Abstract
Severe congenital neutropenia (CN) is a rare bone marrow failure syndrome with a high incidence of acute leukemia. In previous studies, we could show that point mutations in the gene for the granulocyte colony-stimulating factor (G-CSF) receptor CSF3R are a highly predictive marker for leukemic development in CN patients. To find out at which stage of hematopoietic development these mutations emerge and how they are propagated during hematopoietic differentiation, we analyzed single cells of different hematopoietic subpopulations from CN patients with CSF3R mutations. We found that CSF3R mutations are not restricted to the myeloid compartment but are also detectable in lymphoid cells, although at a much lower percentage. From our observations, we conclude that CSF3R mutations are acquired in multipotent hematopoietic progenitor cells in CN patients and that they are clonally expanded in myeloid cells expressing the G-CSF receptor due to the growth advantage mediated by the CSF3R mutation.
Introduction
Severe congenital neutropenia (CN) is a rare disease characterized by an absolute neutrophil count less than 0.2 × 109/L since birth and a predisposition to recurrent and life-threatening infections. CN is a preleukemic syndrome with a cumulative incidence of 21% for the development of acute leukemia or myelodysplastic syndrome after 10 years on granulocyte colony-stimulating factor (G-CSF).1 Acquired mutations in the gene for the G-CSF receptor CSF3R have been implicated in the progression of severe congenital neutropenia (CN) to leukemia. In a previous report, we presented results from a long-term study designed to determine the incidence and the course of the occurrence of mutations in the CSF3R gene and their significance for malignant transformation in patients with CN.2 We showed that CSF3R mutations are a highly predictive marker for leukemic development in CN patients. Moreover, our results from long-term serial analyses of CSF3R mRNA from neutrophil granulocytes of CN patients reveal that the acquisition of CSF3R mutations is an early event in leukemogenesis that occurs prior to malignant transformation rather than a secondary event affecting an already transformed cell clone.
To clarify the involvement of CSF3R mutations in the process of malignant transformation, we asked at which stage of hematopoietic development these mutations emerge and how they are propagated during hematopoietic differentiation. Hence, we analyzed flow cytometrically sorted single cells of different hematopoietic subpopulations from CN patients with CSF3R mutations. The results of the present study show that CSF3R mutations are not restricted to the myeloid compartment but are also detectable in lymphoid cells. However, the percentage of mutated cells was much higher in neutrophils and granulocytic precursors than in lymphoid cells. Therefore, we conclude that CSF3R mutations are acquired in multipotent hematopoietic progenitor cells in CN patients and that they are clonally expanded in G-CSF receptor–bearing cells due to a growth advantage over progenitors expressing the wild-type receptor.
Methods
Patients
We analyzed bone marrow (patient 1 [Pt1] to Pt4) and peripheral blood (Pt4 only) from 4 patients diagnosed with CN. All patients participated in earlier studies where we serially analyzed CSF3R mutations in G-CSF receptor–expressing cells (Pt1 = Pt25, Pt2 = Pt26, Pt3 = Pt14, Pt4 = Pt15).2,3 The study was approved by the IRB at Hannover Medical School. Informed consent was obtained in accordance with the Declaration of Helsinki. Pt1 and Pt2 developed secondary acute myeloid leukemia (AML). Pt1 was analyzed 1 year before development of overt leukemia (AML M5b with monosomy 7) at age 15 10/12; Pt2 was analyzed at time of leukemia (AML M2 with trisomy 8) at age 21 2/12
Pt3 and Pt4 were selected for this study because of a high proportion of different CSF3R mutations in the neutrophils.2 Both underwent hematopoietic stem cell transplantation before any signs of malignant transformation due to a severe course of the disease were stated. Pt3 and Pt4 were analyzed at age 11 5/12 and 11 4/12, respectively.
Methods
We analyzed single cells of flow cytometrically sorted subpopulations of hematopoietic progenitors and mature hematopoietic cells from bone marrow and peripheral blood of 4 CN patients to prove the existence of mutations in the mutation-sensitive region 2342 to 2541 of the CSF3R gene.2 The following subsets were sorted from bone marrow samples of the patients: leukemic blasts (Pt2 only), CD34+ hematopoietic progenitors, myeloid progenitors (Pt3 and Pt4 only), granulocyte progenitors, monocytes, neutrophils (Pt2-Pt4 only), T cells, and B cells. Approximately 100 single cells of each population were analyzed by polymerase chain reaction (PCR) and direct sequencing. Single-cell sorts and subsequent analyses for each of the 4 patients as well as PCR analyses of each single population have been performed independently at different times to avoid the risk of cross contaminations.
RNA analyses from total mononuclear cells (MNCs) at time of AML (Pt1) and from mature neutrophils (Pt2-Pt4) were performed by sequencing at least 20 individual Escherichia coli clones after subcloning reverse-transcription (RT)–PCR products as described earlier.2 The designation of mutations follows accession no. M59818.4 For experimental details see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results and discussion
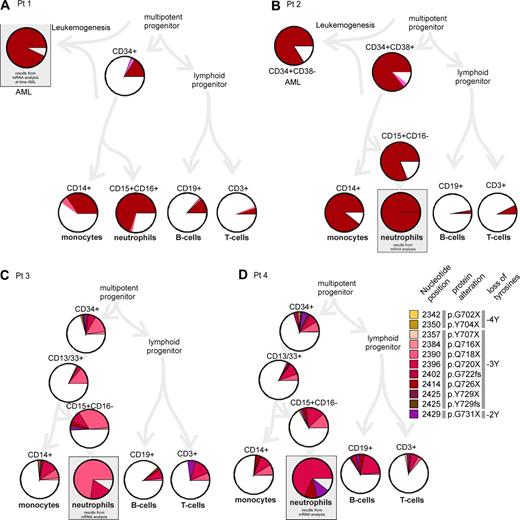
Analysis of single cells from different subpopulations of immature and mature blood cells revealed the fact that CSF3R mutations were present in all analyzed populations including T and B lymphocytes. The frequency of mutated cells, however, varied tremendously among the different patients and the analyzed populations (Figure 1; Table S1). For Pt1 (Figure 1A), we analyzed cells from bone marrow approximately one year before onset of acute monocytic leukemia (AML FAB M5b). The total percentage of mutated cells was low in lymphoid cells (T cells: 6%, B cells: 14% mutated cells), whereas monocytes (40%) and neutrophils (72%) showed a remarkably higher mutation rate. For Pt2 (Figure 1B), we were able to analyze cells at time of acute leukemia (AML FAB M2).3 In comparison with Pt1, we found a notably higher mutation rate in the myeloid compartment (monocytes: 89%, granulocytic precursors: 81%). Mutation rates in lymphocytes were comparatively low as in Pt1 (T cells: 7%, B cells 3%). In Pt3 and Pt4, who were selected for a high number of different mutations in CSF3R mRNA analyses, we also found high mutation rates in the neutrophil granulocytes (∼90% as calculated from mRNA analyses). Mutation rates in monocytes were remarkably lower (26% and 23%, respectively) and in the range of those in lymphocytes (T cells: 27% and 14%, B cells: 12% and 33%, respectively).
Incidence of CSF3R mutations in single cells of the hematopoietic compartment. Single flow cytometrically sorted cells were analyzed for the occurrence of CSF3R mutations. For each population, at least 100 cells were analyzed by PCR and direct sequencing. The diagram summarizes the results. Different mutations are presented in different colors (as in Germeshausen et al2 ). Similar colors symbolize similar effects on the protein as indicated in the color legend on the right side: yellow/orange indicate mutations leading to the loss of 4 tyrosine residues (−4Y); red to brown, −3Y; and violet, −2Y. Nucleotide positions of the mutations refer to Fukunaga et al.4 The percentage of mutated cells is represented by the size of the colored slices. The white portion of the circle represents cells with 2 wild-type alleles. Data for cell types in gray boxes are not derived from single-cell analyses but from mRNA analyses of the respective populations as described previously (data from Germeshausen et al2 ). Percentages of mutated cells were deduced from the percentage of mutated CSF3R mRNA assuming a balanced expression of mutated and wild-type allele.
Incidence of CSF3R mutations in single cells of the hematopoietic compartment. Single flow cytometrically sorted cells were analyzed for the occurrence of CSF3R mutations. For each population, at least 100 cells were analyzed by PCR and direct sequencing. The diagram summarizes the results. Different mutations are presented in different colors (as in Germeshausen et al2 ). Similar colors symbolize similar effects on the protein as indicated in the color legend on the right side: yellow/orange indicate mutations leading to the loss of 4 tyrosine residues (−4Y); red to brown, −3Y; and violet, −2Y. Nucleotide positions of the mutations refer to Fukunaga et al.4 The percentage of mutated cells is represented by the size of the colored slices. The white portion of the circle represents cells with 2 wild-type alleles. Data for cell types in gray boxes are not derived from single-cell analyses but from mRNA analyses of the respective populations as described previously (data from Germeshausen et al2 ). Percentages of mutated cells were deduced from the percentage of mutated CSF3R mRNA assuming a balanced expression of mutated and wild-type allele.
In contrast to previous reports,5,6 we could show that CSF3R mutations are not only restricted to the myeloid compartment of CN patients, but also occurred in lymphoid cells that do not express the G-CSF receptor. Previous attempts to detect CSF3R mutations in lymphocytes failed, most likely due to a less sensitive method for mutation detection.5,6 We found a similar spectrum of different mutations in different populations from a single patient. This makes an independent acquisition of mutations in the lymphoid and myeloid compartment unlikely. We postulate that CSF3R mutations were acquired in early multipotent hematopoietic progenitors as an early event in leukemogenesis and have been clonally expanded by a growth advantage on the level of multipotent progenitor cells. This hypothesis is strongly supported by a recent study by Liu et al: Using mice expressing a mutant Csf3r gene, the authors could demonstrate that the truncated G-CSF receptor confers a strong competitive advantage to hematopoietic stem cells in a strongly G-CSF–dependent fashion via sustained activation of Stat5.7
The acquisition of CSF3R mutations in multipotent hematopoietic progenitors can also explain the development of lymphoblastic leukemia secondary to acquisition of CSF3R mutations recently described for 2 patients with CN.8,9 The proliferative advantage provided by the truncated receptor, however, requires an aberrant expression of the G-CSF receptor on lymphoblastic cells that could indeed be shown for both patients.8,9
The different mutation frequencies between myeloid and lymphoid cells might be due to an expansion of CSF3R mutation expressing cells during hematopoietic development. The results of the analysis of hematopoietic progenitors from the bone marrow of Pt1 to Pt4 confirmed this hypothesis: Mutation rates increased during granulocytic development (CD13/33+ < CD15+CD16− cells < mature granulocytes, Pt3 and Pt4). In contrast, mutation rates in mature lymphocytes were equal to or lower than those in CD34+ progenitors.
Although monocytes express the G-CSF receptor,10 there was no consistent result for a clonal expansion of CSF3R mutation bearing cells during monocyte development: Pt1 and Pt2 demonstrated a mutation rate of monocytes that was notably higher than in the lymphocytes. In contrast, mutation rates of monocytes were in the range of that of lymphocytes in Pt3 and Pt4. Corresponding to these results, Dong et al did not find CSF3R mutations in monocytes, suggesting a low mutation rate in this cell type.5 The higher mutation rate in the neutrophil compartment in comparison with the monocytes may reflect the much higher turnover rate for neutrophils associated with a high proliferative rate of neutrophilic precursors.11 On the other hand, the higher mutation rates in monocytes of Pt1 and Pt2 could be associated with a compensatory up-regulation of monocyte production in these patients.
In our recent report on serial analyses of mutations in granulocytes of patients, we sometimes observed a shift of predominant mutations or the recurrence of apparently lost mutations within a few months.2 The results of the present study suggest that early hematopoietic progenitors may serve as a pool for clonal evolution of different mutations during granulopoiesis. Interestingly enough, all mutations, which have been found during long-term serial analyses in these patients, have been detected at a single date in the CD34+ compartment (Pt3 and Pt4). The predominant expression of one mutation, which was usually found in analyses of CN patients' granulocytes, is most likely due to clonal expansion during granulocytic maturation. The existence of different mutations in the progenitor cell compartment supports the hypothesis of CSF3R mutations as an adaptive response of a defective stem cell pool in congenital neutropenia via activation of antiapoptotic programs.12 The altered signaling in CSF3R-mutated cells that confers a G-CSF–dependent competitive advantage of hematopoietic progenitors may provide the background for the acquisition of additional mutations or chromosomal aberrations that finally lead to acute leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to the patients and their families for participation in our study. We thank Cornelia Zeidler of the Severe Chronic Neutropenia European Registry (SCNIR) and all physicians who referred the patients to our study. We also acknowledge the excellent technical assistance of Gabriele Cleves, Marly Dalton, Sabine Jakobs, and Christina Reimer.
This work was supported through grants from the Federal Ministry of Education and Research (Berlin, Germany; German Network on Congenital Bone Marrow Failure Syndromes) and from the Deutsche Krebshilfe (Bonn, Germany; grant no. 10-1105).
Authorship
Contribution: M.G. and M.B. designed and performed research and wrote the paper; and K.W. initiated and supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Manuela Germeshausen, Pediatric Hematology and Oncology, Medizinische Hochschule Hannover, Carl-Neuberg-Str 1, 30625 Hannover, Germany; e-mail: germeshausen.manuela@mh-hannover.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal