Abstract
Mutations in NOTCH1 are frequently detected in patients with T-cell acute lymphoblastic leukemia (T-ALL) and in mouse T-ALL models. Treatment of mouse or human T-ALL cell lines in vitro with γ-secretase inhibitors (GSIs) results in growth arrest and/or apoptosis. These studies suggest GSIs as potential therapeutic agents in the treatment of T-ALL. To determine whether GSIs have antileukemic activity in vivo, we treated near-end-stage Tal1/Ink4a/Arf+/− leukemic mice with vehicle or with a GSI developed by Merck (MRK-003). We found that GSI treatment significantly extended the survival of leukemic mice compared with vehicle-treated mice. Notch1 target gene expression was repressed and increased numbers of apoptotic cells were observed in the GSI-treated mice, demonstrating that Notch1 inhibition in vivo induces apoptosis. T-ALL cell lines also exhibit PI3K/mTOR pathway activation, indicating that rapamycin may also have therapeutic benefit. When GSIs are administered in combination with rapamycin, mTOR kinase activity is ablated and apoptosis induced. Moreover, GSI and rapamycin treatment inhibits human T-ALL growth and extends survival in a mouse xenograft model. This work supports the idea of targeting NOTCH1 in T-ALL and suggests that inhibition of the mTOR and NOTCH1 pathways may have added efficacy.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is associated with the misexpression of the basic helix-loop-helix protein TAL1/SCL and LIM-domain only proteins LMO1 and LMO2.1-8 These oncogenes are found misexpressed in greater than 60% of human T-ALL patients.3,9 Mouse models of T-ALL recapitulate the disease through ectopic expression of Tal1 in the thymus.10-12 These mice develop respiratory distress due to the presence of large thymic masses and have detectable T-cell blasts in peripheral blood lymphocytes (PBLs), spleen, liver, and kidney.10 Misexpression of Tal1 results in perturbed thymocyte development by interfering with the basic-helix-loop-helix (bHLH) heterodimer E47/HEB that regulates the expression of genes critical for thymocyte differentiation including Rag1/2, Pre-Tα, CD4, CD3, and TCRα/β.13,14 Consistent with this finding, loss of the E2A gene that encodes the E47 and E12 bHLH protein is associated with human B-progenitor ALL.15
Mutations in the Notch1 receptor have been frequently detected in mouse T-ALL models16-19 and importantly in 54% of T-ALL patients.20 These mutations cluster in the heterodimerization domain (HD)20 and the juxtamembrane (JME) region,21 or result in truncation of PEST regulatory sequences.18,20 Mutations in the HD domain result in increased susceptibility to cleavage by the gamma-secretase complex; JME mutations may facilitate metalloprotease cleavage, whereas deletion of PEST regulatory sequences is thought to result in increased Notch1 stability.21-23 Treatment of mouse Tal1 leukemic cell lines in vitro with γ-secretase inhibitors (GSIs) results in cell cycle arrest and apoptosis, revealing that Notch1 signaling is required for leukemic growth/survival.18 Notch1-mediated leukemic growth in mouse and human T-ALL cells is mediated in part by the direct transcriptional activation of c-Myc.24-26 Similarly, Notch1-mediated mammary tumorigenesis in the mouse also appears c-Myc dependent.27 It remains unclear, however, whether c-Myc expression is required for Notch1-mediated leukemogenesis or whether other Notch1 target genes contribute.
Although the majority of mouse Tal1 leukemic cell lines undergo apoptosis upon GSI treatment in vitro, it remains to be tested whether Notch1 can be inhibited for extended periods of time in vivo. An additional concern regarding targeting NOTCH1 in T-ALL is that in contrast to mouse, many human T-ALL lines appear relatively GSI resistant in vitro,20,24,28 raising the possibility that GSIs alone may not prove effective in the treatment of T-ALL patients. Moreover, whether GSIs can be administered in vivo for extended periods of time without associated toxicities remains uncertain.
In this study, we examine the effects of GSI treatment in our mouse T-ALL model. To examine GSI efficacy and to accurately reflect the clinical experience, we treated near-end-stage leukemic mice and found that GSI treatment extends the survival of leukemic mice, but is not sufficient to eliminate disease. Mouse Tal1 leukemic cell lines isolated from GSI-treated mice exhibit mTOR pathway activation, suggesting that mTOR inhibitors may synergize with GSI to limit leukemic growth/survival. Treatment of mouse T-ALL cell lines with GSI or rapamycin reduces mTOR activity; however, combined GSI and rapamycin treatment further ablates mTOR kinase activity, resulting in increased apoptosis. Importantly, GSI and rapamycin treatment reduced human leukemic growth in vivo and significantly increased overall survival. Collectively, this work supports the idea of targeting NOTCH1 in the treatment of T-ALL.
Methods
Mouse and human T-ALL cell lines
Murine T-ALL cell lines were cultured in RPMI with 10% FBS, 1% glutamine, penicillin/streptomycin, 50 μM β mercaptoethanol at 37°C under 5% CO2. Human T-ALL cell lines were cultured in RPMI with 10% FBS, 1% glutamine at 37°C under 5% CO2. To inhibit Notch1 signaling, 106 cells were plated in a 10-cm dish in the presence of either MRK-003 (Merck Research Laboratories) at 1 μM, rapamycin at 10 nM (LC Laboratories, Woburn, MA), or a combination of MRK-003 and rapamycin. Mock-treated cells were cultured with DMSO at a final concentration of 0.01%. All mouse procedures performed in this paper have been approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC).
GSI efficacy studies
A cohort of Tal1/Ink4a/Arf+/− transgenic mice was generated (n = 30) and monitored daily for the onset of leukemia as described previously.29 Once disease became evident (respiratory distress, decrease in activity, weight loss, ruffled coat) leukemic Tal1/Ink4a/Arf+/− mice were randomly assigned to the vehicle or GSI treatment groups (vehicle cohort, n = 14; GSI cohort, n = 16). Leukemic mice were administered either a 150-mg/kg dose of freshly prepared MRK-003 (dissolved in 0.5% methylcellulose) or a comparable volume of 0.5% methylcellulose by oral gavage. Mice were treated for 3 consecutive days followed by a rest period of 4 days. Mice were continuously treated with this regimen throughout the study period and killed when deemed moribund by a third party blinded to the treatment group. Survival data were plotted using Kaplan-Meier survival curves and statistical analysis was performed using SPSS software (Chicago, IL). To examine the effects on human leukemic growth, female CD1 nu/nu mice (Charles River Laboratories, Wilmington, MA) were inoculated with human TALL-1 cells (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ] Braunschweig, Germany) 3 days after an intraperitoneal inoculation of 100 mg/kg cyclophosphamide (no. C0768-1G; Sigma-Aldrich, St Louis, MO) to facilitate tumor growth. TALL-1 cells were implanted at a concentration 5 × 106 cells diluted in 100 μL of 50% matrigel (no. CB-40230C; Fisher Scientific, Pittsburgh, PA)/50% PBS. When tumors reached 250 mm3, mice were randomized into groups (n = 8 mice/treatment group) of equal average tumor volume. MRK-003 was dosed at either 0 or 150 mg/kg by oral gavage once a week. Rapamycin was administered by oral gavage daily at a concentration of either 0 or 20 mg/kg. Tumors were callipered and body weights were recorded twice weekly for the duration of study. Mice were killed when tumors reached the maximum volume allowed by IACUC guidelines. Survival data were plotted as a Kaplan-Meier survival curve using Prism Graphpad software (GraphPad Software, San Diego, CA). Repeated measures analysis of variance was used for the comparison of tumor growth inhibition. MRK-003 levels in plasma and in tumor were measured by high-pressure liquid chromatography using Cohesive Technologies pump and autosampler (Thermo Scientific, Waltham, MA) equipped with a reversed-phase column (Atlantis C18, 3 μm, 3 × 20 mm; Waters, Milford, MA) and linear water/ACN gradient (5%-90% organic in 3 minutes) containing 0.1% formic acid at a flow rate of 850 μL/minute. The protonated molecules were fragmented by collision-induced dissociation with nitrogen as a collision gas. The collision energy voltage was set at 35 V and 48 V for GSI and internal standard, respectively. The data were acquired and processed by Analyst 1.4.1 software (AB/MDS Sciex, Foster City, CA).
TUNEL staining
To quantify apoptotic cells, leukemic mice treated with a 150-mg/kg dose of MRK-003 or 0.5% methylcellulose orally for 3 days, were killed and a postmortem examination was performed. Paraffin-embedded tumor sections were analyzed for apoptosis using a TdT enzyme kit (30 U/slide; Roche, Indianapolis, IN). The staining was automated using a DAB Map kit on the Discovery XT (Ventana Medical Systems, Tucson, AZ). Approximately, 10 fields from each slide were counted and compared with the serial section not treated with the TdT enzyme. The Wilcoxon rank-sum test was used to determine statistical significance.30 Images were taken using an Olympus BX41 microscope (Olympus, Center Valley, PA) with 20× or 40× lenses. The images were acquired with an Evolution MP 5.0 camera (Media Cybernetics, Bethesda, MD) and Q Capture Pro 5.1 acquisition software.
Immunohistochemistry
Paraffin-embedded tumor sections were analyzed for phospho-S6 ribosomal protein expression (1:75; Cell Signaling Technology, Beverly, MA). Automated staining was performed using the ChromoMap Kit on the Discovery XT (Ventana Medical Systems) under standard conditions. Periodic acid Schiff (PAS) staining of mouse duodenum was conducted as previously described.31 Images were taken using a Carl Zeiss (Jena, Germany) Imager, Z1 Plan-Apochromat with a 20× objective lens. The images were acquired with a Carl Zeiss AxioCam HRc camera, and the image acquisition software used was Carl Zeiss AxioVision Rel 4.6.
Flow cytometry
To determine the effects of GSI and/or rapamycin treatment on the cell cycle, mouse and human leukemic cells were pelleted by centrifugation for 10 minutes at 1290g, washed in PBS, and resuspended in 70% ice-cold ethanol. Cells were fixed overnight and then stained with propidium iodide. DNA content was analyzed by flow cytometry (FACScan; BD Biosciences, San Jose, CA). Data were analyzed using FlowJo version 7.0 (TreeStar, Ashland, OR). To quantify the effects of GSI on primary tumors, thymic masses isolated directly from Tal1 transgenic animals were treated ex vivo with GSI or vehicle and then were stained with FITC–annexin V/PI and analyzed by flow cytometry (BD Biosciences).
RNA analysis
RNA was extracted from murine T-ALL cell lines and primary thymic masses treated with vehicle or MRK-003 using Trizol. cDNA was synthesized using Superscript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). To determine the effects of MRK-003 treatment on Notch1 target genes, quantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed using Deltex1 and Hes1 primers as described previously.25 To determine Notch1 target gene expression in primary tumors, quantitative PCR was performed using c-Myc, Hes1, Pre-Tα, Deltex1 primers designed using Primer Express software (Applied Biosystems, Foster City, CA). The primer sequences are as follows: c-Myc forward, 5′-CTGTTTGAAGGCTGGATTTCCT-3′; c-Myc reverse, 5′-CAGCACCGACAGACGCC-3′; Hes1 forward, 5′-AAGACGGCCTCTGAGCACA-3′; Hes1 reverse, 5′-CCTTCGCCTCTTCTCCATGAT-3′; Pre-Tα forward, 5′-CTGCTTCTGGGCGTCAGGT-3′; Pre-Tα reverse, 5′-TGCCTTCCATCTACCAGCAGT-3′; Deltex1 forward, 5′-TGCCTGGTGGCCATGTACT-3′; Deltex1 reverse, 5′-GACACTGCAGGCTGCCATC-3′. The copy number obtained for the gene of interest was normalized to the copy number for β-actin.
Western blot analysis
Murine leukemic cells lines were lysed in 20 mM Tris buffer, pH 7.4, containing 0.14 M NaCl, 1% NP-40, 10% glycerol, 1 mM sodium orthovanadate, and protease inhibitors and protein concentrations determined using the Bradford assay (Bio-Rad, Hercules, CA). Total protein (35 μg) was resolved on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, blocked for 1 hour with a 50 mM Tris buffer, pH 7.5, containing 0.15 M NaCl, 0.05% Tween 20 (TBST), and 5% (wt/vol) nonfat dry milk, and probed overnight at 4°C with buffer containing primary antibodies. After three 10-minute washes in TBST, the filters were incubated for 1 hour in blocking buffer containing horseradish peroxidase–conjugated secondary antibodies. After three 10-minute washes in TBST, proteins were detected by enhanced chemiluminescence (Pierce, Rockford, IL). The following antibodies used for immunoblotting were purchased from Cell Signaling Technology: phospho-p70 S6 kinase T389 (no. 9205), p70 S6 kinase (no. 9202), phospho-Akt S473 (no. 9271), Akt (no. 9272), phospho-S6 ribosomal S240/244 (no. 2215), S6 ribosomal (no. 2217), phospho-4E-BP1 S65 (no. 9451), 4E-BP-1 (no. 9452), PTEN antibody (no. 9552), and NotchIC Val1744 (no. 2421). Blots were stripped and reprobed with β-actin (no. A5441; Sigma-Aldrich) to control for equal loading.
In vitro Vialight assay
The human T-ALL cell line, TALL-1, was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and plated at a concentration of 104 in 80 μL growth media in a flat-bottom 96-well plate. Approximately, 10 μL MRK-003 (at concentrations ranging from 0-50 μM) and 10 μL rapamycin (no. 553210-1 mg; EMD Biosciences, Darmstadt, Germany) at either 0, 1-nM, or 100-nM concentrations were then added to each well. On day 4, new media and drug were added by spinning the plate and replacing 75 μL per well with fresh drug and media. Plates were read 7 days after seeding cells following the Vialight Plus (no. LT07-221; Lonza Bioscience, Basel, Switzerland) manufacturer's protocol.
MTT analysis
In a 96-well flat-bottom plate, approximately 104 cells/200 μL of a cell suspension were plated and treated for 72 hours with vehicle, 1 nM rapamycin, increasing concentrations of MRK-003 (10−5 to 102 μM), or both rapamycin and MRK-003. After treatment, 20 μL of a 5-mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (Sigma-Aldrich) was added and incubated for 4 hours at 37°C. The plate was then spun at 1290g rpm for 5 minutes and the media were removed. The reagent was solubilized with 100 μL dimethyl sulfoxide (Sigma-Aldrich) and incubated for 10 minutes at room temperature. Plates were then analyzed at Α490 wavelength. Data are plotted as percentage of apoptosis. This value was determined by comparing the absorbance reading of each set of test wells to a set of control wells in which no drug was added and baseline proliferation was recorded. The following equation was used to determine effect on growth: ([absorbance of control − absorbance of test]/absorbance of control) × 100.
Results
The GSI MRK-003 represses Notch1 target gene expression and induces apoptosis of mouse T-ALL cell lines and primary Tal1/Ink4a/Arf+/− tumors
The prevalence of mutations that result in activated NOTCH1 in patients with T-ALL raises the possibility that GSIs used to inhibit NOTCH1 and other GS-dependent substrates in vitro may have antileukemic activity in the clinic. To evaluate the ability of MRK-003 to inhibit Notch1, multiple mouse T-ALL cell lines were treated with 1 μM MRK-003 and Notch1 target gene expression was examined. Decreased levels of Hes1 and Deltex1 expression were observed in the MRK-003–treated cultures after treatment for 24 hours (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). We then compared the relative effectiveness of MRK-003 with N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), another GSI known to inhibit mouse leukemic growth.18 Multiple mouse T-ALL cell lines were treated with 1 μM MRK-003 or with 1 μM DAPT for 3 days and cell-cycle analysis was performed. In all 3 of the cell lines tested, increases in the percentage of apoptotic cells were observed in MRK-003–treated cultures compared with treatment with either DMSO (vehicle) or DAPT (Figure S1B). These in vitro data suggested that MRK-003 may be effective at inhibiting Notch1-mediated leukemic growth in vivo. To test this, thymic tumor explants (masses) isolated directly from Tal1/Ink4a/Arf+/− mice were treated with DMSO (vehicle) only, with 1 μM DAPT, or 1 μM MRK-003 for 3 days. The percentage of apoptotic cells was then determined by annexin V/PI staining followed by flow cytometry. We found MRK-003–treated cultures contained significantly more apoptotic cells than were observed in either the DMSO- or DAPT-treated cultures (Figure S1C; P = .027 by Kruskal-Wallis test30 ).
GSI treatment prolongs survival in a mouse T-ALL model
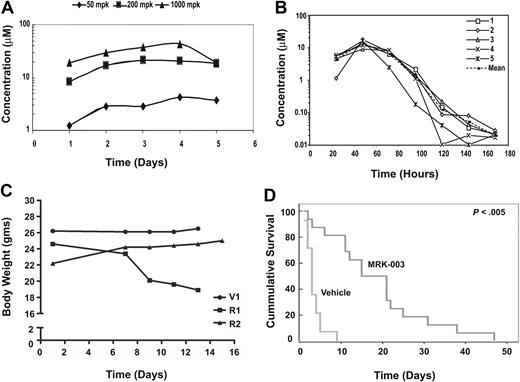
These ex vivo studies indicated that MRK-003 administration to leukemic mice might induce apoptosis in vivo, decrease tumor burden, and increase overall survival. To test this possibility, we treated mice daily with 50 mg/kg, 200 mg/kg, or 1000 mg/kg MRK-003 and determined plasma concentrations of the compound. We found that effective plasma MRK-003 concentrations (1-10 μM) were achieved in mice treated with all 3 doses (Figure 1A). However, when the MRK-003 compound was given daily at 150 mg/kg, the mice began to develop diarrhea and lose weight (Figure 1C; R1 compared with V). Chronic GSI administration is known to result in gastrointestinal (GI) toxicity due to Notch inhibition in the intestinal epithelium, resulting in gut metaplasia.31-34 To assess GSI efficacy on mouse leukemic growth, we adopted an intermittent GSI dosing regimen that achieved effective concentrations of the drug without associated toxicities. Specifically, mice treated with 150 mg/kg MRK-003 for 3 days followed by a 4-day rest period had effective plasma compound levels (Figure 1B), did not lose body weight (Figure 1C, mouse R2), and exhibited limited gut metaplasia (Figure S2). MRK-003 plasma levels peak after administration and effective drug concentrations are detected at 24 hours after treatment. Between 3 to 4 days after MRK-003 administration, compound levels decrease to below detectable levels, and by 96 hours decreases in goblet cells can be observed in the colons of the treated mice (Figure S2). These data indicate that the gut epithelium recovers during the 4-day “off” period. GSI plasma concentrations were also measured each week after 3 full cycles of the intermittent dosing regimen to determine whether the compound accumulates upon successive treatment. We found no significant change in plasma concentration after 3 cycles of intermittent dosing (Figure S3). In addition, in a preclinical mouse model, this intermittent dosing schedule was well tolerated, as no evidence of intestinal effects were detected after a 35-day treatment period.35
GSI treatment prolongs survival in a mouse T-ALL model. (A) Effective plasma compound levels in MRK-003–treated mice. Compound serum levels were analyzed 1 to 5 days after continuous dosing of MRK-003 treatment. Effective and stable compound levels (1-10 μM) were detected in the serum of treated mice after 12 hours at each drug concentration tested. (B) Plasma compound levels decrease during 4-day rest period. Mice were treated for 3 consecutive days with 150 mg/kg MRK-003. After the first treatment, serum was analyzed for compound levels every 24 hours for the duration of the dosing regimen. (C) Intermittent GSI dosing minimizes “on-target” gastrointestinal toxicity. To define a GSI dosing regimen with limited/no associated toxicity, mice were administered vehicle (V) or 150 mg/kg MRK-003 by oral gavage everyday (R1) or for 3 days followed by a 4-day rest period (R2). Mice were monitored daily for loss of body weight and for evidence of diarrhea. (D) Extended survival in MRK-003–treated leukemic mice. Near-end-stage diseased Tal1/Ink4a/Arf+/− mice were treated with 150 mg/kg MRK-003 (n = 16 mice) or 0.5% methylcellulose (n = 14 mice) orally for 3 days and rested for 4 days until mice were deemed moribund. Median survival for T-ALL mice treated with vehicle is 3 days, and 18 days for GSI treated mice (P < .005).
GSI treatment prolongs survival in a mouse T-ALL model. (A) Effective plasma compound levels in MRK-003–treated mice. Compound serum levels were analyzed 1 to 5 days after continuous dosing of MRK-003 treatment. Effective and stable compound levels (1-10 μM) were detected in the serum of treated mice after 12 hours at each drug concentration tested. (B) Plasma compound levels decrease during 4-day rest period. Mice were treated for 3 consecutive days with 150 mg/kg MRK-003. After the first treatment, serum was analyzed for compound levels every 24 hours for the duration of the dosing regimen. (C) Intermittent GSI dosing minimizes “on-target” gastrointestinal toxicity. To define a GSI dosing regimen with limited/no associated toxicity, mice were administered vehicle (V) or 150 mg/kg MRK-003 by oral gavage everyday (R1) or for 3 days followed by a 4-day rest period (R2). Mice were monitored daily for loss of body weight and for evidence of diarrhea. (D) Extended survival in MRK-003–treated leukemic mice. Near-end-stage diseased Tal1/Ink4a/Arf+/− mice were treated with 150 mg/kg MRK-003 (n = 16 mice) or 0.5% methylcellulose (n = 14 mice) orally for 3 days and rested for 4 days until mice were deemed moribund. Median survival for T-ALL mice treated with vehicle is 3 days, and 18 days for GSI treated mice (P < .005).
In an attempt to accurately reflect the clinical experience, near-end-stage leukemic Tal1/Ink4a/Arf+/− mice were treated with either vehicle or 150 mg/kg MRK-003 for 3 days followed by a 4-day recovery period. After the 3 days of GSI treatment, plasma compound levels were determined and an EC50 of 5 to 10 μM MRK-003 was achieved (data not shown). Leukemic mice were treated with vehicle or MRK-003 after the 3 days on, 4 days off treatment regimen throughout the duration of the study. We found that GSI treatment resulted in a statistically significant increase in overall survival of leukemic mice compared with vehicle-treated mice (Figure 1D; P < .005). The median survival period for GSI-treated mice was 18 days compared with 3 days for the vehicle-treated group. In most cases, responses were evident in the MRK-003–treated mice immediately after the 3-day treatment period, as measured by an increase in physical activity and improved rates of respiration. Importantly, body weights are maintained in MRK-003–treated animals after successive cycles of intermittent GSI dosing (Table S1). This pilot study supports the idea that GSI-associated toxicities may be overcome with intermittent dosing and provides evidence that Notch1 inhibition improves mouse leukemic survival in vivo.
GSI treatment induces apoptosis of mouse leukemic cells in vivo
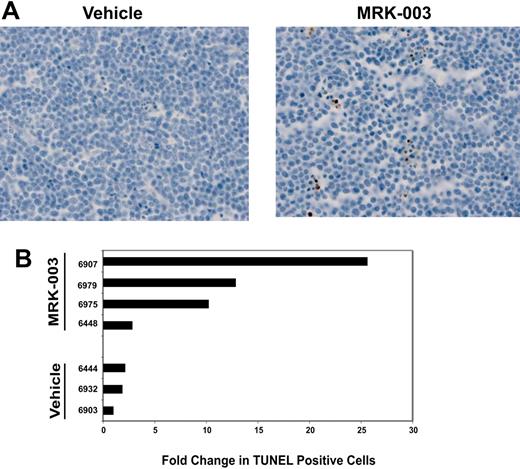
To determine whether GSI treatment induces apoptosis in vivo, we performed TUNEL staining on thymomas isolated from vehicle- or GSI-treated mice. We detected increases in the percentage of apoptotic tumor cells in mice treated with MRK-003, compared with tumors exposed to vehicle only (Figure 2A-B; P = .034 by Wilcoxon rank sum test30 ). However, similar to our in vitro data (Figure S1B), the in vivo tumor response to GSI was variable. Three of the 4 GSI-treated mice examined exhibited an increase in apoptotic leukemic cells, with 2- to 10-fold increases in TUNEL+ tumor cells observed (Figure 2B). An increase in TUNEL+ cells was not observed in 1 of 4 tumors from the GSI-treated group (mouse 6448). However, we were unable to detect a Notch1 mutation in this tumor. The reasons for the variable GSI responses both in vitro and in vivo are unclear. One possibility for the variable in vivo responses may be that some tumors require longer treatment periods. For these studies, leukemic mice were treated with vehicle or with GSI for 3 days and then the mice were humanely killed and tumor sections analyzed for the presence of apoptotic cells. It is conceivable that more consistent responses may be observed in leukemic mice treated with multiple GSI doses. Nonetheless, GSI treatment clearly induced apoptosis and extended the survival of leukemic mice.
GSI treatment induces apoptosis of leukemic cells in vivo.(A) Leukemic mice were treated with a 150-mg/kg dose of MRK-003 or with vehicle for 3 days. Tumor sections were fixed in 10% buffered formalin and number of apoptotic cells was quantified using TUNEL assay (ApoTag Plus peroxidase from Chemicon, Temecula, CA). (B) Bar graph representing the fold change in TUNEL-positive cells compared with vehicle. Ten independent fields/section were counted to obtain the representative value.
GSI treatment induces apoptosis of leukemic cells in vivo.(A) Leukemic mice were treated with a 150-mg/kg dose of MRK-003 or with vehicle for 3 days. Tumor sections were fixed in 10% buffered formalin and number of apoptotic cells was quantified using TUNEL assay (ApoTag Plus peroxidase from Chemicon, Temecula, CA). (B) Bar graph representing the fold change in TUNEL-positive cells compared with vehicle. Ten independent fields/section were counted to obtain the representative value.
Transient GSI responses do not reflect development of GSI resistance
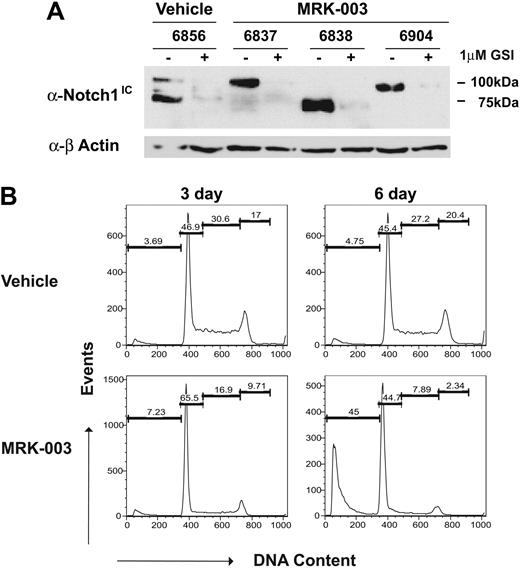
Another potential reason for variable responses may be that the GSI-treated mice develop GSI resistance. To exclude this possibility, T-ALL cell lines were generated from the GSI- and vehicle-treated mice and their response to GSI was quantified in vitro. As expected, truncated Notch1 proteins were detected in the vehicle-treated tumor 6856 and the GSI-treated tumors 6838 and 6904. In contrast, a full-length Notch1 protein appears expressed in tumor 6837, although increased intracellular Notch1 protein levels are observed, suggesting that this tumor may harbor mutations in other regulators of Notch1 protein stability. In all tumors, GSI treatment in vitro significantly reduced Notch1 protein levels, as assessed by immunoblotting with the anti-Notch1 V1744 antibody (Figure 3A). Leukemic cell lines also clearly remained dependent on Notch1 for growth, as G1 arrest and apoptosis were observed upon GSI treatment (Figure 3B). These findings indicate that transient responses to GSI in vivo do not reflect the development of GSI resistance.
GSI-treated tumors do not appear to develop GSI resistance. Thymomas from MRK-003– or vehicle-treated mice were harvested from the animals and converted to in vitro culture. (A) Leukemic cell lines from vehicle- and GSI-treated mice express high levels of intracellular Notch1 and remain GSI responsive. Leukemic cell lines were treated for 48 hours with 1 μM MRK-003 or DMSO carrier. Cell lysates were examined for intracellular Notch1 levels by immunoblotting with an anti-Notch1IC Val1744 (no. 2421; Cell Signaling Technology) and anti–β-actin antibodies. (B) Leukemic growth remains Notch1 dependent. Leukemic cell lines, generated from vehicle- and GSI-treated mice, were treated with vehicle or 1 μM MRK-003 for 3 and 6 days. Cells were then assayed for DNA content by staining with propidium iodide followed by flow cytometry. The figure is a representative experiment using cell line 6838.
GSI-treated tumors do not appear to develop GSI resistance. Thymomas from MRK-003– or vehicle-treated mice were harvested from the animals and converted to in vitro culture. (A) Leukemic cell lines from vehicle- and GSI-treated mice express high levels of intracellular Notch1 and remain GSI responsive. Leukemic cell lines were treated for 48 hours with 1 μM MRK-003 or DMSO carrier. Cell lysates were examined for intracellular Notch1 levels by immunoblotting with an anti-Notch1IC Val1744 (no. 2421; Cell Signaling Technology) and anti–β-actin antibodies. (B) Leukemic growth remains Notch1 dependent. Leukemic cell lines, generated from vehicle- and GSI-treated mice, were treated with vehicle or 1 μM MRK-003 for 3 and 6 days. Cells were then assayed for DNA content by staining with propidium iodide followed by flow cytometry. The figure is a representative experiment using cell line 6838.
Repression of Notch1 target gene expression in GSI-treated mice
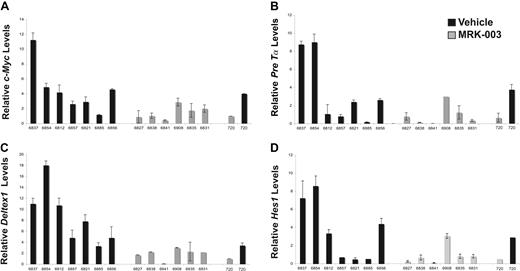
To further understand why GSI treatment prolonged survival but failed to eliminate disease, we examined Notch1 target gene expression (Deltex1, c-Myc, Hes1, and Pre-Tα) in the vehicle- and GSI-treated leukemic cohorts using real-time PCR (Figure 4A-D). We found c-Myc and Deltex1 expression reduced in MRK-003–treated leukemic mice compared with mice treated with vehicle only (Figure 4A,C). The difference in c-Myc and Deltex1 expression in the GSI- versus vehicle-treated leukemic mice was statistically significant (P = .015 and P = .027, respectively by Wilcoxon rank sum test). Moreover, the c-Myc and Deltex1 mRNA levels observed in vivo approximate expression levels observed when the mouse T-ALL cell line 720 was treated with GSI in vitro. In contrast, differences in Hes1 and Pre-Tα expression did not reach statistical significance (Figure 4B,D), suggesting that these genes may also be regulated by other transcription factors in mouse T-ALL cells. This study also raises the possibility that c-Myc and/or Deltex1 expression levels may serve as reliable biomarkers of NOTCH1 activity in T-ALL patients.
Notch1 target gene expression is repressed in GSI-treated leukemic Tal1/Ink4a/Arf+/− mice. At killing, thymomas were harvested from vehicle- and MRK-003–treated mice and c-Myc (A), Pre-Tα (B), Deltex1 (C), and Hes1 (D) expression was quantified using real-time PCR. The copy number for each target gene was normalized to the copy number for β-actin using the ΔΔCT method. The following result is an average of 3 independent experiments. The mouse T-ALL cell line 720 was treated in vitro with 1 μM MRK-003 or DMSO for 72 hours and used as a positive control in these experiments.
Notch1 target gene expression is repressed in GSI-treated leukemic Tal1/Ink4a/Arf+/− mice. At killing, thymomas were harvested from vehicle- and MRK-003–treated mice and c-Myc (A), Pre-Tα (B), Deltex1 (C), and Hes1 (D) expression was quantified using real-time PCR. The copy number for each target gene was normalized to the copy number for β-actin using the ΔΔCT method. The following result is an average of 3 independent experiments. The mouse T-ALL cell line 720 was treated in vitro with 1 μM MRK-003 or DMSO for 72 hours and used as a positive control in these experiments.
mTOR pathway is active in mouse T-ALL cell lines
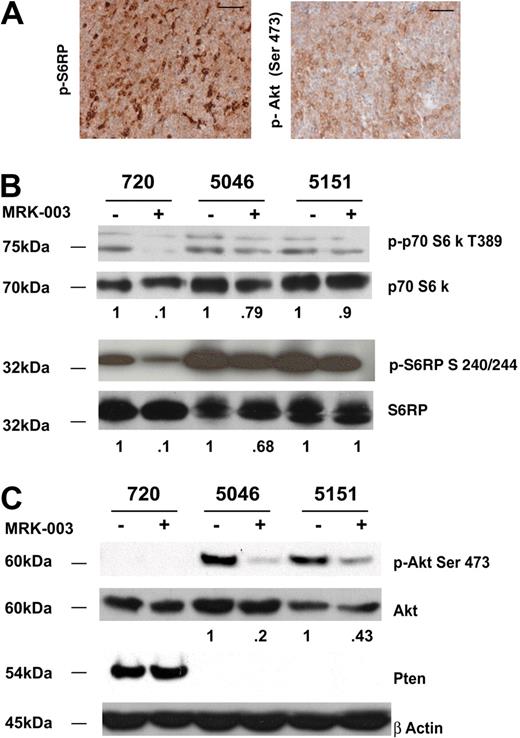
Published work by Chan et al reveals mTOR activation in human T-ALL cell lines and their in vitro studies suggest that GSI and the mTOR inhibitor rapamycin may act synergistically to arrest human leukemic growth in vitro.36 Similarly, active mTOR is detected in a subset of primary mouse T-ALL cells (Figure 5A) and all the mouse T-ALL cell lines we examined exhibited mTOR activation as assayed by phospho-p70 S6 kinase and phospho-S6 ribosomal levels (Figure 5B; Table 1). The 3 mouse T-ALL lines (720, 5046, and 5151) responded to GSI treatment, and reduced phospho-p70 S6 kinase and phospho-S6 ribosomal protein levels were observed (Figure 5B). However, GSI treatment was not sufficient to abolish mTOR activity in any of the mouse T-ALL cell lines examined.
The mTOR pathway is activated in all mouse T-ALL cell lines examined. (A) Primary mouse T-ALL tumors exhibit activation of Akt and mTOR pathways. Thymomas from vehicle-treated mice were harvested at killing and tumor sections were fixed in 10% buffered formalin. Sections were then stained with antibodies to phospho-S6 ribosomal protein (p-S6RP) or phospho-Akt (Ser473). Scale bars represent 50 μM. (B) Reduced levels of mTOR substrates are observed when mouse T-ALL lines are treated with GSI. Primary murine leukemic lines were treated with 1 μM MRK-003 or DMSO for 48 hours. MTOR activity was assayed by immunoblotting cell lysates with anti–phospho-p70 S6 kinase or phospho-S6 ribosomal antibodies (no. 9205, no. 2215; Cell Signaling Technology). p70 S6 kinase and S6 ribosomal were used as loading controls (no. 9202, no. 2217; Cell Signaling Technology). (C) Notch1 regulates Akt activity in some mouse T-ALL lines. Primary murine leukemic lines were treated with 1 μM MRK-003 or DMSO for 48 hours. Akt activity was assayed by immunoblotting cell lysates with anti–phospho-Akt Ser 473 antibody (no. 9271; Cell Signaling Technology). Akt was used as a loading control (no. 9272; Cell Signaling Technology). Fold reductions in kinase activity were determined by densitometry and represent ratios (phospho/total) normalized to DMSO-treated samples.
The mTOR pathway is activated in all mouse T-ALL cell lines examined. (A) Primary mouse T-ALL tumors exhibit activation of Akt and mTOR pathways. Thymomas from vehicle-treated mice were harvested at killing and tumor sections were fixed in 10% buffered formalin. Sections were then stained with antibodies to phospho-S6 ribosomal protein (p-S6RP) or phospho-Akt (Ser473). Scale bars represent 50 μM. (B) Reduced levels of mTOR substrates are observed when mouse T-ALL lines are treated with GSI. Primary murine leukemic lines were treated with 1 μM MRK-003 or DMSO for 48 hours. MTOR activity was assayed by immunoblotting cell lysates with anti–phospho-p70 S6 kinase or phospho-S6 ribosomal antibodies (no. 9205, no. 2215; Cell Signaling Technology). p70 S6 kinase and S6 ribosomal were used as loading controls (no. 9202, no. 2217; Cell Signaling Technology). (C) Notch1 regulates Akt activity in some mouse T-ALL lines. Primary murine leukemic lines were treated with 1 μM MRK-003 or DMSO for 48 hours. Akt activity was assayed by immunoblotting cell lysates with anti–phospho-Akt Ser 473 antibody (no. 9271; Cell Signaling Technology). Akt was used as a loading control (no. 9272; Cell Signaling Technology). Fold reductions in kinase activity were determined by densitometry and represent ratios (phospho/total) normalized to DMSO-treated samples.
Absence of Pten expression in mouse T-ALL cell lines does not predict GSI resistance
| Cell line . | Genotype . | Pten expression . | p-Akt detected . | p-p70 S6 k detected . | Notch status . | ICN detected . | GSI . |
|---|---|---|---|---|---|---|---|
| 130 | Tal1/HEB+/− | + | – | + | PEST insertion | Truncated | Sensitive |
| 135 | Tal1/HEB+/− | – | + | + | PEST insertion | Truncated | Sensitive |
| 720 | Tal1/HEB+/− | + | – | + | PEST insertion (2361) | Truncated | Sensitive |
| 2869 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | Full length | Sensitive |
| 2912 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | ND | Sensitive |
| 2913 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | ND | Sensitive |
| 3569 | Tal1/Ink4a/Arf+/− | – | + | + | HD mutation (L1699P) | Full length | Sensitive |
| 5010 | Tal1/+ | – | + | + | No mutation | Full length | Resistant |
| 5046 | Tal1/+ | – | + | + | PEST Insertion (2420) | Truncated | Sensitive |
| 5151 | Tal1/+ | – | + | + | PEST deletion (2492) | Truncated | Sensitive |
| Cell line . | Genotype . | Pten expression . | p-Akt detected . | p-p70 S6 k detected . | Notch status . | ICN detected . | GSI . |
|---|---|---|---|---|---|---|---|
| 130 | Tal1/HEB+/− | + | – | + | PEST insertion | Truncated | Sensitive |
| 135 | Tal1/HEB+/− | – | + | + | PEST insertion | Truncated | Sensitive |
| 720 | Tal1/HEB+/− | + | – | + | PEST insertion (2361) | Truncated | Sensitive |
| 2869 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | Full length | Sensitive |
| 2912 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | ND | Sensitive |
| 2913 | Tal1/Ink4a/Arf+/− | + | – | + | No mutation | ND | Sensitive |
| 3569 | Tal1/Ink4a/Arf+/− | – | + | + | HD mutation (L1699P) | Full length | Sensitive |
| 5010 | Tal1/+ | – | + | + | No mutation | Full length | Resistant |
| 5046 | Tal1/+ | – | + | + | PEST Insertion (2420) | Truncated | Sensitive |
| 5151 | Tal1/+ | – | + | + | PEST deletion (2492) | Truncated | Sensitive |
GSI indicates γ-secretase inhibitors; and ND, not detected.
Notch1 has been thought to mediate thymocyte survival by direct activation of Akt,37 and based on these findings we examined the phosphorylation of Akt using an anti–phospho-Akt Ser473 antibody (Figure 5A,C). In mouse leukemic cells where Pten is not expressed, GSI treatment resulted in reductions in phospho-Akt levels (Figure 5C), however, Akt phosphorylation could not be measured in leukemic cell lines where Pten was expressed. Irrespective of the Pten status, all mouse leukemic lines exhibited increased amounts of phosphorylated mTOR substrates (Table 1). Collectively, these data indicate that mTOR activation is observed in all mouse leukemic cell lines examined (Table 1; n = 10), and that at least in Pten-negative mouse leukemic cells, Notch1 signaling clearly stimulates Akt activity (Figure 5C). In addition, GSI treatment reduces but does not eliminate mTOR activity, indicating that GSIs and the mTOR inhibitor rapamycin may be required to completely inactivate mTOR in leukemic cells.
GSI and rapamycin treatment ablates mTOR kinase activity and induces apoptosis
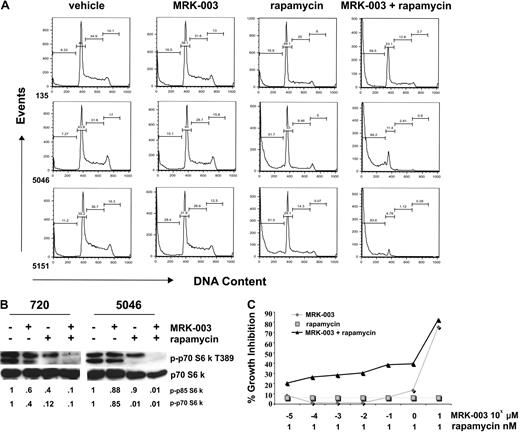
These signaling data suggested that rapamycin treatment may inhibit leukemic growth and that GSI and rapamycin treatment may prove more effective in inducing leukemic cell death. We tested this possibility by treating our mouse leukemic cell lines with vehicle, 1 μM MRK-003, or 10 nM rapamycin, or with 1 μM MRK-003 and 10 nM rapamycin and quantified the apoptotic leukemic cells by PI staining followed by flow cytometry. In all cases, treatment with GSI and rapamycin induced more cell death than treatment with GSI or rapamycin alone (Figure 6A). At the concentrations tested, the effects of MRK-003 and rapamycin treatment on leukemic apoptosis appeared, at a minimum, additive.
GSI and rapamycin treatment in vitro induces massive apoptosis of mouse T-ALL cells and cooperates to suppress mTOR activity. (A) Mouse T-ALL cell lines, 135, 5046, and 5151, were treated with DMSO, 1 μM MRK-003, 10 nM rapamycin, or 1 μM MRK-003 and 10 nM rapamycin for 24 hours. Cells were assayed for DNA content by staining with PI followed by flow cytometry. (B) mTOR activity is ablated when mouse T-ALL lines are treated with GSI and rapamycin. Mouse T-ALL lines, 720 and 5046, were treated with DMSO, 1 μM MRK-003, 10 nM rapamycin, or 1 μM MRK-003 and 10 nM rapamycin, and mTOR kinase activity was assayed by immunoblotting the cell lysates with phospho-p70 S6 kinase antibody (no. 9205; Cell Signaling Technology) after 18 hours. Total p70 S6 kinase was used as a loading control (no. 9202; Cell Signaling Technology). Fold reductions in kinase activity were determined by densitometry and represent ratios (phospho/total) normalized to DMSO-treated samples. (C) At low pharmacologic doses, MRK-003 and rapamycin may have synergistic effects on mouse leukemic growth. Mouse T-ALL lines, 720 and 5046, were treated for 72 hours with 1 nM rapamycin, increasing concentrations of MRK-003 (10−5 to 101 μM), or rapamycin and MRK-003, and growth was assayed by MTT analysis. The figure is a representative of 3 independent experiments using mouse T-ALL cell line 720.
GSI and rapamycin treatment in vitro induces massive apoptosis of mouse T-ALL cells and cooperates to suppress mTOR activity. (A) Mouse T-ALL cell lines, 135, 5046, and 5151, were treated with DMSO, 1 μM MRK-003, 10 nM rapamycin, or 1 μM MRK-003 and 10 nM rapamycin for 24 hours. Cells were assayed for DNA content by staining with PI followed by flow cytometry. (B) mTOR activity is ablated when mouse T-ALL lines are treated with GSI and rapamycin. Mouse T-ALL lines, 720 and 5046, were treated with DMSO, 1 μM MRK-003, 10 nM rapamycin, or 1 μM MRK-003 and 10 nM rapamycin, and mTOR kinase activity was assayed by immunoblotting the cell lysates with phospho-p70 S6 kinase antibody (no. 9205; Cell Signaling Technology) after 18 hours. Total p70 S6 kinase was used as a loading control (no. 9202; Cell Signaling Technology). Fold reductions in kinase activity were determined by densitometry and represent ratios (phospho/total) normalized to DMSO-treated samples. (C) At low pharmacologic doses, MRK-003 and rapamycin may have synergistic effects on mouse leukemic growth. Mouse T-ALL lines, 720 and 5046, were treated for 72 hours with 1 nM rapamycin, increasing concentrations of MRK-003 (10−5 to 101 μM), or rapamycin and MRK-003, and growth was assayed by MTT analysis. The figure is a representative of 3 independent experiments using mouse T-ALL cell line 720.
To obtain a molecular readout of the combination treatment, we measured phospho-p70 and -p85 S6 kinase levels in leukemic cell lines treated with vehicle, GSI, or rapamycin alone or with both GSI and rapamycin. We found that treatment with either GSI or rapamycin reduced phospho-p70 S6 kinase activity, but treatment with either single agent failed to ablate mTOR activation (Figure 6B). In contrast, phospho-p70 S6 kinase and the hyperphosphorylated-p85 S6 kinase were undetectable when leukemic cell lines were treated with both GSI and rapamycin (Figure 6B). These findings indicate that the suppression of the mTOR and Notch1 pathways results in extensive apoptosis of mouse leukemic cells, thereby supporting the idea that GSI and rapamycin may be efficacious in the clinic.
Moreover, our in vitro studies and work done by Chan et al36 support the idea that MRK-003 and rapamycin may have synergistic effects on mouse leukemic growth. To test this possibility, mouse leukemic cells were cultured with 1 nM rapamycin and increasing concentrations of MRK-003 (10−5 to 101 μM) for 72 hours and leukemic growth was assayed using MTT assay. Treatment with 1 nM rapamycin resulted in modest growth inhibition and as expected, MRK-003 caused extensive cell death between the 1- and 10-μM concentrations (Figure 6C). Treatment with both drugs, specifically at low pharmacologic doses, appeared to have synergistic effects, resulting in a greater than additive amount of growth inhibition detected. These results support the idea that reduced concentration of GSIs may be used when administered in combination with rapamycin to increase the therapeutic window.
GSI and rapamycin treatment inhibits human T-ALL growth and prolongs survival
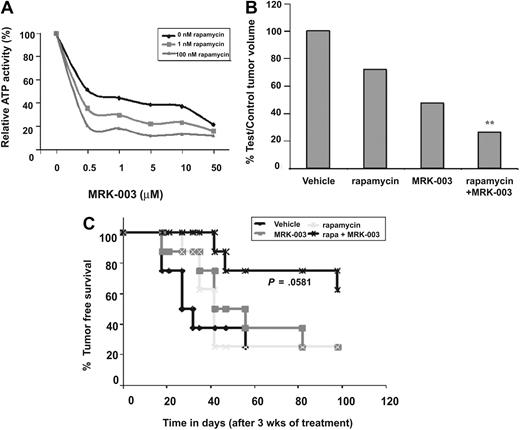
One potential limitation of these studies is that mouse leukemic cell lines appear more sensitive to GSI treatment than established human T-ALL cell lines.20,24,28 GSI treatment of mouse leukemic cell lines induces apoptosis by 48 to 72 hours (Figure 6A and Figure S1B), whereas human T-ALL cell lines require 6 or more days of GSI treatment to induce, at best, a modest G1 arrest.20,24,28 The reasons for the differential response to GSI remain unclear and may reflect activation of additional antiapoptotic pathways in the human T-ALL. Alternatively, mouse leukemic cell line sensitivity to GSIs may reflect inherent differences between the species. To test the effects of GSI and rapamycin treatment on human leukemic growth and survival, human T-ALL cell lines were treated in vitro with increasing concentrations of MRK-003 in the presence of 1 nM or 100 nM rapamycin and ATP activity was measured using a Vialight assay kit. As expected, increasing amounts of GSI reduced human leukemic growth, however the addition of rapamycin enhanced the effects of GSI treatment, resulting in greater inhibition of growth (Figure 7A). Thus, reduced concentrations of GSI may be used in vivo in combination with rapamycin to minimize on target GSI-associated toxicities and to maximize efficacy.
GSI and rapamycin treatment inhibits human T-ALL growth and extends survival. (A) GSI and rapamycin treatment inhibits human T-ALL growth in vitro. The human T-ALL cell line (TALL-1) was treated with 0, 0.5, 1, 5, 10, or 50 μM MRK-003 in addition to 0, 1, or 100 nM rapamycin and ATP activity quantified using the Vialight assay kit. (B) The combination treatment (GSI and rapamycin) inhibits human T-ALL growth in vivo more effectively than treatment with either single agent. CD1 nu/nu mice were injected with human T-ALL cell line, TALL-1. When tumors reached 250 mm3, xenograft mice were treated with vehicle, rapamycin, MRK-003, or a combination of MRK-003 and rapamycin for 3 weeks. MRK-003 was dosed at either 0 or 150 mg/kg by mouth once a week. Rapamycin was dosed at either 0 or 20 mg/kg by mouth daily. After treatment, tumors were callipered and body weight was recorded. Bar graph indicates relative tumor volumes at killing. The following statistics were analyzed by a t test; vehicle versus rapamycin (P = .001), vehicle versus MRK-003 (P = .325), rapamycin versus rapamycin + MRK-003 (P = .002), MRK-003 versus rapamycin + MRK-003 (P = .001). (C) GSI and rapamycin treatment inhibits human leukemic growth in vivo and increases overall survival. After 3 weeks of treatment with vehicle, MRK-003 (150 mg/kg per week), rapamycin (20 mg/kg daily), or MRK-003 and rapamycin, T-ALL-1 xenograft mice were monitored for tumor recurrence. Tumor-free survival was compiled on a Kaplan-Meier survival plot. Data were analyzed by a log rank test (P = .058).
GSI and rapamycin treatment inhibits human T-ALL growth and extends survival. (A) GSI and rapamycin treatment inhibits human T-ALL growth in vitro. The human T-ALL cell line (TALL-1) was treated with 0, 0.5, 1, 5, 10, or 50 μM MRK-003 in addition to 0, 1, or 100 nM rapamycin and ATP activity quantified using the Vialight assay kit. (B) The combination treatment (GSI and rapamycin) inhibits human T-ALL growth in vivo more effectively than treatment with either single agent. CD1 nu/nu mice were injected with human T-ALL cell line, TALL-1. When tumors reached 250 mm3, xenograft mice were treated with vehicle, rapamycin, MRK-003, or a combination of MRK-003 and rapamycin for 3 weeks. MRK-003 was dosed at either 0 or 150 mg/kg by mouth once a week. Rapamycin was dosed at either 0 or 20 mg/kg by mouth daily. After treatment, tumors were callipered and body weight was recorded. Bar graph indicates relative tumor volumes at killing. The following statistics were analyzed by a t test; vehicle versus rapamycin (P = .001), vehicle versus MRK-003 (P = .325), rapamycin versus rapamycin + MRK-003 (P = .002), MRK-003 versus rapamycin + MRK-003 (P = .001). (C) GSI and rapamycin treatment inhibits human leukemic growth in vivo and increases overall survival. After 3 weeks of treatment with vehicle, MRK-003 (150 mg/kg per week), rapamycin (20 mg/kg daily), or MRK-003 and rapamycin, T-ALL-1 xenograft mice were monitored for tumor recurrence. Tumor-free survival was compiled on a Kaplan-Meier survival plot. Data were analyzed by a log rank test (P = .058).
We then tested whether GSI and rapamycin in combination could have additional efficacy against human leukemic growth in vivo. To perform this study, approximately 5 × 106 human T-ALL cells were implanted subcutaneously into nude mice and once the tumors reached 250 mm3, mice were randomized into 4 treatment groups of 8 mice each. Mice were then treated with vehicle, a nonefficacious dose of GSI (MRK-003 150 mg/kg once per week), rapamycin (20 mg/kg daily), or GSI and rapamycin and time to disease progression was determined. Mice treated with GSI or rapamycin exhibited inhibition of tumor progression, as measured by percentage of test normalized to control tumor volume (Figure 7B), however, overall survival was not affected (Figure 7C). In contrast, mice treated with GSI and rapamycin survived on average longer than vehicle-treated mice or mice treated with either single agent (GSI or rapamycin alone; Figure 7C). The administration of GSI and rapamycin resulted in an increase in the percentage of mice surviving (60% vs 25%) as well as an increase in the mean survival period of more than 50 days (P < .058 by log rank test). Mice treated with either GSI or rapamycin succumbed to disease rapidly, with an average latency of approximately 40 days. Collectively, these in vivo data suggest that GSI in combination with the mTOR inhibitor rapamycin may have added efficacy in the treatment of T-ALL patients who exhibit NOTCH1 activation.
Discussion
The prevalence of NOTCH1 and FBXW7 mutations in human T-ALL20,21,38-41 and in mouse T-ALL models16-18 prompted several laboratories to ask whether leukemic growth remained NOTCH1 dependent. GSI treatment of mouse and human T-ALL cell lines and primary mouse tumors expressing mutated Notch1 proteins (Figure S1C) revealed that sustained Notch1 signals appear required for continued growth and survival in vitro.18,20,24,28,42 These findings raise the exciting possibility that Notch1 inhibition may prove effective in treating T-ALL patients and led to the opening of a phase 1 clinical trial involving 8 relapsed leukemia and lymphoma patients. Most of the patients were dosed with a continuous GSI dosing schedule, which was poorly tolerated, and the trial was closed before being able to demonstrate clinical activity of GSIs in T-ALL.43 A well-tolerated intermittent GSI dosing regimen for patients has since been identified, thereby facilitating future clinical evaluations of GSIs in T-ALL and potentially other cancers.
Here we examined the effect of GSI treatment in our mouse T-ALL model where the effects of Notch1 inhibition could be addressed on primary tumors opposed to patients with relapsed or refractory disease. Seventy-four percent of Tal1/Ink4a/Arf+/− transgenic mice develop a T-ALL–like disease due to mutations that result in premature truncation of the Notch1 receptor.18 To investigate whether Notch1 could be targeted in vivo, we developed an intermittent GSI dosing regimen (150 mg/kg, 3 days on and 4 days off) that resulted in minimal toxicity and then tested whether this regimen had any effect on the survival of leukemic Tal1/Ink4a/Arf+/− mice. We found that GSI-treated mice survived on average 15 days longer than leukemic mice treated with vehicle only. The expression of Notch1 target genes Deltex1 and c-Myc was repressed, and increased numbers of apoptotic cells were detected in the thymic masses of diseased mice treated with GSI. These data support the idea that Notch can be successfully targeted in vivo and that treatment with GSI as a single agent has efficacy and extends the survival of leukemic mice.
Despite the expression of other T-ALL–associated oncogenes such as Tal1 and Lmo1/2 and the loss of tumor suppressors (Ink4a/Arf, Pten), mouse T-cell leukemic growth appears preferentially “addicted” to the Notch1 proliferative signal(s). Although GSI treatment significantly increased the overall survival period, it was not sufficient to cure the mice and 100% of the GSI-treated mice eventually succumbed to disease. The transient GSI response was not due to the development of GSI resistance, as all mouse tumors tested remained GSI responsive when reexamined in vitro. One possibility is that the transient response to GSI may reflect the intermittent dosing regimen, and long-term suppression of Notch1 activity may require more frequent GSI administration or combination therapies. Alternatively, chronic GSI treatment may promote reliance on other growth and survival pathways. For example, imatinib mesylate administration to mice with chronic myeloid leukemia (CML)–like disease shifts the reliance from activated ABL toward a greater dependence on the IL-7 signaling pathway.44
As in human T-ALL,45 PTEN loss of expression is frequently observed in our mouse T-ALL model (Table 1). In human T-ALL cell lines and primary samples, loss of PTEN has been associated with resistance to NOTCH1 inhibition.45 Interestingly, 2 of 6 mouse T-ALL cell lines examined harbor Notch1 mutations and express Pten, whereas the remaining 4 mutant Notch1 mouse T-ALL cell lines do not express Pten (Table 1). Yet, all 6 mouse T-ALL cell lines remain GSI responsive, irrespective of their Pten status (Table 1). Unlike the reported role of PTEN in GSI resistance in human T-ALL cell lines,45 we observe no correlation between Pten status and GSI responsiveness. However, it is important to note that GSI-resistant mouse T-ALL cell lines are relatively rare and, thus far, we have isolated only 2 GSI-resistant mouse T-ALL cell lines; one of them retains Pten, whereas the other does not. Nonetheless, it is clear in our mouse T-ALL model, Pten loss does not result in GSI resistance and these mouse T-ALL cell lines remain dependent on Notch1 for their growth and survival.
Although the results from this mouse GSI study are promising, many human T-ALL cell lines are far less responsive to GSI treatment in vitro. Even at increased concentrations and extended incubation periods, only modest amounts of cell death are observed when human T-ALL cell lines or primary leukemic samples are treated with GSIs.20,24,28 These findings cast doubt as to whether GSI or other Notch1 inhibitors will be effective in T-ALL patients. Collectively, these data suggest that combination therapies may be needed to successfully treat T-ALL patients. We provide evidence that combining GSI treatment with the mTOR inhibitor rapamycin limits human leukemic growth in vivo and increases the overall survival. Moreover, the combination treatment is effective after only one GSI dose, indicating that lower concentrations of GSI may prove effective when administered in combination with other antileukemic agents. Similarly, work by Chan et al shows that GSI and rapamycin synergize to limit human T-ALL cell line growth in vitro.36 We extend their in vitro findings and show that Notch and mTOR pathway inhibition limits human leukemic growth in vivo, when tested in a mouse xenograft setting. Thus, whether GSI and rapamycin will prove efficacious remains to be tested in mouse T-ALL models.
Notch receptors and ligands are found up-regulated in multiple human cancers46 and expression of activated Notch alleles results in T-ALL–like disease47-49 and induction of mammary tumors in mice.27 In addition to cell-autonomous effects, Notch pathway inhibition may also have antiangiogenic activity50-53 and thereby render a “double hit” to the malignant cell. Thus, Notch inhibitors may have enormous potential in targeted cancer therapy.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge Jun Zhao of Merck Research Laboratories (Boston, MA) for technical support for PK analysis and Xiaoli Shirley Hou of Merck Research Laboratories (West Point, PA) for statistical analysis. We thank the University of Massachusetts Medical School Flow Cytometry Core for help with FACS analyses and the University of Massachusetts Medical School Morphology Core for assistance with tissue preparation. Core facilities used in this study were supported in part by Program Project Grant P30DK32529 from the National Institute of Diabetes and Digestive and Kidney Diseases.
This research was supported in part by Merck Research Laboratories and by a National Institutes of Health/National Cancer Institute grant (CA096899; M.A.K.). V.M.S. is supported by a Lymphoma Research Foundation Fellowship. M.A.K. is a Leukemia & Lymphoma Society Scholar and a 2009 Stohlman Scholar.
National Institutes of Health
Authorship
Contribution: K.C. provided the data in Figures 1B and 3 through 6 and Figures S1B and 2; K.M.D. provided the data shown in Figures 1C and D and 2B; N.H. assisted with data shown in Figure 1D; V.K. and V.M.S. performed research presented in Figures S1C and 1A, respectively; C.W. performed gut metaplasia, and phospho-S6 and TUNEL stainings; J.T. performed experiments presented in Figures 1A and 7; P.K.M. and G.N. provided the pharmacokinetic analyses shown in Figure 1A and B and the xenograft study shown in Figure 7; P.K.M. and M.A.K. designed the research and analyzed the data; and M.A.K. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michelle A. Kelliher, University of Massachusetts Medical School, Department of Cancer Biology and the UMass Cancer Center, 364 Plantation St, Worcester, MA 01605; e-mail: michelle.kelliher@umassmed.edu; or Pradip K. Majumder, Department of Oncology/Pharmacology, Merck Research Laboratories, 33 Ave Louis Pasteur, Boston, MA-02445; e-mail: pradip_majumder@merck.com.