Abstract
Chromosomal abnormalities are frequent in myeloid malignancies, but in most cases of myelodysplasia (MDS) and myeloproliferative neoplasms (MPN), underlying pathogenic molecular lesions are unknown. We identified recurrent areas of somatic copy number–neutral loss of heterozygosity (LOH) and deletions of chromosome 4q24 in a large cohort of patients with myeloid malignancies including MDS and related mixed MDS/MPN syndromes using single nucleotide polymorphism arrays. We then investigated genes in the commonly affected area for mutations. When we sequenced TET2, we found homozygous and hemizygous mutations. Heterozygous and compound heterozygous mutations were found in patients with similar clinical phenotypes without LOH4q24. Clinical analysis showed most TET2 mutations were present in patients with MDS/MPN (58%), including CMML (6/17) or sAML (32%) evolved from MDS/MPN and typical MDS (10%), suggesting they may play a ubiquitous role in malignant evolution. TET2 mutations affected conserved domains and the N terminus. TET2 is widely expressed in hematopoietic cells but its function is unknown, and it lacks homology to other known genes. The frequency of mutations in this candidate myeloid regulatory gene suggests an important role in the pathogenesis of poor prognosis MDS/MPN and sAML and may act as a disease gene marker for these often cytogenetically normal disorders.
Introduction
Identification of recurrent pathogenic mutations and genomic aberrations has greatly advanced our understanding of the molecular mechanisms of leukemia evolution and has led to the development of therapeutics and diagnostic tests. Of particular importance for explaining leukemia evolution is loss of heterozygosity (LOH) which arises either via hemizygous deletion, where a DNA segment is lost from one homolog while the other remains at one copy per cell, or uniparental disomy (UPD), wherein the retained homolog is duplicated so as to preserve 2 total copies per cell at the locus. Both types of somatic LOH have been observed in studies of various cancer types and may explain some of the mechanisms by which tumor suppressor genes are inactivated or activating mutations duplicated. Thus, analysis of recurrent LOH may point toward the presence of important recurrent mutations.
Metaphase cytogenetics (MC) has an established role in the diagnosis and prognosis of hematologic malignancies including myelodysplastic syndrome (MDS), acute myelogenous leukemia (AML), and associated myelodysplastic/myeloproliferative neoplasms (MDS/MPN). Using this technique, several recurrent chromosomal abnormalities have been described and minimal affected chromosomal regions delineated, pointing toward potentially pathogenic genes. For the purposes of gene discovery and genomic characterization, the diagnostic yield of routine MC can now be enhanced by single nucleotide polymorphisms array (SNP-A)–based karyotyping, allowing for a better resolution of chromosomal defects and thereby identification of previously cryptic unbalanced lesions including microduplications and deletions.1 In particular, one unique feature of SNP-A is its ability to identify UPD.2 We and others have recently shown that somatic UPD affecting various chromosomes can be frequently found in MDS, MDS/MPN, and sAML and have identified several recurrent areas of UPD. Initially, identification of UPD9p was instrumental in the discovery of the JAK2 mutation, and further work has explored the clinical relevance of homozygosity for the JAK2V617F mutation in MPN often associated with UPD.3-6 Subsequent application of SNP-A illustrated that somatic UPD affecting various chromosomes is frequent in myeloid malignancies and allowed for identification of a new recurrent lesions.7-9 We and others demonstrated that other invariant somatic UPD can be associated with homozygous mutations including, for example, UPD13p (FLT-3ITD) and UPD1p (MPLW515L).10,11 Based on this paradigm, we have recently delineated a novel recurrent area of UPD11q present in chronic myelomonocytic leukemia (CMML) and sAML and identified pathogenic homozygous c-Cbl mutations.11 We also observed another frequent area of somatic UPD at 4q24, most frequently encountered in patients with CMML, mixed MDS/MPN, and in some typical MDS and sAML cases. Further investigation revealed microdeletions of the corresponding region in other cases with a similar clinical phenotype.
Consequently, in this study, we set out to more completely define a minimally affected genomic region at 4q24, identify mutations of genes within this interval, and correlate clinical phenotypes with any lesions present as an approach to the identification of novel mutated genes that participate in the pathogenesis of poor prognosis myeloid disorders, in particular MDS, MDS/MPN, and sAML.
Methods
Patients
Bone marrow aspirates were collected for SNP-A analysis from patients with MDS (n = 235), MDS/MPN (n = 80), and sAML (n = 81). Of the patients with sAML, 71 had antecedent MDS, and 10 had antecedent MDS/MPN. Among patients with MDS/MPN, 46, 21, and 13 had CMML-1/2, MDS/MPN-U, and RARS-T, respectively. Clinical features of patients are summarized in Table 1. Sequencing was performed on 68 patients (59 from the cohort tested by SNP-A). Informed consent for sample collection was obtained according to protocols approved by the institutional review boards (IRBs) of the Cleveland Clinic and Johns Hopkins University and in accordance with the Declaration of Helsinki. Diagnosis was assigned according to World Health Organization (WHO) classification criteria.12,13
Baseline patient characteristics
| Characteristic . | SNP-A (TET2 seq), n . |
|---|---|
| Median age, y (range) | 68 (19-92) |
| Risk stratification by WHO*† | |
| Low risk | 227 (28) |
| High risk | 169 (31) |
| IPSS risk category† | |
| Low/Int-1 | 199 (27) |
| High/Int-2 | 82 (15) |
| Unclassified | 114 (17) |
| No growth by MC | 19 (5) |
| Clinical data not available | 23 (2) |
| Others‡ | 73 (10) |
| WHO classification | |
| MDS | 235 (14) |
| RA/RCMD/5q− syndrome/MDS-U | 117 (4) |
| RARS/RCMD-RS | 51 (6) |
| RAEB-1/2 | 67 (4) |
| MDS/MPN overlap | 80 (30) |
| CMML-1/2 | 46 (17) |
| MDS/MPN-U§ | 21 (5) |
| RARS-T | 13 (3) |
| JMML | 0 (5) |
| AML | 81 (20) |
| 2° to MDS‖ | 71 (7) |
| 2° to MDS/MPN | 10 (14) |
| 2° to MPN | 1 (1) |
| MPN | 2 (2) |
| Characteristic . | SNP-A (TET2 seq), n . |
|---|---|
| Median age, y (range) | 68 (19-92) |
| Risk stratification by WHO*† | |
| Low risk | 227 (28) |
| High risk | 169 (31) |
| IPSS risk category† | |
| Low/Int-1 | 199 (27) |
| High/Int-2 | 82 (15) |
| Unclassified | 114 (17) |
| No growth by MC | 19 (5) |
| Clinical data not available | 23 (2) |
| Others‡ | 73 (10) |
| WHO classification | |
| MDS | 235 (14) |
| RA/RCMD/5q− syndrome/MDS-U | 117 (4) |
| RARS/RCMD-RS | 51 (6) |
| RAEB-1/2 | 67 (4) |
| MDS/MPN overlap | 80 (30) |
| CMML-1/2 | 46 (17) |
| MDS/MPN-U§ | 21 (5) |
| RARS-T | 13 (3) |
| JMML | 0 (5) |
| AML | 81 (20) |
| 2° to MDS‖ | 71 (7) |
| 2° to MDS/MPN | 10 (14) |
| 2° to MPN | 1 (1) |
| MPN | 2 (2) |
An additional 9 patients screened for TET2 mutation but no SNP-A analysis.
AML indicates acute myelogenous leukemia; FAB, French-American-British; IPSS, International Prognostic Scoring System; JMML, juvenile myelomonocytic leukemia; MC, metaphase cytogenetics; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; MDS/MPN-U, MDS/MPN unclassifiable; MDS-U, MDS unclassified; RA, refractory anemia; RAEB, refractory anemia with excess blasts; RAEB-T, refractory anemia with excess blasts in transformation; RARS, refractory anemia with ringed sideroblasts; RARS-T, refractory anemia with ringed sideroblasts associated with marked thrombocytosis; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, refractory cytopenia with multilineage dysplasia and ringed sideroblasts; and WHO, World Health Organization.
Low risk includes RA, RARS, RCMD, RCMD-RS, CMML-1, MDS/MPN-U < 5% blast, RARS-T, MDS-U, and 5q- syndrome. High risk includes RAEB-1/2, CMML-2, MDS/MPN-U ≥ 5% blasts, AML.
Patients with MPN and JMML are excluded.
AML with ≥ 30% BM blast or previously treated prior to sample collection.
One patient is classified as aCML.
Twenty-nine patients are classified as RAEB-T by FAB classification.
SNP-A analysis
DNA was extracted from patient marrow specimens and, when sample material allowed, CD3+ cell fractions were isolated using RoboSep (StemCell Technologies, Vancouver, BC). The Affymetrix GeneChip Human Mapping 250K Array and Genome-Wide Human SNP Array 6.0 (n = 398; Affymetrix, Santa Clara, CA) were used for SNP-A analysis as previously described.14 Signal intensity was analyzed, and SNP calls determined using GeneChip Genotyping Analysis software, version 4.0 (GTYPE). Copy number variations (CNVs) and areas of UPD were investigated using a Hidden Markov Model and CN Analyzer for Affymetrix GeneChip Mapping 250K arrays (CNAG version 3.0).8,15 Genotyping console version 2.0 (Affymetrix) was used for analysis of 6.0 arrays. As shown previously through mixing studies, an admix of 30% of clonal cells can be detected by 250K arrays.8 We excluded germline–encoded CNVs and nonclonal areas of UPD from further analysis by using a bioanalytic algorithm based on lesions identified by SNP-A in an internal control series (n = 713) and reported in the Database of Genomic Variants (http://projects.tcag.ca/variation/). In brief, cases with concordant results between MC and SNP-A did not require further confirmation. Through analysis of a large number of controls, we have determined the size of homozygous stretches of DNA and, based on the size and location criterion, excluded upfront all cases with UPD less than 25.8 Mb (95% confidence interval [CI] of nonclonal UPD seen in controls) and interstitial stretches of LOH, as these type of LOH were never found to be clonal in all instances tested. We excluded microduplications and microdeletions when they showed an overlap greater than 50% with known CNV. The remaining microdeletions and gains and some important areas of UPD including those on 4q24 were confirmed by array analysis of germline DNA derived from sorted CD3+ cells (Figure 1B) or microsatellite analysis whenever possible. This has been done in 86 patients.
TET2 and JAKV617F mutational screening
Screening for mutations in TET2 was carried out using direct genomic sequencing. PCR primers were designed to amplify and sequence all coding exons 3-11 including TET2 isoform A (NM_001127208-2002 amino acids) and TET2 isoform B (NM_017628-1165 amino acids) and are available on request. When needed, multiple set of primers overlapping by 100 bp were used to ensure complete coverage (exon 3-10 sets, exon 11-3 sets). For each polymerase chain reaction (PCR), 40 ng genomic DNA was used for PCR amplification followed by purification using Montage Cleanup kit (Millipore, Billerica, MA). Sequencing was performed using ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). All TET2 mutations were detected by bidirectional sequencing and scored as pathogenic mutations on the basis of the observation that they were not detected in normal samples or nonclonal CD3+ cells. For germline confirmation, only exons containing mutations were tested. All mutations were first compared with published SNP data (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP) and annotated according to the coding sequence using TET2 isoform A as the predominant isoform. Mutations within introns were not scored. Screening for JAK2V617F mutation was performed as previously described.16
TET2 mRNA expression
Fractions of CD34+, CD33+, CD14+, CD61+, and glycophorin A+ cells were isolated from healthy bone marrow mononuclear cells by magnetic bead separation (Miltenyi Biotec, Auburn, CA). Total RNA was extracted using RNeasy Mini kit (QIAGEN, Valencia, CA), and cDNA was generated using SuperScript III RT kit (Invitrogen, Carlsbad, CA). TET2 mRNA expression was detected with TaqMan TET2 Gene Expression Mix (hs00325999_m1-isoform A) according to the manufacturer's instructions (Applied Biosystems). Values obtained for the target gene expression in patients' CD34+ cells were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and expressed in relation to the expression in control samples.
Methylation assay
The Infinium Human Methylation 27k BeadChip (Illumina, San Diego, CA) was used for methylation detection per manufacturer's protocol as previously described.17 Methylation status of the interrogated CpG sites (106286401 and 106286501) was calculated as a ratio of the fluorescence signals corresponding to methylated and unmethylated status (β-value). The β-value provides a continuous measure of DNA methylation at a CpG site, ranging from 0 in the case of completely unmethylated sites to 1 in completely methylated sites. Image extraction and statistical analysis were performed using BeadStudio Methylation Module (Illumina). Results were normalized to the background fluorescence for each array. Statistical significance of the difference of methylation level between different groups was assessed using Student t test.
Statistical analysis
When appropriate, Kaplan-Meier statistics were applied to assess survival. For comparison of the frequency of mutation or other clinical features between disease groups, categorical variables were analyzed using the Fisher exact test, and continuous variables were tested using the Mann-Whitney U test.
Results
Recurrent areas of LOH4q24 in patients with myeloid malignancies
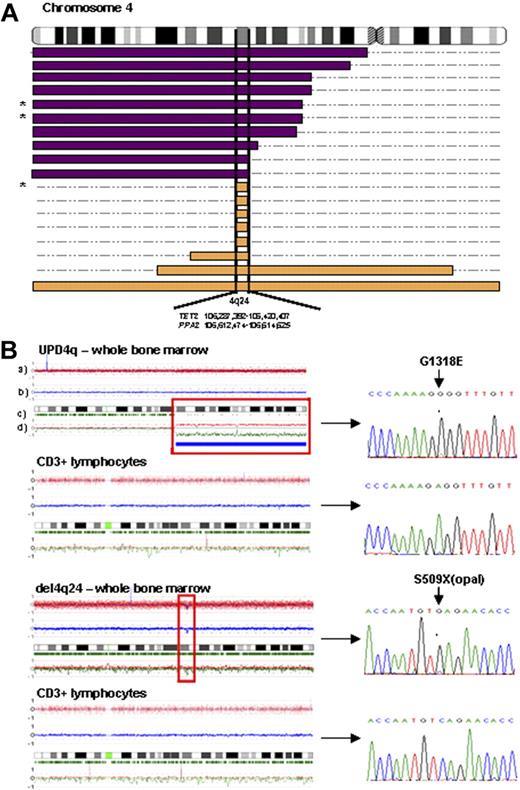
First, we examined patients with closely related myeloid malignancies and identified LOH represented by either somatic UPD4q or 4q deletions (Table 1 and Figure 1). In total we have identified 10 cases with UPD4q and 8 with deletions involving 4q24. UPD4q was most commonly encountered in MDS/MPN patients and in those with antecedent MDS/MPN progressing to AML (7/90), including CMML (3/46). We also found microdeletions of 4q24 in patients with typical MDS (5/235), including RCMD and RCMD-RS (Figure 1A). LOH4q was not found in RARS-T (n = 13) or atypical CML (n = 1). Although UPD4q was mostly associated with myeloproliferative features, deletions coincided with various forms of MDS and sAML. We also noted that LOH4q24 was mostly present as a sole abnormality or was accompanied by a solitary clonal defect. The chromosomal lesions found in patients were mapped and, because of the presence of 4q24 microdeletions, a commonly affected area containing TET2 was identified (Figure 1A).
LOH 4q24 mapping and TET2 sequencing. (A) Topographical map illustrating overlapping regions of UPD4q24 (purple bars) and del4q24 (orange bars) in patients with myeloid malignancies. A minimally affected region on 4q24 (0.35 Mb), containing TET2 and PPA2, was identified in 18 of 398 patients studied (* indicates that some patients' DNA was not available for mutational screening; n = 3). (B) Representative examples of SNP-A karyograms (CNAG version 3.0) of chromosome 4 of both whole bone marrow cells and paired CD3+ lymphocytes in 2 patients, demonstrating the somatic nature of acquired LOH4q24 and TET2 mutations, which are shown in corresponding ferrograms in the right portion of the figure. SNP-A karyograms: rows a and b correspond to the copy number as measured by the intensity of individual hybridization signals; row c represents frequency of heterozygous calls; and row d indicates the allelic imbalance with parental allele copy numbers.
LOH 4q24 mapping and TET2 sequencing. (A) Topographical map illustrating overlapping regions of UPD4q24 (purple bars) and del4q24 (orange bars) in patients with myeloid malignancies. A minimally affected region on 4q24 (0.35 Mb), containing TET2 and PPA2, was identified in 18 of 398 patients studied (* indicates that some patients' DNA was not available for mutational screening; n = 3). (B) Representative examples of SNP-A karyograms (CNAG version 3.0) of chromosome 4 of both whole bone marrow cells and paired CD3+ lymphocytes in 2 patients, demonstrating the somatic nature of acquired LOH4q24 and TET2 mutations, which are shown in corresponding ferrograms in the right portion of the figure. SNP-A karyograms: rows a and b correspond to the copy number as measured by the intensity of individual hybridization signals; row c represents frequency of heterozygous calls; and row d indicates the allelic imbalance with parental allele copy numbers.
TET2 gene mutations
Based on the paradigm that areas of somatic UPD contain homozygous mutations,18-20 we stipulated that TET2 carried mutations and proceeded with sequencing. For the initial sequencing analysis, we used the index cases selected based on the presence of LOH4q. SNP-A identified 10 patients with UPD4q, but DNA was available for sequencing in 8 of these patients. Homozygous TET2 mutations were identified in all 8 cases. Hemizygous mutations were found in 2 of 7 patients with 4q24 microdeletions for whom sequencing was performed (Figure 2A and Table 2). It is possible that the remaining patients with del4q24 (n = 5) had promoter, regulatory region, or intronic mutations, or gene haploinsufficiency was present, playing a role in the pathophysiology of the corresponding malignancy.21 The somatic nature of the lesions was confirmed by SNP-A and sequencing of lymphocyte DNA showing absence of LOH4q and a wild-type (WT) constellation of germline TET2 (Figure 1B). When we serially studied a patient with sAML evolved from MDS/MPN, a mutation seen at initial testing disappeared when remission was achieved after bone marrow transplantation and through induction chemotherapy.
Identification of mutations in TET2 gene (4q24). (A) Genomic sequencing of all coding TET2 exons revealed the presence of nonsense, missense, and frameshift mutations. These mutations were homozygous and associated with UPD4q (top left), hemizygous associated with deletion (top right), or heterozygous associated with no chromosomal lesion (bottom right). Unique compound heterozygosity was found in 1 patient (bottom left). Detailed description of the SNP-A karyograms is provided in Figure 1B. (B) Schematic representation of the topographic distribution of the individual mutations in the TET2 protein (Isoform A NM_001127208). Amino acid sequences of TET family members are aligned (TET2, TET1, and TET3). *Highly conserved residues among all 3 proteins (LCXH domains). Most mutations were found in 2 conserved domains, LCXH 1 and 2 (50%, missense changes, frameshifts and codon stop changes are in red and underlined) while nonsense and frameshift mutations were more prominent in the N terminus (62%). Q1084P and Y867H are novel, unannotated SNPs (see “Note added in proof”).
Identification of mutations in TET2 gene (4q24). (A) Genomic sequencing of all coding TET2 exons revealed the presence of nonsense, missense, and frameshift mutations. These mutations were homozygous and associated with UPD4q (top left), hemizygous associated with deletion (top right), or heterozygous associated with no chromosomal lesion (bottom right). Unique compound heterozygosity was found in 1 patient (bottom left). Detailed description of the SNP-A karyograms is provided in Figure 1B. (B) Schematic representation of the topographic distribution of the individual mutations in the TET2 protein (Isoform A NM_001127208). Amino acid sequences of TET family members are aligned (TET2, TET1, and TET3). *Highly conserved residues among all 3 proteins (LCXH domains). Most mutations were found in 2 conserved domains, LCXH 1 and 2 (50%, missense changes, frameshifts and codon stop changes are in red and underlined) while nonsense and frameshift mutations were more prominent in the N terminus (62%). Q1084P and Y867H are novel, unannotated SNPs (see “Note added in proof”).
Characteristics of TET2-mutated patients
| Patient no. . | Age, y . | Initial Dx . | Current Dx . | Cytogenetics . | SNP-A karyotyping . | TET2 mutation . | JAK2V617F . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gain . | Loss . | UPD . | Exon . | Consequence . | Type . | State . | ||||||
| 1 | 59 | RCMD-RS | RCMD-RS | 45,X,-Y[17]/46,XY[3] | N | N | 4q24-qter | 11 | G1913D | missense | homozygous | negative |
| 2 | 76 | CMML-1 | CMML-1 | 46,XY[20] | N | N | 4q21.21-qter | 7 | insT1310 | frameshift | homozygous | negative |
| 3 | 60 | CMML-1 | CMML-1 | 45,X,-Y[20] | N | N | 4q23-qter, 7q11.23-qter | 7 | E1318G | missense | homozygous | negative |
| 4 | 77 | CMML-1 + CLL | CMML-1 | 46,XY[20] | 16p11.2,21p11.2-q11.2 | N | 4q12-qter | 3 | delG2 | frameshift | homozygous | negative |
| 5 | 69 | CMML-2 | sAML | 46,XY[20] | 22q11.1 | 7q22.1 | 4q21.21-qter, 11q13.4-qter | 3 | S835X | nonsense | homozygous | negative |
| 6 | 78 | CMML-1 | sAML | 46,XY[20] | N | N | 4q13.1-qter, 13q12.11-q12.13 | 4 | R1167T | missense | homozygous | negative |
| 7 | 60 | MDS/MPN | sAML | 47,XY,+8[4]/46,XY[16] | 8 | N | 4q24qter | 10 | delG1439 | frameshift | homozygous | negative |
| 8 | 70 | CMML-2 | sAML | 46,XY[20] | N | 4q21.23-qter | 3 | L1040X | nonsense | homozygous | negative | |
| 9 | 74 | CMML-1 | CMML-1 | 45,X,-Y[20] | N | 4q24 | N | 9 | P1367S | missense | hemizygous | negative |
| 10 | 80 | RCMD-RS | RCMD-RS | 46,XY[20] | N | 4q24; 7p21.3 | N | 3 | S509X | nonsense | hemizygous | negative |
| 11 | 76 | MDS/MPN-U ≥ 5% | MDS/MPN-U ≥ 5% | 46,XY,del(7) (q11.2), del(20)(q11q13), −21,+r[cp20] | N | 7p12.3-p14.1, 7q11.22-qter, 11p15.4-p15.5, 20q11.23-q12, 20q13.12-q13.2 | 22q11.21-qter | 7 | F1287L | missense | heterozygous | negative |
| 12 | 71 | CMML-2 | CMML-2 | 46,XY[20] | N | N | 7q11.21-qter | 7 | K1299N | missense | heterozygous | negative |
| 13 | 58 | CMML-2 | CMML-2 | 46,XX,del(17)(q24)[6]/46,XX[24] | 21q22.13-q22.2 | 2q24.3-q32.1 | 1p36.13-pter | 3 | Q1084P* | missense | heterozygous | negative |
| 14 | 68 | CMML-1 | sAML | 47,XY,+19[20] | 19, 21q22.2 | 3q28-q29 | 1p; 2p22.1-p22.3, 4q28.1-qter, 6p22.1-24.1, 13q12.12-q12.3, 16p12.3-p13.13, 17q22-q23.2 21q21.1 | 3 | A308T | missense | heterozygous | negative |
| 15 | 50 | CMML-1 | sAML | 46,XY[20] | N | N | N | 7 11 | R1302G S1638X | missense nonsense | heterozygous | negative |
| 16 | 45 | CMML-1 | sAML | N/A | 7q21.13 20q11.21-pter, 21p11.2-q21.3, 21q22.13-q22.3 | 3p14.3-pter, 5p12-q12.1, 5q12.3-q14.1, 5q14.3-qter, 12p13.1-p13.31, 12p11.1-p12.1, 16p11.1-q24.3 | 17p12-pter | 3 | Y867H* | missense | heterozygous | negative |
| 17 | 73 | CMML-1 | CMML-1 | 46,XX[20] | N | N | N | 11 | I1873T | missense | heterozygous | negative |
| 18 | 74 | RARS-T | RARS-T | 47,XY,+8[20] | 8 | N | N | 11 | V1718L | missense | heterozygous | negative |
| 19 | 82 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 46,XY[20] | N | 3p14.2 | 11q13.5-qter | 3 | S631X | nonsense | heterozygous | negative |
| 20 | 78 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 46,XY[20] | 2p16.1 | 6q16.1 | N | 3 | S817T | missense | heterozygous | negative |
| 21 | 80 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 47,XY,+8[7]/46,XY[13] | 8 | N | 8q24.21-qter | 3 | P399L | missense | heterozygous | positive |
| Patient no. . | Age, y . | Initial Dx . | Current Dx . | Cytogenetics . | SNP-A karyotyping . | TET2 mutation . | JAK2V617F . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gain . | Loss . | UPD . | Exon . | Consequence . | Type . | State . | ||||||
| 1 | 59 | RCMD-RS | RCMD-RS | 45,X,-Y[17]/46,XY[3] | N | N | 4q24-qter | 11 | G1913D | missense | homozygous | negative |
| 2 | 76 | CMML-1 | CMML-1 | 46,XY[20] | N | N | 4q21.21-qter | 7 | insT1310 | frameshift | homozygous | negative |
| 3 | 60 | CMML-1 | CMML-1 | 45,X,-Y[20] | N | N | 4q23-qter, 7q11.23-qter | 7 | E1318G | missense | homozygous | negative |
| 4 | 77 | CMML-1 + CLL | CMML-1 | 46,XY[20] | 16p11.2,21p11.2-q11.2 | N | 4q12-qter | 3 | delG2 | frameshift | homozygous | negative |
| 5 | 69 | CMML-2 | sAML | 46,XY[20] | 22q11.1 | 7q22.1 | 4q21.21-qter, 11q13.4-qter | 3 | S835X | nonsense | homozygous | negative |
| 6 | 78 | CMML-1 | sAML | 46,XY[20] | N | N | 4q13.1-qter, 13q12.11-q12.13 | 4 | R1167T | missense | homozygous | negative |
| 7 | 60 | MDS/MPN | sAML | 47,XY,+8[4]/46,XY[16] | 8 | N | 4q24qter | 10 | delG1439 | frameshift | homozygous | negative |
| 8 | 70 | CMML-2 | sAML | 46,XY[20] | N | 4q21.23-qter | 3 | L1040X | nonsense | homozygous | negative | |
| 9 | 74 | CMML-1 | CMML-1 | 45,X,-Y[20] | N | 4q24 | N | 9 | P1367S | missense | hemizygous | negative |
| 10 | 80 | RCMD-RS | RCMD-RS | 46,XY[20] | N | 4q24; 7p21.3 | N | 3 | S509X | nonsense | hemizygous | negative |
| 11 | 76 | MDS/MPN-U ≥ 5% | MDS/MPN-U ≥ 5% | 46,XY,del(7) (q11.2), del(20)(q11q13), −21,+r[cp20] | N | 7p12.3-p14.1, 7q11.22-qter, 11p15.4-p15.5, 20q11.23-q12, 20q13.12-q13.2 | 22q11.21-qter | 7 | F1287L | missense | heterozygous | negative |
| 12 | 71 | CMML-2 | CMML-2 | 46,XY[20] | N | N | 7q11.21-qter | 7 | K1299N | missense | heterozygous | negative |
| 13 | 58 | CMML-2 | CMML-2 | 46,XX,del(17)(q24)[6]/46,XX[24] | 21q22.13-q22.2 | 2q24.3-q32.1 | 1p36.13-pter | 3 | Q1084P* | missense | heterozygous | negative |
| 14 | 68 | CMML-1 | sAML | 47,XY,+19[20] | 19, 21q22.2 | 3q28-q29 | 1p; 2p22.1-p22.3, 4q28.1-qter, 6p22.1-24.1, 13q12.12-q12.3, 16p12.3-p13.13, 17q22-q23.2 21q21.1 | 3 | A308T | missense | heterozygous | negative |
| 15 | 50 | CMML-1 | sAML | 46,XY[20] | N | N | N | 7 11 | R1302G S1638X | missense nonsense | heterozygous | negative |
| 16 | 45 | CMML-1 | sAML | N/A | 7q21.13 20q11.21-pter, 21p11.2-q21.3, 21q22.13-q22.3 | 3p14.3-pter, 5p12-q12.1, 5q12.3-q14.1, 5q14.3-qter, 12p13.1-p13.31, 12p11.1-p12.1, 16p11.1-q24.3 | 17p12-pter | 3 | Y867H* | missense | heterozygous | negative |
| 17 | 73 | CMML-1 | CMML-1 | 46,XX[20] | N | N | N | 11 | I1873T | missense | heterozygous | negative |
| 18 | 74 | RARS-T | RARS-T | 47,XY,+8[20] | 8 | N | N | 11 | V1718L | missense | heterozygous | negative |
| 19 | 82 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 46,XY[20] | N | 3p14.2 | 11q13.5-qter | 3 | S631X | nonsense | heterozygous | negative |
| 20 | 78 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 46,XY[20] | 2p16.1 | 6q16.1 | N | 3 | S817T | missense | heterozygous | negative |
| 21 | 80 | MDS/MPN-U < 5% | MDS/MPN-U < 5% | 47,XY,+8[7]/46,XY[13] | 8 | N | 8q24.21-qter | 3 | P399L | missense | heterozygous | positive |
Novel, unannotated SNPs (see “Note added in proof”).
In some genes located in areas of somatic UPD, both homozygous and heterozygous mutations can be found (eg, JAK2 or FLT-3), whereas in other instances, mutations occur mostly in the context of UPD and are homozygous (eg, c-Cbl), suggesting that the remaining WT allele is protective.3,11,18 In addition, most deletions involving 11q do not harbor mutations in c-Cbl.11 We therefore proceeded to analyze a series of patients in which chromosome 4q was identified as unaffected by SNP-A analysis (n = 53). We focused our study on pathomorphologic subgroups containing a majority of the UPD4q or 4q deletions and homozygous TET2 mutations. This approach was highly effective, and 10 heterozygous mutations were found. One unique patient was a compound heterozygote, displaying 2 different TET2 mutations. Consequently, we detected TET2 mutations in a total of 20 of 68 patients sequenced. TET2 mutations were spread over all exons; a portion resulted in either stop codon (5/20) or frameshift (3/20) changes; 8 of 20 TET2 mutations were found in 2 conserved domains (LCHX1 and 2), and 4 of 5 stop codons and 1 of 3 frameshifts were present in the N terminus (Figure 2B). The clinical associations of patients with mutant TET2 are described in Tables 2 and 3.
Comparison of clinical variables at presentation between WT and mutant TET2*
| Variable† . | TET2 mutant . | TET2 WT . | P . |
|---|---|---|---|
| Age (n = 60) | |||
| ≥ 60 y | 17 | 31 | .21 |
| < 60 y | 2 | 10 | |
| Sex‡ (n = 61) | |||
| Male | 18 | 24 | .003 |
| Female | 1 | 18 | |
| Hemoglobin§ (n = 61) | |||
| ≥ 10 g/dL | 7 | 22 | .38 |
| < 10 g/dL | 11 | 21 | |
| WBC count (n = 61) | |||
| ≥ 10 × 109/L | 13 | 12 | .01 |
| < 10 × 109/L | 8 | 28 | |
| Platelet count (n = 62) | |||
| ≥ 450 × 109/L | 1 | 3 | .65 |
| 150-450 × 109/L | 5 | 7 | |
| < 150 × 109/L | 13 | 33 | |
| Absolute monocyte count (n = 62) | |||
| ≥ 1 × 109/L | 12 | 15 | .03 |
| < 1 × 109/L | 7 | 28 | |
| Number of cytopenias (n = 61) | |||
| 0 | 4 | 6 | .74 |
| 1 | 5 | 16 | |
| 2 | 7 | 14 | |
| 3 | 2 | 7 | |
| Palpable splenomegaly (n = 62) | |||
| Yes | 9 | 19 | .82 |
| No | 10 | 24 | |
| Metaphase cytogenetics§ (n = 60) | |||
| Normal | 11 | 12 | .07 |
| Abnormal | 8 | 24 | |
| Bone marrow blasts§ (n = 61) | |||
| ≥ 5% | 6 | 21 | .17 |
| < 5% | 13 | 21 | |
| Bone marrow cellularity (n = 56) | |||
| Hypercellular | 16 | 28 | .44 |
| Normocellular | 3 | 6 | |
| Hypocellular | 0 | 3 | |
| No. dysplastic BM cell lines (n = 59) | |||
| 0 | 5 | 18 | .09 |
| 1 | 7 | 4 | |
| 2 | 4 | 10 | |
| 3 | 3 | 8 | |
| Bone marrow megakaryocytes (n = 57) | |||
| Dysplastic | 10 | 22 | .94 |
| Normal | 8 | 17 | |
| Progression to AML (n = 62) | |||
| Yes | 14 | 21 | .06 |
| No | 5 | 22 |
| Variable† . | TET2 mutant . | TET2 WT . | P . |
|---|---|---|---|
| Age (n = 60) | |||
| ≥ 60 y | 17 | 31 | .21 |
| < 60 y | 2 | 10 | |
| Sex‡ (n = 61) | |||
| Male | 18 | 24 | .003 |
| Female | 1 | 18 | |
| Hemoglobin§ (n = 61) | |||
| ≥ 10 g/dL | 7 | 22 | .38 |
| < 10 g/dL | 11 | 21 | |
| WBC count (n = 61) | |||
| ≥ 10 × 109/L | 13 | 12 | .01 |
| < 10 × 109/L | 8 | 28 | |
| Platelet count (n = 62) | |||
| ≥ 450 × 109/L | 1 | 3 | .65 |
| 150-450 × 109/L | 5 | 7 | |
| < 150 × 109/L | 13 | 33 | |
| Absolute monocyte count (n = 62) | |||
| ≥ 1 × 109/L | 12 | 15 | .03 |
| < 1 × 109/L | 7 | 28 | |
| Number of cytopenias (n = 61) | |||
| 0 | 4 | 6 | .74 |
| 1 | 5 | 16 | |
| 2 | 7 | 14 | |
| 3 | 2 | 7 | |
| Palpable splenomegaly (n = 62) | |||
| Yes | 9 | 19 | .82 |
| No | 10 | 24 | |
| Metaphase cytogenetics§ (n = 60) | |||
| Normal | 11 | 12 | .07 |
| Abnormal | 8 | 24 | |
| Bone marrow blasts§ (n = 61) | |||
| ≥ 5% | 6 | 21 | .17 |
| < 5% | 13 | 21 | |
| Bone marrow cellularity (n = 56) | |||
| Hypercellular | 16 | 28 | .44 |
| Normocellular | 3 | 6 | |
| Hypocellular | 0 | 3 | |
| No. dysplastic BM cell lines (n = 59) | |||
| 0 | 5 | 18 | .09 |
| 1 | 7 | 4 | |
| 2 | 4 | 10 | |
| 3 | 3 | 8 | |
| Bone marrow megakaryocytes (n = 57) | |||
| Dysplastic | 10 | 22 | .94 |
| Normal | 8 | 17 | |
| Progression to AML (n = 62) | |||
| Yes | 14 | 21 | .06 |
| No | 5 | 22 |
Q1084P and Y86H SNPs are excluded (see “Note added in proof”).
Some clinical data are not available.
Median WBC count for TET2 mutant is 18 × 109/L and for TET2 wild-type is 6 × 109/L.
Median platelet count for TET2 mutant is 72 × 109/L and for TET2 wild-type is 88 × 109/L.
TET2 gene methylation/expression status
When we analyzed the levels of TET2 mRNA using TaqMan PCR, the highest expression was found in myeloid cells selected based on myelomonocytic (CD33) or monocytic surface antigens (CD14); however, expression was also present in immature CD34+ cells derived from healthy bone marrow (n = 5). In addition to inactivating mutations, decreased TET2 expression may be present in patients with corresponding pathomorphologic phenotypes. There were no differences in TET2 mRNA expression in CD34+ cells derived from patients with MDS/MPN (n = 16), including those with WT and mutant TET2 (Figure 3). Similarly, when we studied whether TET2 expression can be down-modulated through promoter methylation, the average β-value (measure of methylation) at 2 CpG sites (106286401 and 106286501) did not differ significantly between the patients (n = 18) and controls (n = 32; P = .28 and P = .13), or between cases with TET2 mutant (n = 8) or TET2 WT (n = 10) and controls (P = .11, P = .23 and P = .93, P = .26, respectively).
TET2 mRNA expression. (A) TET2 mRNA expression in fractions of healthy bone marrows (n = 5). Error bars represent SD of 3 replicates used for each group. (B) TET2 mRNA expression in CD34+ cells derived from controls (n = 6) and patients with TET2 WT and TET2 mutants (n = 16). ● represent TET2 WT patients; ○, TET2-mutated patients; and —, mean for each group.
TET2 mRNA expression. (A) TET2 mRNA expression in fractions of healthy bone marrows (n = 5). Error bars represent SD of 3 replicates used for each group. (B) TET2 mRNA expression in CD34+ cells derived from controls (n = 6) and patients with TET2 WT and TET2 mutants (n = 16). ● represent TET2 WT patients; ○, TET2-mutated patients; and —, mean for each group.
Clinical features/outcomes of patients with TET2 mutations
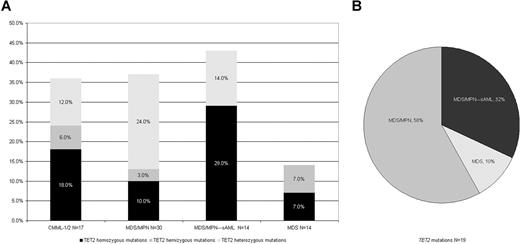
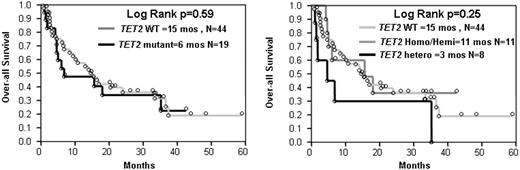
In total, we sequenced 68 patients (Table 1). When individual pathomorphologic groups were analyzed, we found TET2 mutations in 2 of 14 (14%) patients with MDS, 11 of 30 (37%) with MDS/MPN (including 6 of 17 [35%] with CMML-1/2). TET2 mutations were found in 6 of 14 patients with antecedent MDS/MPN progressing to AML (Figure 4). Among RARS-T patients analyzed in whom no JAK2 or c-MPL mutations were found (n = 3), one patient had a heterozygous TET2 mutation. No mutations were found in JMML (n = 5), sAML that evolved from MDS (n = 7) and healthy controls (n = 4). Among JAK2V617F positive patients present among MDS/MPN patients (n = 5), in only one case was a TET2 mutation found. The presence of a TET2 mutation correlated with elevated white blood cell (WBC) count, monocytosis, and male sex (Table 3). In addition, a trend toward association with malignant transformation was noted. However, Kaplan-Meier curves did not reveal an impact of TET2 mutations on survival, and there was also no difference between the homo/hemizygous and heterozygous cases (Figure 5).
Frequency of TET2 mutations in studied patients. (A) Frequency of TET2 mutations among various clinical subtypes of myeloid disorders. The frequencies of TET2 mutations are calculated for each disease category including CMML-1/2 (n = 17), MDS/MPN-sAML (n = 14), MDS/MPN (n = 30), and MDS (n = 14). (B) Distribution of various disease subtypes among patients with TET2 mutation (n = 19 or 100% TET2 mutations).
Frequency of TET2 mutations in studied patients. (A) Frequency of TET2 mutations among various clinical subtypes of myeloid disorders. The frequencies of TET2 mutations are calculated for each disease category including CMML-1/2 (n = 17), MDS/MPN-sAML (n = 14), MDS/MPN (n = 30), and MDS (n = 14). (B) Distribution of various disease subtypes among patients with TET2 mutation (n = 19 or 100% TET2 mutations).
Survival of TET2-mutated and WT patients with myeloid malignancies. Each dot represents 1 patient.
Survival of TET2-mutated and WT patients with myeloid malignancies. Each dot represents 1 patient.
Discussion
Identification of pathogenic mutations or other chromosomal defects allows for improved diagnostics and ultimately the design of targeted molecular therapies. The most illustrative examples include the discovery of BCR/ABL in CML, PML/RARa in PML, and more recently, JAK2V617F mutations in MPN. However, to date for most cases of MDS and atypical MDS/MPN including CMML, pathogenic molecular lesions have not been identified. The current pathomorphologic WHO classification separates classical MDS entities from the related yet distinct MDS/MPN overlap syndromes, which include CMML,22 previously considered a subentity of MDS.23 Both MDS and MDS/MPN can evolve into sAML, clearly a part of a pathologic continuum, but not included into the MDS category under the WHO classification. Due to the imprecise distinction and pathophysiologic continuum of these diseases, common genetic defects have to be analyzed in the context of the broader nosologic categories. Recently, we have shown that somatic areas of UPD are common in patients with MDS/MPN, in particular CMML.8,11 One of most frequently recurring lesions found in these patients was UPD11q, and we have shown that this defect indicates the presence of homozygous c-Cbl mutations, also particularly frequent in patients with CMML.11
Here we focused on another area of UPD frequently occurring in MPN. SNP-A-based karyotyping allowed for identification of recurrent LOH located on the long arm of chromosome 4, frequently affected by microdeletions and copy-number neutral LOH. Both UPD4q and deletions were somatic as confirmed by analysis of the corresponding nonclonal CD3+ cells. Among patients studied, these lesions were most prevalent in CMML, closely related MDS/MPN, and AML with antecedent history of these conditions. This is not surprising as there is a great deal of overlap between more advanced forms of CMML (CMML-2) and sAML, as well as some forms of AML with monocytoid differentiation, explaining why a common lesion may be found in these conditions. Previously, 4q24 deletion was described in 4 cases of hematopoietic malignancy,24 and we and others observed several cases of UPD4q in the initial studies describing the application of SNP-A in MDS.7,8 Theoretically it is possible that some microdeletions may have not been detected by 250K SNP-A, but no gain in diagnostic yield has been observed by application of very high-density arrays, including Affymetrix Genome-Wide Human SNP Array 6.0 containing more than 900K SNP and 900K CNV probes.25
Fine-scale mapping of the commonly affected region was facilitated by the presence of recurrent microdeletions in multiple patients. This 0.35 Mb region spanned 2 genes, PPA2 and TET2. Sequencing of both genes revealed the presence of missense, stop codon, and frameshift mutations in TET2. No recurrent mutations were found, but in the recent report of TET2 mutations in classical MPN,26 3 mutations were identical to those found in our study.
While for precise estimation of frequencies of TET2 mutations in morphologic subentities, a much larger population needs to be studied, our results demonstrate that TET2 mutations are common in myeloid malignancies, in particular CMML and sAML with proliferative, myelomonocytic features. The function of this novel gene is not clear, and no clues can be derived from its structure, with homology only to TET family members and orthologs in other species.27,28 The structurally related TET1 has been previously described as a fusion partner in an invariant translocation t(10;11) in AML.28 The presence of nonsense and frameshift mutations suggests that TET2 lesions result in inactivation, consistent with a putative tumor suppressor function, while heterozygous mutations indicate that WT allele is not completely protective.
Testing of cases of LOH on 4q revealed homozygous mutations in UPD4q and hemizygous mutations in patients with del4q. However, sequencing of patients without LOH4q revealed the presence of heterozygous mutations occurring at a similar frequency as those in the context of LOH. The pathogenic nature of TET2 mutations was also demonstrated by the detection of compound heterozygous mutations. UPD and deletions affecting 4q, as well as TET2 mutations, were somatic as determined by analysis of germ line DNA. Heterozygous mutations may represent a low clonal burden, with dilution of a homozygous clone with a similar number of WT cells. According to the 2-hit-hypothesis, TET2 mutation may represent either a first or second hit. This theory is well illustrated by the association of TET2 mutations with JAK2-positive MPN.26,29 Possibly, the heterozygous cases require another molecular event such as JAK2 mutation, while UPD4q or deletions 4q represent another type of the second event. Thus, theoretically, the “first hit” affecting TET2 may not be pathogenic and does not constitute a primary selecting event, rather representing a passenger lesion. Only after the second allele is lost or inactivated do patients progress. Studies of expression levels of TET2 transcripts and methylation analyses of the TET2 promoter did not show any significant differences between the patients, suggesting that down-modulation of this gene that could substitute for inactivating mutations is unlikely involved in the pathogenesis of malignant evolution or could produce a similar clinical phenotype.
Analysis of a spectrum of closely related MPN showed that TET2 mutations are associated with CMML, MDS/MPN, and sAML, while no mutations were found in JMML. Patients with MDS less frequently had TET2 mutations. The pathomorphologic classification of these disorders is based on the subjective evaluation of marrow specimens, and there is a great deal of overlap. It is possible that morphologic features only imprecisely reflect molecular pathogenesis, well exemplified by JAK2; mutations of this gene are found in a proportion of cases with MDS/MPN as well as morphologically distinct cases of polycythemia rubra vera, myelofibrosis, or essential thrombocytosis.4,5 Common features observed among patients with TET2 mutations included predominantly high WBC count and the presence of monocytosis. Progression to AML seems to be associated with the presence of TET2 mutation, but this preliminary observation has to be confirmed in a larger cohort of patients.
Distribution of TET2 mutations among morphologic subtypes of leukemia suggests this gene plays an important role in the regulation of myeloid proliferation. Similarly, it is expressed in hematopoietic cells and leukemic cell lines.28 The types of mutations encountered suggest that TET2 may serve as a tumor suppressor gene, while high frequency of heterozygous mutations may imply dominant negative effects exerted by the mutant TET2.
In sum, we have identified a novel tumor suppressor gene strongly associated with proliferative MDS/MPN cases. These results are consistent with a recent abstract, identifying TET2 mutations in JAK2V617F positive and negative MPN.29 Further identification of intrinsic TET2 function and pathogenic networks will reveal mechanistic insight into the pathogenesis of specific subtypes of myeloid malignancies, while screening a large number of tumors for mutations of this ubiquitously expressed gene will likely also provide insights into its potential role in carcinogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof.
Since initial submission of this report, a subsequent publication on TET230 indicated that nucleotide variants resulting in amino acid changes to Q1084P and Y867H constitute previously unannotated SNPs rather than mutations. These findings do not significantly impact our conclusions, and appropriate annotations have been made in the text.
Acknowledgments
This work was supported by National Institutes of Health (Bethesda, MD; NIH) grants RO1HL-082983 (J.P.M.), U54 RR019391 (J.P.M.), K24 HL-077522 (J.P.M.), and Department of Defense grant DOD MPD510343 (M.A.M.), and a grant from the AA & MDS International Foundation (Rockville, MD), and the Robert Duggan Charitable Fund (Cleveland, OH; J.P.M.).
National Institutes of Health
Authorship
Contribution: A.M.J., H.S., and H.M. performed the majority of the experimental work; A.M.J., H.S., and C.L.O. compiled the manuscript; C.L.O. and J.H. performed SNP-A and cytogenetic analyses; R.V.T. and M.A. performed clinical analyses; R.G. performed methylation studies; M.A.M. selected some of the patients and contributed to the design and analysis of the study; and J.P.M. conceptualized the project, supervised the experimentation, analyzed the results, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Center/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.