels have been detected in the CD34+ granulocyte-macrophage progenitor (GMP) subpopulation from patients with BP CML relative to patients with CP CML.48 These cells can be found expanded in BP CML.48 Alternatively, it can be hypothesized that the BCR-ABL1 oncoprotein promotes the expression of BCR-ABL1 transcripts and/or that the degradation of BCR-ABL1 transcripts may be down-regulated selectively in CD34+ CML progenitors.49 Despite the importance of BCR-ABL1 expression, BCR-ABL1–independent mechanisms may play an important role in CML progression. For instance, overexpression and/or activation of the SFKs HCK, LYN, and FYN has linked to CML progression and imatinib resistance.50-53 BCR-ABL1 retrovirus-transduced bone marrow from LYN−/−HCK−/−FGR−/− mice efficiently induced CML but not B-ALL in recipients, indicating that SFKs may be important in the pathogenesis of Ph+ B-ALL and lymphoid BP CML.54,55 BCR-ABL1–independent activation of LYN has been also shown in imatinib-resistant patients expressing unmutated BCR-ABL1.56 Anti-SFK therapy may therefore be beneficial in advanced-phase CML.55
Arrest of differentiation
Gradual disruption of the differentiation program is a characteristic feature of CML progression. BCR-ABL1 modulates the activity of transcription factors that regulate the expression of several differentiation-related genes.57,58 The transcription factor CCAAT/enhancer-binding protein-α (CEBPα) is essential for normal granulocyte differentiation and is found expressed in normal bone marrow cells and in CP CML samples. Conversely, CEBPα expression is lost in BP CML.59 CEBPα down-regulation is mediated by BCR-ABL1 at the translational level through the stabilization of the poly(rC)-binding protein heterogeneous nuclear ribonucleoprotein E2 (hnRNP E2).59 Expression of hnRNP E2 is low or undetectable in CP CML but becomes readily detectable in BP CML samples. Transplantation of BCR-ABL1–expressing CEBPα−/− fetal liver cells failed to induce a myeloid disease in transplanted mice but instead induced an immature, lethal transplantable erythroleukemia, whereas recipients of CEBPα-transduced cells consistently phenocopied CP CML disease.60 These data suggest that lack of CEBPα expression prevents cell commitment toward a myeloid cell fate. In the same model, residual levels of CEBPα were not sufficient to induce normal differentiation but appeared to be required for the development of a myeloid BP CML phenotype.60 Recently, deletions of IKZF1 gene, which encodes Ikaros, a transcription factor needed for early lymphoid lineage commitment, have been associated with CML progression, particularly to lymphoid BP CML.61 In addition, 84% of patients with BCR-ABL1–positive ALL exhibited loss of Ikaros expression, typically due to monoallelic deletion of exons 3 to 6 of IKZF1.61 Given that B lymphocytes from mice engineered to express low Ikaros levels arrest their maturation at the pro–B-cell stage,62 it is tempting to speculate that Ikaros loss contributes to the arrested maturation of B cells in BCR-ABL1–positive ALL and lymphoid BP CML.61
Occasionally, the development of a BP CML phenotype involves the interaction of BCR-ABL1 with translocations, resulting in the formation of dominant-negative transcription factors such as NUP98-HOXA963 or AML1-EVI1,64 following a 2-hit paradigm. Although AML1-EVI1 blocks differentiation, NUP98-HOXA9 subverts the normal balance of symmetric and asymmetric renewal division, favoring the former and causing preferential growth of immature precursors and facilitating the transition to BP CML.65 It is tempting to hypothesize that other genetic aberrations encountered in patients with CML may alter the renewal division pattern to promote the proliferation of immature leukemic progenitors to promote progression to BP CML.
Genomic instability and DNA repair
BCR-ABL1 has been shown to induce endogenous reactive oxygen species (ROS) that result in chronic oxidative DNA damage, double-strand breaks (DSBs) in S and G2/M cell-cycle phases, and mutagenesis.66 Moreover, DNA damage surveillance is impaired in CML because of inhibited ataxia telangiectasia and RAD3-related nuclear protein kinase signaling, which results in reduced activation of checkpoint kinase 1 and abrogation of the intra-S-phase cell-cycle checkpoint.67 Nonhomologous end-joining and homologous recombination as well as nucleotide excision repair exhibit unfaithful repair of DSBs induced by ROS66 and γ-irradiation68 as a consequence of BCR-ABL1 kinase activity.66 As a result, G/C to A/T transitions and G/C to T/A transversions in the coding regions of multiple genes (including the kinase domain of BCR-ABL1) have been demonstrated in CML cells but not in BCR-ABL1–negative cells.66,69 Collectively, these results suggest that BCR-ABL1–mediated ROS generation in combination with aberrant regulation of DNA repair pathways contributes to a mutator phenotype in CML cells and results in genomic instability that results in point mutations and cytogenetic abnormalities.69,70
Additional chromosomal abnormalities
Approximately 80% of patients with CML develop additional nonrandom cytogenetic aberrancies in Ph+ cells, an occurrence known as “clonal evolution,” which is a reflection of the genetic instability that characterizes the transition to advanced-phase CML.71 The most frequent cytogenetic abnormalities encountered in patients with clonal evolution are trisomy 8 (34%), isochromosome 17 (20%), and duplicate Ph chromosome (38%),72 which have been associated with MYC overexpression, loss of 17p, and BCR-ABL1 overexpression, respectively.44,73 Other aberrancies, such as trisomy 19, trisomy 21, trisomy 17, and deletion 7, have been described in less than 10% of cases of clonal evolution.1 These genetic lesions are more frequently associated with myeloid than with lymphoid BP CML. A causative role in the development of these abnormalities of therapies used before or concomitantly with imatinib (ie, IFNα, cytoreductive agents) cannot be excluded. However, almost 50% of patients with BP CML receiving imatinib therapy develop new cytogenetic abnormalities in Ph+ cells.74 Moreover, 10% to 15% of patients with CML present with deletions of the derivative chromosome 9, which may be a reflection of genomic instability leading to more rapid progression to BP.75 The detrimental prognosis conferred by deletions of the derivative chromosome 9 may be offset by imatinib therapy.76 Cytogenetic abnormalities have also been detected in Ph-negative metaphases of 2% to 17% of patients with CML and have been occasionally linked with development of myelodysplasia or AML.77 Recently, by using single-nucleotide polymorphism (SNP) arrays, researchers have shown that patients with CP CML harbor 0.47 copy number alterations per case (range, 0-8), whereas 7.8 copy number alterations (range, 0-28) per BP CML case were detected,61 further supporting the notion that multiple genomic aberrations accumulate during progression to BP CML.
Inactivation of tumor suppressor genes
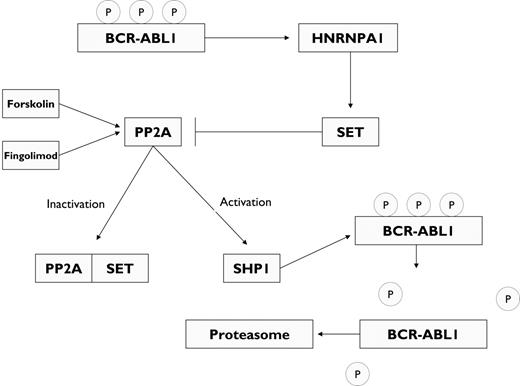
One of the most common mutations associated with CML progression involves the tumor suppressor p53, which is mutated in 25% to 30% of patients with myeloid BP. In addition, exon 2 of the INK4A/ARF locus is found deleted in 50% of cases of lymphoid BP.78 This latter event results in loss of p16 and p14/ARF expression, which regulate the G1/S checkpoint by inhibiting the G1-phase cyclin D-Cdk4/Cdk6 and p53 down-regulation, respectively.73 Because ARF enhances p53 levels by interfering with the activity of MDM2, the principal negative regulator of p53, homozygous deletion at the p16/ARF locus might represent a functional equivalent of p53 mutation in myeloid BP.73 In fact, retroviral transduction of BCR-ABL1 into ARF-null murine bone marrow cells rapidly generates polyclonal cellular populations of continuously self-renewing pre-B cells.79 The p53 pathway also plays a key role in response to imatinib therapy. Treatment of BCR-ABL1–expressing cells with imatinib results in selective activation of p53 as a result of BCR-ABL1 kinase inhibition.80 By contrast, p53 inactivation, which frequently accompanies CML progression, blocks the response to imatinib in vitro and in vivo without preventing BCR-ABL1 kinase inhibition. Therefore, mutations affecting the p53 pathway may contribute to imatinib resistance in CML-BP.80 The RUNX family of transcription factors has also been implicated in imatinib response and disease persistence in patients with CML.81 In mice transplanted with bone marrow cells retrovirally infected with BCR-ABL1 and subsequently treated with imatinib to select for leukemic cells in which the proviral integration had affected genes modulating imatinib response, it was demonstrated that clonal outgrowth of cells carry similar integration sites. Proviral integration near the RUNX3 promoter induced expression of RUNX3, and BCR-ABL1–positive cell lines with stable or inducible expression of RUNX1 or RUNX3 were protected from apoptosis induced by imatinib treatment.81 Imatinib therapy also selected for RUNX1-expressing cells in vivo after infection of primary bone marrow cells with both BCR-ABL1 and RUNX1,81 suggesting that RUNX1 contributes to disease persistence. The serine/threonine phosphatase 2A (PP2A) functions as a tumor suppressor by antagonizing BCR-ABL1 (Figure 4).82 BCR-ABL1 kinase inhibits PP2A by up-regulating SET, a phosphoprotein that inhibits PP2A.82 In turn, PP2A activates protein tyrosine phosphatase 1 (SHP1), which catalyzes BCR-ABL1 dephosphorylation and proteosomal degradation.82 Both SET inhibition and PP2A activation are therefore potential strategies for CML therapy. Silencing of SET by siRNA or treatment of BCR-ABL1–positive cells with forskolin, a pharmacologic activator of PP2A, results in decreased BCR-ABL1 expression.82 Fingolimod (FTY720), another PP2A activator, used as immunosuppressive therapy for patients with multiple sclerosis,83 induces caspase-dependent apoptosis and impaired clonogenic potential of the imatinib- and dasatinib-resistant 32D-p210(T315I)BCR-ABL1 cell line and of primary bone marrow cells from patients with BP CML and Ph+ B-ALL.84 Fingolimod suppressed leukemogenesis in SCID mice transplanted with myeloid or lymphoid progenitors transformed with p210BCR-ABL1 or p190BCR-ABL1, respectively. More importantly, 80% and 90% of p210 and p190 mice, respectively, attained molecular remission after 11 weeks of fingolimod therapy, as well as 50% of those transplanted with progenitors expressing the T315I p210BCR-ABL1 mutant.84
BCR-ABL1–induced inactivation of PP2A. BCR-ABL1 mediates the post-transcriptional up-regulation of SET, a phosphoprotein that functions as an inhibitor of the serine/threonine phosphatase PP2A. This activity is mediated via the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1). The activation of PP2A recruits the SH2 domain–containing protein tyrosine phosphatase 1 (SHP1), which dephosphorylates BCR-ABL1, resulting in BCR-ABL1 down-regulation through proteosomal degradation. Pharmacologic activation of PP2A with forskolin or fingolimod (FTY720) results in abrogation of CML cell proliferation, including cells expressing the pan-resistant mutation T315I.
BCR-ABL1–induced inactivation of PP2A. BCR-ABL1 mediates the post-transcriptional up-regulation of SET, a phosphoprotein that functions as an inhibitor of the serine/threonine phosphatase PP2A. This activity is mediated via the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1). The activation of PP2A recruits the SH2 domain–containing protein tyrosine phosphatase 1 (SHP1), which dephosphorylates BCR-ABL1, resulting in BCR-ABL1 down-regulation through proteosomal degradation. Pharmacologic activation of PP2A with forskolin or fingolimod (FTY720) results in abrogation of CML cell proliferation, including cells expressing the pan-resistant mutation T315I.
In summary, multiple lines of evidence suggest that the increased survival, proliferation, and differentiation arrest of BP CML cells relies upon the intimate cooperation of BCR-ABL1 with an array of genes deregulated during disease progression. A better understanding of these abnormalities, which will undoubtedly be facilitated by the development of more faithful CML animal models, is warranted to improve currently available therapies for BP CML.
Gene-expression profiling in CML
Genome-wide analysis of gene expression profiles may aid in characterizing gene candidates responsible for disease progression. Unfortunately, most studies failed to provide clear-cut answers, probably because of differences in types of samples, array platforms, and statistical methods used.85-90 A genome-wide screening at a resolution of 1 Mb in 54 samples from patients with CML in all phases as well as in 12 CML cell lines revealed that CML progression was associated with a spectrum of recurrent genome imbalances, frequently involving losses at 1p36, 5q21, 9p21, and 9q34 and gains at 1q, 8q24, 9q34, 16p, and 22q11, in patients with AP or BP but not CP CML.91 A recent analysis used DNA microarrays to compare gene expression in 91 cases of CML in all phases. The correlation of expression level between AP and BP gene expression was strong (r = 0.81), suggesting that CML might follow a biphasic rather than a classic triphasic model of progression.89 Genes potentially involved in advanced disease compared with CP included cytokines (IL3RA, SOCS2), alternative RAS pathways (Rras2), and genes involved in cell adhesion (WNT/β-catenin), among others such as JUNB, FOS, PRAME, MXF1, and EF1δ.89
CML stem cells
BCR-ABL1, unlike other fusion oncogenes involved in human leukemia, such as MLL-ENL or MOZ-TIF2, can transform HSCs but is not sufficient to transform committed myeloid progenitors lacking inherent self-renewal capacity.92 In CML, CD34+CD38−Lin− leukemic stem cells (LSCs) express high BCR-ABL1 transcript levels.48 During the transition from CP to BP, LSCs acquire additional genetic and/or epigenetic abnormalities that provide survival advantage, resistance to programmed cell death, and extended replicative lifespan (Figure 5).93,94 Mice genetically lacking JUNB43 or ICSBP39 have been shown to induce an MPD that mimics CML, including progression to BP. Whether BCR-ABL1 abrogates the expression of these transcription factors and whether these events are related or constitute stochastic events during disease progression are currently unknown. Nonetheless, in addition to increased proliferative potential, progression to BP CML requires either the expansion of LSCs or the acquisition of self-renewal properties by a subset of committed progenitors. In a mouse model, expression of the hMRP8p210BCR-ABL1 transgene targeted to GMPs and their myelomonocytic progeny, but not to HSCs, renders a phenotype that resembles human CML, including progression to AP and BP. Enforced expression of hMRP8p210BCR-ABL1 synergizes with expression of BCL-2 (resulting in blockade of programmed cell death), but not MYC or N-RAS, in myeloid progenitors to induce BP.95 These data suggest that BP may result from the progressive acquisition of genetic alterations within progenitors downstream of the hematopoietic stem cell (HSC) that acquire self-renewal properties.95
Theoretical model of chronic myeloid leukemia. BCR-ABL1 fails to transform cells lacking inherent self-renewal potential, thus supporting the notion of CP CML as an HSC disorder. HSCs, through successive accumulation of preleukemic genetic abnormalities (eg, BCR-ABL1 expression, BCL-2 overexpression, or loss of JUNB or ICSBP expression), acquire a proliferative and survival advantage and lose the ability to undergo apoptosis. Further genetic and/or epigenetic events in BCR-ABL1–positive committed myeloid cells (eg, common myeloid progenitors [CMP] and granulocyte-monocyte progenitors [GMP]) endow the latter with self-renewal potential (eg, aberrant β-catenin activation) and arrest myeloid differentiation (eg, loss of CEBPα or IKZF1 expression), thus facilitating the emergence of a leukemic stem cell (LSC) clone driving the transition to blast-phase CML (partially adapted from Weissman94 ).
Theoretical model of chronic myeloid leukemia. BCR-ABL1 fails to transform cells lacking inherent self-renewal potential, thus supporting the notion of CP CML as an HSC disorder. HSCs, through successive accumulation of preleukemic genetic abnormalities (eg, BCR-ABL1 expression, BCL-2 overexpression, or loss of JUNB or ICSBP expression), acquire a proliferative and survival advantage and lose the ability to undergo apoptosis. Further genetic and/or epigenetic events in BCR-ABL1–positive committed myeloid cells (eg, common myeloid progenitors [CMP] and granulocyte-monocyte progenitors [GMP]) endow the latter with self-renewal potential (eg, aberrant β-catenin activation) and arrest myeloid differentiation (eg, loss of CEBPα or IKZF1 expression), thus facilitating the emergence of a leukemic stem cell (LSC) clone driving the transition to blast-phase CML (partially adapted from Weissman94 ).
HSC self-renewal in mice requires the activation of the β-catenin pathway, which results in the translocation of β-catenin to the nucleus, where it interacts with lymphoid enhancer factor/T-cell factor (LEF/TCF) and regulates the transcription of genes such as MYC and Cyclin D1.96 In humans, progression to BP is associated with a 6- to 10-fold expansion of GMPs, rather than expansion of the pool of HSCs. Furthermore, transfection of axin, a specific inhibitor of the β-catenin pathway, blocked leukemic GMP replating, and no difference in the levels of β-catenin and its transcriptional coactivator LEF/TCF in HSCs was observed between healthy control subjects and patients with CP or BP CML. However, increased levels of both (as well as of BCR-ABL1 transcripts) were readily detected in GMPs isolated from patients in BP CML, thus endowing this subset of cells with “stemness” properties.48 BP CML GMPs could replate self-renewing cells in vitro, unlike normal GMPs. Thus, hypothetically, CML could be considered an HSC-derived but progenitor-driven disorder in which 2 distinct LSC populations with self-renewal capability may coexist in patients with BP CML: one chiefly involving GMPs expressing high BCR-ABL1 transcripts and enhanced nuclear β-catenin responsible for rapid expansion in BP CML, and another one mainly involving quiescent BCR-ABL1–positive HSCs responsible for maintaining a CP CML phenotype (Figure 5).
Quiescent BCR-ABL1–expressing LSCs account for approximately 0.5% of the CD34+ population and, unlike their progeny, are refractory to chemotherapeutic agents, radiation,97 and BCR-ABL1 TKIs.98-100 The clearance of BCR-ABL1 transcripts during imatinib therapy follows a biphasic decline, reflecting the fact that differentiated cells are readily cleared by TKIs, whereas CML LSCs are not affected by virtue of its quiescent status, leading to persistence of residual disease.101,102 An array of phenotypic and functional characteristics underlie the refractoriness to TKI therapy exhibited by BCR-ABL1–positive LSCs, including (1) enhanced expression of interleukin-3 (IL-3) and granulocyte colony-stimulating factor, which is believed to be tightly regulated by BCR-ABL1 in a dose-dependent fashion and to correlate with the primitive status of LSCs; (2) down-regulation of human organic cation transporter-1 and up-regulation of the adenosine triphosphate (ATP)–binding cassette (ABC) transporter ABCB1 (MDR-1) and ABCG2, responsible for the influx and efflux of imatinib, respectively49 ; (3) increased expression of BCR-ABL1 mRNA, protein, and kinase activity, which suggests that TKI-induced killing of CML LSCs will require much higher doses that those required to eliminate more mature BCR-ABL1–positive progenitors. It is noteworthy that neither imatinib nor the potent TKIs nilotinib and dasatinib are significantly active against quiescent CML progenitor cells.103-105 In these cells, mitogen-activated protein kinase (MAPK), PI3K, and STAT5 remain active despite BCR-ABL1 kinase inhibition105 ; (4) increased instability of the BCR-ABL1 oncogene, which has been linked to increased levels of ROS and oxidative DNA damage resulting in a higher rate of BCR-ABL1 kinase domain mutations in BCR-ABL1–positive LSCs even before exposure to therapy,106,107 thus setting the stage for TKI resistance and CML relapse.
BCR-ABL1 kinase domain mutations and TKI resistance
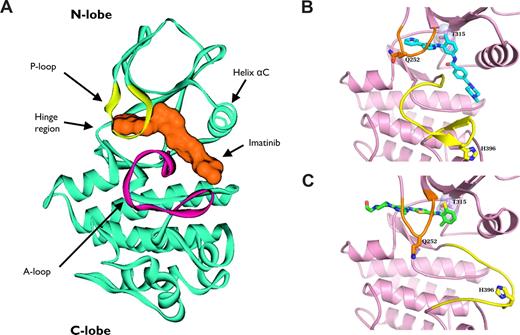
The development of mutations within the kinase domain of BCR-ABL1 represents the most frequent mechanism of resistance to TKI therapy in CML, being detected in 40% to 90% of patients who are resistant to imatinib.108-113 Imatinib and nilotinib bind to the “DFG-out” conformation (catalytically inactive) of ABL1 kinase, in which the highly conserved Asp-Phe-Gly (DFG) residues are displaced out of the catalytic groove.114,115 X-ray crystallography116 and NMR spectroscopy117 have shown that dasatinib binds the active “DFG-in” conformation. Imatinib extends deeply into the catalytic domain, and its pyridinyl group is located underneath the αC helix in the NH2-terminal lobe of ABL1 kinase (Figure 6A).114 Thr315, also known as the gatekeeper residue, is located at the periphery of the nucleotide-binding site of ABL1 and forms a key H-bond interaction with imatinib and dasatinib (Figure 6B,C).118 Mutation of Thr315 to isoleucine (T315I) disrupts this H-bond interaction, which, in addition to the steric hindrance imposed by the isoleucine side chain, impairs imatinib binding and causes complete insensitivity to this compound as well as to the second-generation TKIs dasatinib, nilotinib, bosutinib, and INNO-406.2,110,115,119-122 Recent reports have shown that T315I mutation can be found in approximately 15% of patients after failure of imatinib therapy.123
Ribbon representation of ABL1 kinase in complex with tyrosine kinase inhibitors. (A) Ribbon representation of 3-dimensional structure of ABL1 kinase domain (blue) in complex with imatinib (orange). The ATP-binding site in the ABL1 kinase domain is located between the activation loop (A-loop; magenta) and the phosphate-binding loop (P-loop; yellow). The A-loop controls the ABL1 catalytic activity by switching between different states in a phosphorylation-dependent manner. Imatinib inserts its pyridinyl group underneath the helix αC in the NH2-terminal lobe of ABL1 kinase, displacing ATP and trapping the kinase in its inactive conformation. (B,C) The imatinib:ABL1 (blue) and the dasatinib:ABL1 (green) complexes are shown. The A-loop (yellow) adopts diverging positions in the 2 complexes. Conformational changes in the A-loop prevent imatinib binding to the active form of the enzyme. By contrast, dasatinib binds ABL1 in its active conformation and is not involved in critical interactions with most mutated residues involved in imatinib resistance. For instance, the H396 residue is involved in a hydrogen bond that stabilizes the inactive conformation of the A-loop in the imatinib:ABL1 complex (B), whereas no discernible interactions are observed between His396 and ABL1 or dasatinib (C). Thr315 makes a critical hydrogen bond with dasatinib. T315I mutation disrupts this interaction and causes steric clash, impairing the activity of dasatinib, as well as of imatinib and nilotinib, against this mutant.
Ribbon representation of ABL1 kinase in complex with tyrosine kinase inhibitors. (A) Ribbon representation of 3-dimensional structure of ABL1 kinase domain (blue) in complex with imatinib (orange). The ATP-binding site in the ABL1 kinase domain is located between the activation loop (A-loop; magenta) and the phosphate-binding loop (P-loop; yellow). The A-loop controls the ABL1 catalytic activity by switching between different states in a phosphorylation-dependent manner. Imatinib inserts its pyridinyl group underneath the helix αC in the NH2-terminal lobe of ABL1 kinase, displacing ATP and trapping the kinase in its inactive conformation. (B,C) The imatinib:ABL1 (blue) and the dasatinib:ABL1 (green) complexes are shown. The A-loop (yellow) adopts diverging positions in the 2 complexes. Conformational changes in the A-loop prevent imatinib binding to the active form of the enzyme. By contrast, dasatinib binds ABL1 in its active conformation and is not involved in critical interactions with most mutated residues involved in imatinib resistance. For instance, the H396 residue is involved in a hydrogen bond that stabilizes the inactive conformation of the A-loop in the imatinib:ABL1 complex (B), whereas no discernible interactions are observed between His396 and ABL1 or dasatinib (C). Thr315 makes a critical hydrogen bond with dasatinib. T315I mutation disrupts this interaction and causes steric clash, impairing the activity of dasatinib, as well as of imatinib and nilotinib, against this mutant.
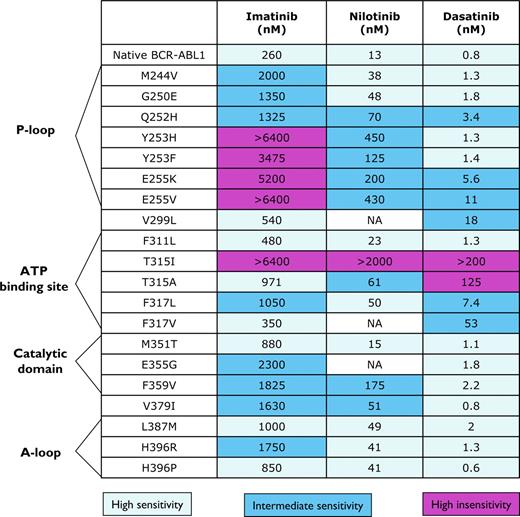
So far, more than 100 different point mutations encoding for single amino acid substitutions in the kinase domain of BCR-ABL1 have been reported in patients with imatinib-resistant CML,16,109,114,118,124,125 and others have been generated in vitro by random mutagenesis of BCR-ABL1.124,126 Imatinib-resistant mutations are frequently mapped to the P-loop region (residues 244 to 255) of the kinase domain, which serves as a docking site for phosphate moieties of ATP.127-129 Some studies have linked P-loop mutations to a poorer clinical outcome,130,131 but others have not confirmed this observation.132 Second-generation TKIs (eg, nilotinib, dasatinib) are active against P-loop mutations (Figure 7). Other areas within ABL1 kinase frequently affected by mutations include the activation (A) loop (residues 381-402), which prevent the kinase from adopting the inactive conformation to which imatinib binds, and the catalytic (C) domain (residues 350-363). A series of mutations (Gln253, Tyr253, Glu255, Thr315, Glu459, and Phe486) have been detected more frequently in patients with advanced phases of CML.133 Despite the wide variety of point mutations found in BCR-ABL1, most mutants are rare, with mutations involving residues Gly250, Tyr253, Glu255, Thr315, Met351, and Phe359 accounting for 60%-70% of all mutations.134
Activity of imatinib mesylate and the second generation tyrosine kinase inhibitors nilotinib and dasatinib against a selection of BCR-ABL1 mutants found in patients with CML. All concentrations are shown in nanomoles per milliliter and represent IC50 values.
Activity of imatinib mesylate and the second generation tyrosine kinase inhibitors nilotinib and dasatinib against a selection of BCR-ABL1 mutants found in patients with CML. All concentrations are shown in nanomoles per milliliter and represent IC50 values.
Different mutations are endowed with different transforming capabilities. For instance, in a pre–B-cell transformation assay, T315I (which has weaker kinase activity than p210BCR-ABL1) and E255K consistently showed a 10% to 20% increase in oncogenic potency relative to p210BCR-ABL1, whereas Y253F and E255V displayed potencies similar to those of p210BCR-ABL1, and Y253H, T315A, F317L, and M351T were markedly weaker.135 Mass spectroscopy associated distinct phosphorylation signatures to different mutants, confirming that point mutations determine substrate specificity and activation of different downstream pathways.136
Some patients with CML have been found to carry more than one mutation within the same BCR-ABL1 molecule (polymutants) after failure of sequential TKI therapy, which was associated with increased oncogenic potency compared with each individual mutation in transformation assays.137 A recent mutational analysis of 61 patients with CML after failure of imatinib therapy revealed that 90% of patients harbored BCR-ABL1 kinase domain mutations, and polymutants were detected in 57% of patients.125 The treatment of patients carrying polymutants might require the use of a combination of TKIs with activity against all single-point mutations contained in the compound mutation.
Conclusions
BCR-ABL1 plays a causal role in the pathogenesis of CML. BCR-ABL1–transformed cells exhibit deregulated cell proliferation, growth-factor independence, and reduced apoptosis, which result from the activation of an intricate network of signaling pathways and override the tightly regulated homeostatic molecular circuits that govern the growth and differentiation of hematopoietic progenitors. However, it is clear that BCR-ABL1 kinase acts in concert with multiple other cellular and genetic events that accumulate over time to inexorably drive the disease into BP. The reliance of CML cells on the constitutive activation of BCR-ABL1 kinase has provided the opportunity to develop highly efficacious targeted agents against this oncogenic kinase. Although highly manageable, CML remains currently incurable outside the realm of allogeneic SCT. A better understanding of the mechanisms that regulate progression to BP, TKI resistance, stem cell quiescence, tumor suppressor inactivation, as well as genomic instability and faulty DNA repair, is essential to develop definitive curative strategies for patients with CML.
Acknowledgments
A.Q.-C. is indebted to Sean Post, PhD, for invaluable daily doses of “selective pressure,” to the reviewers of this manuscript for constructive criticism, and to Mark Potter and David Liu for providing materials to elaborate Figure 6.
A.Q.-C. is a recipient of an American Society of Clinical Oncology Young Investigator Award 2006-2008.
Authorship
Contribution: A.Q.-C. designed and performed research, analyzed data, and wrote the manuscript; and J.C. designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alfonso Quintás-Cardama, M.D., M. D. Anderson Cancer Center, Department of Leukemia, Unit 428, 1515 Holcombe Boulevard, Houston, TX 77030; e-mail: aquintas@mdanderson.org.

![Figure 5. Theoretical model of chronic myeloid leukemia. BCR-ABL1 fails to transform cells lacking inherent self-renewal potential, thus supporting the notion of CP CML as an HSC disorder. HSCs, through successive accumulation of preleukemic genetic abnormalities (eg, BCR-ABL1 expression, BCL-2 overexpression, or loss of JUNB or ICSBP expression), acquire a proliferative and survival advantage and lose the ability to undergo apoptosis. Further genetic and/or epigenetic events in BCR-ABL1–positive committed myeloid cells (eg, common myeloid progenitors [CMP] and granulocyte-monocyte progenitors [GMP]) endow the latter with self-renewal potential (eg, aberrant β-catenin activation) and arrest myeloid differentiation (eg, loss of CEBPα or IKZF1 expression), thus facilitating the emergence of a leukemic stem cell (LSC) clone driving the transition to blast-phase CML (partially adapted from Weissman94).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/8/10.1182_blood-2008-03-144790/5/m_zh80240828250005.jpeg?Expires=1770996442&Signature=Vpou~g1gapEVKGUUT12t4O-OjOLsoMQiASYtvjixjvtNNtN4gpcB73LAj~n-p5FwggBxj6g4~5pEX~~zoTR8OyrhHNUHQfl3ISIqe1PCVzU69bkh5Lf7sQLwTaZz5AZxoIVraOLqUogarm4MLelr0ww4G9ee06PiugccxLokdBQxFjpsJs6yh0DGQBGs7ErZyCxhM1GM9tHglRoGaXt7L2Bg7k-lmX9vAlIt~0KY57uEcg1dIB9VyJhC5Ia-AbDe93cScsQdqa5NvCPXjKm3M2s-MkURwmP2NF90iSUMmJ8JkwyukuQWdZkXu9rwtrLOWo-RwkBw9YE8A7FDJLiDiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal