Abstract
Folylpoly-γ-gluatamate synthetase (FPGS) catalyzes the polyglutamylation and thus intracellular retention of folates and antifolates (eg, methotrexate; MTX) through the addition of multiple glutamate equivalents to their γ-carboxyl residue. Since polyglutamylation of antifolates is crucial for their pharmacological activity in leukemia, loss of FPGS function results in decreased cellular levels of polyglutamylation-dependent antifolates and consequent drug resistance. Whereas resistance to pulse exposure to antifolates is frequently associated with loss of FPGS activity, the underlying molecular mechanism remains elusive. Here we explored the molecular basis of antifolate resistance in human MTX-resistant leukemia cell lines displaying marked loss of FPGS activity. We demonstrate that these MTX-resistant cells exhibit impaired splicing of FPGS mRNA based on intron retention and/or exon skipping, thereby resulting in loss of FPGS function due to premature translation termination. Furthermore, analysis of FPGS transcripts in blood or bone marrow specimens from patients with acute lymphoblastic leukemia revealed exon 12 skipping, both at diagnosis and at relapse, the latter of which occurs after high-dose MTX-containing chemotherapy. These results constitute the first demonstration of the loss of FPGS function via aberrant mRNA splicing, thereby resulting in loss of antifolate retention and drug resistance. The clinical ramifications of these novel findings are discussed.
Introduction
Folates are essential vitamins that serve as one-carbon donors in a variety of biosynthetic pathways including the de novo biosynthesis of purines and thymidylate, mitochondrial protein synthesis, and amino acid conversion.1 Folate analogues (ie, antifolates) exert their cytotoxic activity via potent inhibition of various folate-dependent enzymes that mediate de novo nucleotide biosynthesis, block DNA replication, and hence induce cell death.2 Consequently, antifolates are currently used as important anticancer drugs for the treatment of various human malignancies; in this respect, methotrexate (MTX), the first antifolate introduced nearly 60 years ago, achieved remarkable remissions in acute lymphoblastic leukemia (ALL).3 MTX and novel antifolates including pemetrexed (Alimta) and raltitrexed (Tomudex) are currently used for the treatment of various human cancers including ALL, lymphomas, breast cancer, osteosarcoma, colorectal cancer, malignant pleural mesothelioma, and choriocarcinoma. Antifolates are also used, particularly upon a low-dose regimen, for the treatment of nonneoplastic disorders including rheumatoid arthritis, psoriasis, and Crohn disease. Currently, antifolates are also used for the treatment of parasitic diseases and fungal diseases such as malaria and Pneumocystis carinii pneumonia, respectively
Folate cofactors and antifolates are divalent anions and hence require specific membrane transporters and receptors for their translocation across biologic membranes.4 These transport routes include the proton-coupled folate transporter (PCFT/SLC46A1), the reduced folate carrier (RFC/SLC19A1), and folate receptors (summarized in Assaraf5 and Zhao and Goldman6 ). Once taken up into cells, folates and antifolates undergo polyglutamylation catalyzed by folylpolyglutamate synthetase (FPGS), which adds up to 10 glutamate residues, one at a time, to the γ-carboxyl residue of folates and antifolates.7,8 This unique metabolism renders folates and antifolates polyanions that can no longer be effluxed out of cells, thereby resulting in enhanced intracellular retention. Since polyglutamylation plays a key role in cellular retention of antifolates and thus increases their cytotoxic activity, loss of FPGS activity is an established mechanism of resistance to polyglutamylation-dependent antifolates in vitro and in vivo.9-20 This phenomenon was observed in various malignant cell lines selected for resistance to intermittent exposure to high-dose polyglutamatable antifolates including MTX, raltitrexed, and pemetrexed.9-17,20 Using this modality of pulse drug exposure, the vast majority of antifolate-resistant tumor cell lines displayed a 90% to 99% loss of FPGS activity, frequently in the absence of any substantial decrease in FPGS mRNA levels. Although inactivating FPGS mutations may result in the loss of FPGS activity,10,21 they are relatively infrequent.
In the clinic, high-dose (HD)–MTX is a major component in all contemporary treatment protocols for ALL and much is known about the cellular and molecular pharmacology of MTX in ALL. Impaired polyglutamylation is an established mechanism of resistance to MTX in T-lineage ALL as opposed to c/preB-ALL.18 The ability of lymphoblasts to form and accumulate MTX polyglutamates is a well-recognized determinant of the antileukemic effects of MTX in vivo,19,22 and was therefore shown to be an important prognostic factor positively predicting curative chemotherapy that contains MTX.19 Moreover, Kager et al23 showed that 42 to 44 hours after administration of MTX (0.8 g/m2), T-ALL bone marrow cells displayed a poor formation of long-chain (Glu4-7) MTX polyglutamates (355 pmol/109 cells), whereas MTX polyglutamylation in hyperdiploid (lower than 50 chromosomes) B-cell leukemia was 9-fold higher (3170 pmol/109 cells). Hence, the initial aim of the current study was to explore the posttranscriptional mechanisms involved in the frequent loss of FPGS activity in antifolate-resistant tumor cell lines, particularly when no decrease in FPGS mRNA levels is observed. Toward this end, we here discovered a novel molecular mechanism underlying loss of FPGS activity in antifolate-resistant human ALL cell lines that is based on impaired FPGS splicing including intron retention and/or exon skipping. Moreover, defective FPGS splicing was also identified in bone marrow and peripheral blood samples obtained from ALL patients at diagnosis and at relapse, the latter of which occurs after HD-MTX–containing chemotherapy. This novel modality of antifolate resistance is based upon impaired FPGS splicing as reflected in the retention of introns and/or skipping of exons, thereby resulting in a frameshift in the open reading frame and premature translation termination leading to loss of FPGS function.
Methods
Tissue culture
The human T-cell lymphocytic leukemia lines CCRF-CEM and Molt-4 as well as their antifolate-resistant sublines CEM-MTXR5P, CEM-MTXR10, CEM-7OH MTX, and Molt-4-7OH MTX were maintained in RPMI-1640 medium (GIBCO, Carlsbad, CA) containing 2.3 μM folic acid supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin sulfate (Biological Industries, Beth-Haemek, Israel). The MTX-resistant sublines MTXR5P and MTXR10 were established by exposure of parental CCRF-CEM cells to 6 cycles each of 24-hour pulse of 5 or 10 μM MTX, respectively (Sigma, St Louis, MO) as follows: after an initial 24-hour pulse exposure of parental CCRF-CEM cells to 5 or 10 μM MTX at 37°C, cells were washed once with drug-free growth medium, adjusted to 3 × 105 cells/mL, and allowed to grow for 1 to 2 weeks. Thereafter, a maintenance treatment of 24-hour pulses of MTX was performed once a month. CEM-7OH MTX and Molt-4-7OH MTX (generously provided by Dr F. Albertioni from Karolinska Institute, Stockholm, Sweden) were established and maintained as previously described.14
Antifolate growth inhibition assay
Details are available in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
[3H]MTX transport assay
Details are available in Document S1.
FPGS activity assay
Details are available in Document S1.
Semiquantitative reverse-transcription–polymerase chain reaction analysis of various genes
Cells (107) from the mid-log phase of growth were harvested by centrifugation and washed with phosphate-buffered saline (PBS), and total RNA was isolated using the Tri-Reagent kit according to the instructions of the manufacturer (Sigma) and treated with DNase (Promega, Madison, WI). A portion of total RNA (12 μg in a total volume of 50 μL) was reverse transcribed using Moloney murine leukemia virus (M-MLV; 400 units; Promega) in a reaction buffer containing random hexamer primers (Promega), dNTPs (Larova, Teltow, Germany), and a ribonuclease inhibitor Rnasin (TaKaRa, Kyoto, Japan). Polymerase chain reaction (PCR) was performed using 10 pmol of each primer (Tables 1,Table 2–3) in 2xReddyMix PCR master mix reaction buffer according to the instructions of the manufacturer (ABgene, Epsom, United Kingdom). Then, the PCR products were resolved on 1% agarose gels containing ethidium bromide.
Primer pairs used to screen the entire FPGS gene (accession no. NM_004957)
| Primer . | Position on cDNA . | Length, bp . | Position on gDNA . | Length, bp . |
|---|---|---|---|---|
| EX1-up 5′-CGCGGCATAACGACCCAG-3′ | 108-125 | Exon 1 | ||
| EX3-dw 5′-TTCCCCTTCGTCCCAGTGAC-3′ | 351-370 | 263 | Exon 3 | 1570 |
| EX3-up 5′-CCGGCTGAACATCATCCA-3′ | 332-349 | Exon 3 | ||
| EX10-dw 5′-AGCATCGGACACAGGTATAGA-3′ | 878-898 | 567 | Exon 10 | 4071 |
| EX8-up 5′-TCTCCTCTCTTGGCATCGA-3′ | 712-730 | Exon 8 | ||
| EX13-dw 5′-CGGTCCCCGGTAGCATTGA-3′ | 1288-1306 | 594 | Exon 13 | 2452 |
| EX11-up 5′-CAAAGGCATCCAGGCCAGG-3′ | 1033-1051 | Exon 11 | ||
| EX15-dw 5′-TGCTCTTCGTCCAGGTGGTT-3′ | 1473-1492 | 460 | Exon 15 | 4471 |
| EX15-up 5′-TTCACAGTGACACTGGACCA-3′ | 1416-1435 | Exon 15 | ||
| EX15b-dw 5′-AGTATGTAAGTTCATGGGGAG-3′ | 1862-1882 | 467 | Exon 15 | 467 |
| Primer . | Position on cDNA . | Length, bp . | Position on gDNA . | Length, bp . |
|---|---|---|---|---|
| EX1-up 5′-CGCGGCATAACGACCCAG-3′ | 108-125 | Exon 1 | ||
| EX3-dw 5′-TTCCCCTTCGTCCCAGTGAC-3′ | 351-370 | 263 | Exon 3 | 1570 |
| EX3-up 5′-CCGGCTGAACATCATCCA-3′ | 332-349 | Exon 3 | ||
| EX10-dw 5′-AGCATCGGACACAGGTATAGA-3′ | 878-898 | 567 | Exon 10 | 4071 |
| EX8-up 5′-TCTCCTCTCTTGGCATCGA-3′ | 712-730 | Exon 8 | ||
| EX13-dw 5′-CGGTCCCCGGTAGCATTGA-3′ | 1288-1306 | 594 | Exon 13 | 2452 |
| EX11-up 5′-CAAAGGCATCCAGGCCAGG-3′ | 1033-1051 | Exon 11 | ||
| EX15-dw 5′-TGCTCTTCGTCCAGGTGGTT-3′ | 1473-1492 | 460 | Exon 15 | 4471 |
| EX15-up 5′-TTCACAGTGACACTGGACCA-3′ | 1416-1435 | Exon 15 | ||
| EX15b-dw 5′-AGTATGTAAGTTCATGGGGAG-3′ | 1862-1882 | 467 | Exon 15 | 467 |
Diagnostic PCR primers used to identify splicing alterations in the FPGS gene (accession no. NM_004957)
| Primer . | Position on gDNA . | Length, bp . |
|---|---|---|
| INT10 5′-GAGTGGGCAGCTGAGTGGG-3′* | Intron 10 | 1368 |
| ex9-10 5′-GCCCAGCAGATCTCATGTCC-3′* | 3′ of exon 9+ 5′ of exon 10 | 449 |
| ex9-11 5′-CAGATCTCATGCTGGGGAGC-3′* | 3′ of exon 9+ 5′ of exon 11 | 294 |
| ex10in10 5′-AGGACCGCCATGGTGAGTG-3′* | 3′ of exon 10+ 5′ of intron 10 | 393 |
| ex11-12 5′-GCTCGGGCTTCGGAACAC-3′† | 3′ of exon 11+ 5′ of exon 12 | 387 |
| ex11-13 5′-CCCACATGCGGCTCGTG-3′† | 3′ of exon 11+ 5′ of exon 13 | 245 |
| Primer . | Position on gDNA . | Length, bp . |
|---|---|---|
| INT10 5′-GAGTGGGCAGCTGAGTGGG-3′* | Intron 10 | 1368 |
| ex9-10 5′-GCCCAGCAGATCTCATGTCC-3′* | 3′ of exon 9+ 5′ of exon 10 | 449 |
| ex9-11 5′-CAGATCTCATGCTGGGGAGC-3′* | 3′ of exon 9+ 5′ of exon 11 | 294 |
| ex10in10 5′-AGGACCGCCATGGTGAGTG-3′* | 3′ of exon 10+ 5′ of intron 10 | 393 |
| ex11-12 5′-GCTCGGGCTTCGGAACAC-3′† | 3′ of exon 11+ 5′ of exon 12 | 387 |
| ex11-13 5′-CCCACATGCGGCTCGTG-3′† | 3′ of exon 11+ 5′ of exon 13 | 245 |
PCR primer pairs used to examine the splicing of various genes
| Gene . | Accession no. . | Primer . | Position on cDNA . | Length, bp . | Position on gDNA . | Length, bp . |
|---|---|---|---|---|---|---|
| β-actin | CR624852 | β-actin-up 5′-CCGTCTTCCCCTCCATCGTG-3′ | 160-179 | 576 | Exon 2 | 1151 |
| β-actin-dw 5′-GGGCGACGTAGCACAGCTTCT-3′ | 715-735 | Exon 4 | ||||
| RNA Polymerase I D | NM_015972 | RNA-up 5′-TGGAAGAGGATCAGGAGCTGG-3′ | 94-114 | 310 | Exon 1 | 1176 |
| RNA-dw 5′-CATGAGCTCATTCAGGCCTCTC-3′ | 383-404 | Exon 2 | ||||
| MRP1 | NM_004996 | MRP-EX23 5′-GTGCGGCTGGAGTGTGTGG-3′ | 3776-3794 | 198 | Exon 23 | 1092 |
| MRP-EX24 5′-CTCCTTGAGCCTCTCCACGG-3′ | 3954-3973 | Exon 24 | ||||
| TS | NM_001071 | TS-U5 5′-CCTGAATCACATCGAGCCAC-3′ | 879-898 | 142 | Exon 6 | 1550 |
| TS-D4 5′-TGGATGCGGATTGTACCCTT-3′ | 1001-1020 | Exon 7 | ||||
| DHFR | NM_000791 | DHFR-EX1 5′-TCGCTAAACTGCATCGTCGC-3′ | 502-521 | 115 | Exon 1 | 460 |
| DHFR-D1 5′-AGGTTGTGGTCATTCTCTGG-3′ | 597-616 | Exon 2 | ||||
| RFC | U19720.1 | RFC2-up 5′-GGGCGTGTTCACCAGCTC-3′ | 581-598 | 576 | Exon 3 | 993 |
| RFC2-dw 5′-GCCAGAAGGAAGACCAGCC-3′ | 1138-1156 | Exon 4 | ||||
| PDGF | M63193 | TP02-up 5′-GCTTCGTGGCCGCTGTGGTG-3′ | 290-309 | 350 | Exon 2 | 1030 |
| TP02-dw 5′-CTGCTCTGGGCTCTGGATGA-3′ | 620-639 | Exon 4 | ||||
| GAPDH | NM_002046 | GAP-up 5′-AGGGGGGAGCCAAAAGGG-3′ | 440-457 | 541 | Exon 6 | 826 |
| GAP-up 5′-GAGGAGTGGGTGTCGCTGTTG-3′ | 960-980 | Exon 8 | ||||
| TNFα | NM_000594 | TNFaG-up 5′-GATCATCTTCTCGAACCCCG-3′ | 402-420 | 301 | Exon 3 | 602 |
| TNFaG-dw 5′-TGGCAGGGGCTCTTGATG-3′ | 685-702 | Exon 4 | ||||
| UBTF | NM_014233 | UBTF-up 5′-TGGTCCCAGGAAGACATGCTG-3′ | 131-151 | 356 | Exon 2 | 991 |
| UBTF-dw 5′-AGGTTGCTCATCTCAGGGTGG-3′ | 466-486 | Exon 4 | ||||
| Plexin A1 | NM_032242 | Plex-up 5′-AGCTCCCCACTCATCCTCAAG-3′ | 3431-3451 | 362 | Exon 17 | 847 |
| Plex-dw 5′-TCCATCTGGAGCTGCAGCC-3′ | 3774-3792 | Exon 19 |
| Gene . | Accession no. . | Primer . | Position on cDNA . | Length, bp . | Position on gDNA . | Length, bp . |
|---|---|---|---|---|---|---|
| β-actin | CR624852 | β-actin-up 5′-CCGTCTTCCCCTCCATCGTG-3′ | 160-179 | 576 | Exon 2 | 1151 |
| β-actin-dw 5′-GGGCGACGTAGCACAGCTTCT-3′ | 715-735 | Exon 4 | ||||
| RNA Polymerase I D | NM_015972 | RNA-up 5′-TGGAAGAGGATCAGGAGCTGG-3′ | 94-114 | 310 | Exon 1 | 1176 |
| RNA-dw 5′-CATGAGCTCATTCAGGCCTCTC-3′ | 383-404 | Exon 2 | ||||
| MRP1 | NM_004996 | MRP-EX23 5′-GTGCGGCTGGAGTGTGTGG-3′ | 3776-3794 | 198 | Exon 23 | 1092 |
| MRP-EX24 5′-CTCCTTGAGCCTCTCCACGG-3′ | 3954-3973 | Exon 24 | ||||
| TS | NM_001071 | TS-U5 5′-CCTGAATCACATCGAGCCAC-3′ | 879-898 | 142 | Exon 6 | 1550 |
| TS-D4 5′-TGGATGCGGATTGTACCCTT-3′ | 1001-1020 | Exon 7 | ||||
| DHFR | NM_000791 | DHFR-EX1 5′-TCGCTAAACTGCATCGTCGC-3′ | 502-521 | 115 | Exon 1 | 460 |
| DHFR-D1 5′-AGGTTGTGGTCATTCTCTGG-3′ | 597-616 | Exon 2 | ||||
| RFC | U19720.1 | RFC2-up 5′-GGGCGTGTTCACCAGCTC-3′ | 581-598 | 576 | Exon 3 | 993 |
| RFC2-dw 5′-GCCAGAAGGAAGACCAGCC-3′ | 1138-1156 | Exon 4 | ||||
| PDGF | M63193 | TP02-up 5′-GCTTCGTGGCCGCTGTGGTG-3′ | 290-309 | 350 | Exon 2 | 1030 |
| TP02-dw 5′-CTGCTCTGGGCTCTGGATGA-3′ | 620-639 | Exon 4 | ||||
| GAPDH | NM_002046 | GAP-up 5′-AGGGGGGAGCCAAAAGGG-3′ | 440-457 | 541 | Exon 6 | 826 |
| GAP-up 5′-GAGGAGTGGGTGTCGCTGTTG-3′ | 960-980 | Exon 8 | ||||
| TNFα | NM_000594 | TNFaG-up 5′-GATCATCTTCTCGAACCCCG-3′ | 402-420 | 301 | Exon 3 | 602 |
| TNFaG-dw 5′-TGGCAGGGGCTCTTGATG-3′ | 685-702 | Exon 4 | ||||
| UBTF | NM_014233 | UBTF-up 5′-TGGTCCCAGGAAGACATGCTG-3′ | 131-151 | 356 | Exon 2 | 991 |
| UBTF-dw 5′-AGGTTGCTCATCTCAGGGTGG-3′ | 466-486 | Exon 4 | ||||
| Plexin A1 | NM_032242 | Plex-up 5′-AGCTCCCCACTCATCCTCAAG-3′ | 3431-3451 | 362 | Exon 17 | 847 |
| Plex-dw 5′-TCCATCTGGAGCTGCAGCC-3′ | 3774-3792 | Exon 19 |
DNA sequencing
Details are available in Document S1.
Expression vectors
pcDNA3/hFPGS harboring the full-length human FPGS cDNA was generously provided by Prof R. G. Moran from Virginia Commonwealth University School of Medicine (Richmond, VA). To construct a vector containing the FPGS gene lacking exon 10, the FPGS cDNA was PCR amplified with PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) using the following primers: Nhe/FPGS 5′-tatagctagccaccatggagtaccaggatg-3′ and FPGS/EcoRI 5′-ttaagaattcgccttggctactgggac-3′; the PCR product was then digested with NheI and HindIII (Fermentas, Hanover, MD) and cloned into pIRES2-EGFP (Clontech, Palo Alto, CA) upstream of the IRES element. Deletion of exon 10 was performed using the Site-Directed Mutagenesis kit (Stratagene) and the new vector was termed pIRES/FPGSΔexon10.
Stable transfections with hFPGS expression constructs
Exponentially growing cells (ie, CCRF-CEM and CEM-MTXR10) were harvested by centrifugation and stably transfected by electroporation (1000 μF, 234 V) with 10 μg of the vectors pcDNA3.1, pIRES2-EGFP, pcDNA3/hFPGS, or pIRES/FPGSΔexon10. After 24 hours of growth at 37°C, cells were exposed to 450 to 600 μg/mL active G-418 and single clones were then picked and expanded. Stable transfectants obtained after 4 weeks of G-418 selection were analyzed for FPGS mRNA levels by Northern blots as detailed in Document S1 and used for further analyses.
Northern blot analysis
Details are available in Document S1.
Leukemia patient specimens
Analysis of FPGS splicing defects was performed on stored RNA samples previously obtained from adult ALL patients who were treated according to the UKALL12/ECOG 2993 protocol24 at the Department of Hematology, Rambam Medical Center. The samples were previously derived as part of the routine clinical management and were used in the current study after obtaining approval from the local IRB committee (study no. 2902) at the Rambam Medical Center and informed consent in accordance with the Declaration of Helsinki. We studied 13 RNA samples derived from 8 ALL patients, 5 of which were matched samples both at the time of first diagnosis and relapse, as well as 3 unpaired diagnosis samples. The samples had been originally used (ie, irrespective of the current study) for RNA extraction; leukocytes were isolated from either peripheral blood or bone marrow by a standard Ficoll-Hypaque density centrifugation and total RNA was purified using the Tri-Reagent kit according to the instructions of the manufacturer (Sigma). Aliquots of RNA stored at − 80°C were reverse transcribed as described under “Semiquantitative reverse-transcription–polymerase chain reaction analysis of various genes.” The entire FPGS gene was amplified by PCR, screened for either intron retention and/or exon skipping using primers from Tables 1 and 2 as well as from Liani et al.10
Results
Resistance to polyglutamatable antifolates in MTXR5P leukemia cells is associated with a marked loss of FPGS activity
In order for polyglutamatable antifolates to exert their cytotoxic activity, they must first undergo polyglutamylation catalyzed by FPGS. Hence, loss of FPGS activity results in resistance to polyglutamylation-dependent antifolates.5,10-16 Consistently, MTXR5P leukemia cells were found here to display less than 1% of FPGS activity relative to their parental CCRF-CEM cells; this profound loss of FPGS activity was comparable with that of another FPGS-deficient cell line CEM/MTXR10 that we also established (Table 4). Moreover, RFC (SLC19A1), the predominant transporter mediating the uptake of reduced folates and antifolates, retained as much as 50% of its [3H]MTX transport activity, compared with parental cells (Table 4). Hence, a substantial preservation of RFC activity was observed in MTXR5P cells. The major loss of FPGS activity in MTXR5P cells resulted in a 50 000-fold resistance to raltitrexed (ZD1694), a polyglutamylation-dependent antifolate that exerts its cytotoxic activity via potent inhibition of thymidylate synthase (TS; Table 4). In contrast, compared with their parental cells, MTXR5P cells were 2-fold more sensitive to the polyglutamylation-independent antifolate, ZD9331, that also potently inhibits TS activity. Thus, these results suggest that the mechanism underlying antifolate resistance in MTXR5P cells is loss of FPGS activity.
Values for growth inhibition, MTX influx and FPGS activity assays
| Cell line/antifolate . | ZD1694, IC50, nM . | ZD9331, IC50, nM . | MTX, IC50, nM . | MTX influx, % . | FPGS activity, % . |
|---|---|---|---|---|---|
| CCRF-CEM WT | 6.06 ± 0.42 | 28.6 ± 2.3 | 25.7 ± 2.1 | 100 | 100 |
| CEM-MTXR5P | > 300 000 | 13.9 ± 1.7 | 23.1 ± 2 | 50 ± 5 | 0.8 ± 0.2 |
| CEM-MTXR10 | 110.1 ± 9.8 | 40.8 ± 4.1 | 28.2 ± 2.9 | 916 + 6 | 1.2 ± 0.4 |
| Cell line/antifolate . | ZD1694, IC50, nM . | ZD9331, IC50, nM . | MTX, IC50, nM . | MTX influx, % . | FPGS activity, % . |
|---|---|---|---|---|---|
| CCRF-CEM WT | 6.06 ± 0.42 | 28.6 ± 2.3 | 25.7 ± 2.1 | 100 | 100 |
| CEM-MTXR5P | > 300 000 | 13.9 ± 1.7 | 23.1 ± 2 | 50 ± 5 | 0.8 ± 0.2 |
| CEM-MTXR10 | 110.1 ± 9.8 | 40.8 ± 4.1 | 28.2 ± 2.9 | 916 + 6 | 1.2 ± 0.4 |
MTXR5P cells display retention of introns in the mRNA of FPGS and other genes
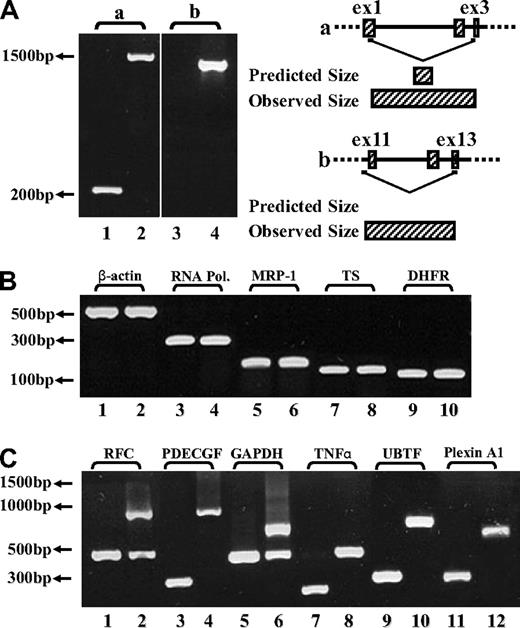
To explore the molecular basis of the loss of FPGS function in MTXR5P cells, the entire FPGS coding region was screened for inactivating mutations using overlapping reverse transcription (RT)–PCR primers (Table 1). While screening for mutations in MTXR5P cells, we surprisingly obtained markedly longer FPGS PCR fragments than expected (Figure 1A lanes 2,4), whereas parental cells showed a PCR product with the expected size or no product at all (Figure 1A lanes 1 and 3, respectively). DNA sequencing of these long PCR products from MTXR5P cells revealed that introns 1, 2, 10, 11, and 12 were retained in the FPGS mRNA (Figure 1A; introns 1,2 and 10-12: lanes 2 and 4, respectively). These findings were indicative of defective splicing that could constitute the molecular basis for the loss of FPGS activity in these antifolate-resistant cells. To examine the status of global splicing in MTXR5P cells, we undertook an RT-PCR analysis on an assortment of genes using primers positioned within neighboring exons, thereby allowing for the formation of 2 distinct fragments that are diagnostic of intron retention. The different primers used for this PCR analysis and the various possible product sizes are depicted in Table 3 and the PCR products are shown in Figure 1B and C. Interestingly, although essential genes such as RNA polymerase ID and β-actin exhibit normal size PCR products (Figure 1B), a variety of other genes showed defective splicing; the latter genes included upstream binding transcription factor, UBTF; tumor necrosis factor α, TNFα; glyceraldehyde-3-phosphate dehydrogenase, GAPDH; platelet-derived growth factor, PDGF; RFC; as well as plexin A1. Therefore, at least 50% of the transcripts encoded by these genes contained unspliced introns, thereby corroborating the prominent splicing defect affecting various genes in MTXR5P cells (Figure 1C). To verify the presence and identity of the unspliced introns, DNA sequencing of all aberrant RT-PCR products was undertaken. Sequencing corroborated the presence of the expected introns in the aberrant unspliced PCR products (Table 3). These results establish that antifolate resistance in MTXR5P cells is associated with a splicing defect that impairs the splicing of multiple genes.
RT-PCR analysis reveals the presence of introns in various cDNAs. (A) (Left panel) PCR was performed on the FPGS gene using 2 sets of forward and reverse primers. (i) Primers residing within the first and third exons (EX1-up and EX3-dw). RT-PCR on cDNA from CCRF-CEM cells produced the expected product size (eg, 263 bp, lane 1), whereas the MTXR5P cells cDNA yielded a markedly longer product of 1570 bp (lane 2). (ii) PCR was performed to confirm the presence of intron 10 in cDNA from MTXR5P cells using a forward primer (INT10) residing within this intron and EX13-dw (lane 4). The white vertical line has been inserted to indicate repositioned gel lanes. (Right panel) Scheme illustrating the positions of the primers as well as the predicted and observed PCR products. (B,C) PCR was performed on various genes using a panel of forward and reverse primers (Table 3) residing within neighbor exons to obtain different product sizes revealing normal and unspliced cDNAs. (B) RT-PCR on cDNA from parental and MTXR5P cells produced the expected product sizes. (C) RT-PCR on WT cDNA produced normal size products, whereas MTXR5P cell's cDNA revealed longer fragments as predicted for unspliced cDNA containing introns; all sizes of PCR fragments are depicted in Table 3.
RT-PCR analysis reveals the presence of introns in various cDNAs. (A) (Left panel) PCR was performed on the FPGS gene using 2 sets of forward and reverse primers. (i) Primers residing within the first and third exons (EX1-up and EX3-dw). RT-PCR on cDNA from CCRF-CEM cells produced the expected product size (eg, 263 bp, lane 1), whereas the MTXR5P cells cDNA yielded a markedly longer product of 1570 bp (lane 2). (ii) PCR was performed to confirm the presence of intron 10 in cDNA from MTXR5P cells using a forward primer (INT10) residing within this intron and EX13-dw (lane 4). The white vertical line has been inserted to indicate repositioned gel lanes. (Right panel) Scheme illustrating the positions of the primers as well as the predicted and observed PCR products. (B,C) PCR was performed on various genes using a panel of forward and reverse primers (Table 3) residing within neighbor exons to obtain different product sizes revealing normal and unspliced cDNAs. (B) RT-PCR on cDNA from parental and MTXR5P cells produced the expected product sizes. (C) RT-PCR on WT cDNA produced normal size products, whereas MTXR5P cell's cDNA revealed longer fragments as predicted for unspliced cDNA containing introns; all sizes of PCR fragments are depicted in Table 3.
DNA sequencing reveals alternatively spliced FPGS transcripts in CEM-7OH-MTX cells
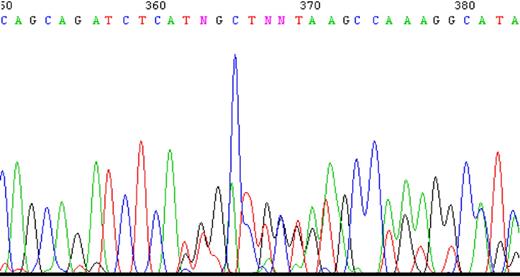
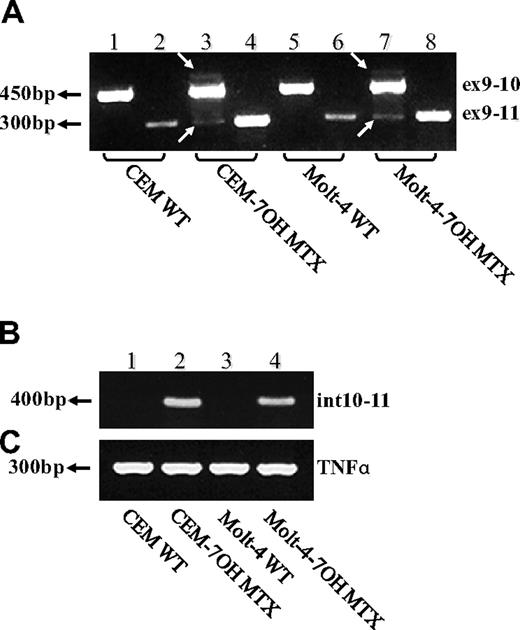
To determine whether impaired splicing of the FPGS gene occurs in other antifolate-resistant human leukemia cell lines, we used CEM-7OH MTX and Molt-4-7OH MTX cells that were recently shown to display 98% loss of FPGS activity.14 These cell lines acquired resistance to 7-OH-MTX, the primary metabolite of MTX. Thus, FPGS cDNAs from these cell lines were first screened for point mutations and splicing alterations by PCR (primers are depicted in Table 1) and then sequenced. DNA sequencing of the PCR product obtained with primers EX8-up and EX13-dw (Table 1) revealed the existence of 2 different transcripts differing in the presence or absence of exon 10 (Figure 2). Since the nucleotide signal peaks of the aberrant transcript (ie, that lacks exon 10) were either equivalent or higher than those of the expected transcript, we hypothesized that this was a major phenotype of alternative splicing. Following this observation, diagnostic primers were specifically designed to PCR amplify FPGS transcripts that contain or lack exon 10. Thus, half of the forward primer resides in the 3′-end of exon 9, whereas the other half maps to the beginning of either exon 10 or exon 11 (ie, primers ex9-10 and ex9-11, respectively; Table 2). PCR performed on both CEM-7OH MTX and Molt-4-7OH MTX cells resulted in high levels of PCR products lacking exon 10 (Figure 3A lanes 3 and 4 as well as 7 and 8), whereas parental cells exhibited residual levels of this exon 10–free product (Figure 3A lanes 2 and 6). PCR performed on these mutants' cDNA resulted in multiple bands (Figure 3A lanes 3 and 7, white arrows), thereby suggesting the presence of additional splicing alterations in the FPGS transcript. The upper most band containing an additional approximately 100 bp may be indicative of the presence of intron 10, which is 95 bp long. To confirm the presence of intron 10 in some of the FPGS transcripts in these splicing defective cell lines, we designed a diagnostic primer, half of which resides in the 3′-end of exon 10 and the other half on the 5′-end of intron 10 (ie, primer ex10-int10; Table 2). As evident from Figure 3B, no visible product is obtained in parental cell lines (Figure 3B lanes 1 and 3), whereas antifolate-resistant cells exhibit the predicted product containing intron 10 (Figure 3B lanes 2 and 4). DNA sequencing performed on all PCR products confirmed the presence of intron 10. Although FPGS splicing is not completely deficient, we PCR amplified the TNFα gene to explore whether other genes remain intact. Using the same primer set as in the MTXR5P cells, we obtained a normal PCR product size of TNFα (Figure 3C).
A sample of the tracing of DNA sequencing illustrating the existence of 2 separate sequences obtained with 2 primers. PCR product was produced using primers EX8-up and Ex13-dw and then sequenced. Two nucleotides appear at the same position starting at nucleotide 362, one sequence matching the expected exon 10, whereas the other matches exon 11 ahead of its time.
A sample of the tracing of DNA sequencing illustrating the existence of 2 separate sequences obtained with 2 primers. PCR product was produced using primers EX8-up and Ex13-dw and then sequenced. Two nucleotides appear at the same position starting at nucleotide 362, one sequence matching the expected exon 10, whereas the other matches exon 11 ahead of its time.
RT-PCR analysis reveals exon skipping and intron inclusion in the FPGS cDNA. PCR was performed on the FPGS gene using 3 sets of forward primers with EX13-dw. (A) Primer ex9-10 residing in the junction between exons 9 and 10 (lanes 1, 3, 5, 7), primer ex9-11 residing in the 3′-end of exon 9 and 5′-end of exon 11 and thus being diagnostic of exon 10 skipping (lanes 2, 4, 6, 8). (B) PCR was performed to corroborate the presence of intron 10 in CEM-7OH MTX and Molt-4-7OH MTX cDNA using a forward primer (ex10int10) on the junction between exon 10 and intron 10. (C) TNFα as a control gene for normal splicing.
RT-PCR analysis reveals exon skipping and intron inclusion in the FPGS cDNA. PCR was performed on the FPGS gene using 3 sets of forward primers with EX13-dw. (A) Primer ex9-10 residing in the junction between exons 9 and 10 (lanes 1, 3, 5, 7), primer ex9-11 residing in the 3′-end of exon 9 and 5′-end of exon 11 and thus being diagnostic of exon 10 skipping (lanes 2, 4, 6, 8). (B) PCR was performed to corroborate the presence of intron 10 in CEM-7OH MTX and Molt-4-7OH MTX cDNA using a forward primer (ex10int10) on the junction between exon 10 and intron 10. (C) TNFα as a control gene for normal splicing.
Northern blot analysis reveals the absence of a principal FPGS transcript in CEM-7OH MTX cells
We have recently studied the mechanism underlying antifolate resistance in CEM-7OH MTX and Molt-4-7OH MTX leukemia cells14 ; in these cell lines, FPGS activity was decreased by at least 98%, whereas no quantitative alteration was observed at the FPGS mRNA level. Based on our current findings regarding the defective splicing of the FPGS gene in MTXR5P cells, we hypothesized that RT-PCR may misrepresent the actual FPGS mRNA levels as a result of the specific position of the primers on the FPGS cDNA. We therefore performed Northern blot analysis to determine quantitatively and qualitatively the actual levels and sizes of the FPGS transcripts in these 7OH MTX-resistant mutants. As evident from Figure 4A, no discrete FPGS transcript could be detected in CEM-7OH MTX cells (lane 2); instead, a smear spanning several hundreds of base pairs was apparent. Moreover, MTXR10 cells, which retained only 1.2% of parental FPGS activity (Table 4), displayed an extended smear as well as a band, though being slightly lower in size than the expected principal FPGS transcript (Figure 4A lane 3).
Northern blot analysis of FPGS mRNA levels. RNA was blotted onto Zeta-Probe nylon membrane and hybridized with a [32P]-labeled FPGS probe as detailed under “Northern blot analysis.” (A) RNA from parental CCRF-CEM and Molt-4 cells (lanes 1 and 4, respectively) as well as antifolate-resistant CEM-7OH MTX, CEM MTXR10, and Molt-4-7OH MTX cells (lanes 2, 3, and 5, respectively). A discrete band with an expected size of 2400 bp can be observed for both parental cell lines (lanes 1 and 4), whereas the antifolate-resistant cells CEM-7OH MTX and MTXR10 exhibit a smear spanning several hundreds of base pairs (lanes 2 and 3, respectively). (C) RNA from parental CCRF-CEM cells and their pIRES/FPGSΔexon10 transfectant (lanes 1 and 2, respectively) as well as the antifolate-resistant cell line CEM MTXR10, its pcDNA3/hFPGS transfectant and its pIRES/FPGSΔexon10 transfectant (lanes 3 and 4, respectively). pIRES/FPGSΔexon10 is transcribed to yield an approximately 3550-bp-long operon including the FPGS gene, the IRES translation initiation sequence, and the EGFP gene. (B,D) The intensity of the FPGS transcript was normalized to the methylene blue staining of the 18S ribosomal RNA band.
Northern blot analysis of FPGS mRNA levels. RNA was blotted onto Zeta-Probe nylon membrane and hybridized with a [32P]-labeled FPGS probe as detailed under “Northern blot analysis.” (A) RNA from parental CCRF-CEM and Molt-4 cells (lanes 1 and 4, respectively) as well as antifolate-resistant CEM-7OH MTX, CEM MTXR10, and Molt-4-7OH MTX cells (lanes 2, 3, and 5, respectively). A discrete band with an expected size of 2400 bp can be observed for both parental cell lines (lanes 1 and 4), whereas the antifolate-resistant cells CEM-7OH MTX and MTXR10 exhibit a smear spanning several hundreds of base pairs (lanes 2 and 3, respectively). (C) RNA from parental CCRF-CEM cells and their pIRES/FPGSΔexon10 transfectant (lanes 1 and 2, respectively) as well as the antifolate-resistant cell line CEM MTXR10, its pcDNA3/hFPGS transfectant and its pIRES/FPGSΔexon10 transfectant (lanes 3 and 4, respectively). pIRES/FPGSΔexon10 is transcribed to yield an approximately 3550-bp-long operon including the FPGS gene, the IRES translation initiation sequence, and the EGFP gene. (B,D) The intensity of the FPGS transcript was normalized to the methylene blue staining of the 18S ribosomal RNA band.
The truncated form of FPGS due to exon 10 skipping has no dominant negative effect on the activity of the wt FPGS
The absence of exon 10 from the FPGS transcript in CEM-7OH MTX and Molt-4-7OH MTX cells introduces both a frameshift and a premature stop codon, which may result in a truncated FPGS protein containing only 334 amino acids. Since we found equal levels of FPGS transcripts that contain or lack exon 10, we further determined whether this truncated protein is capable of modulating the activity of the normal FPGS protein. We therefore generated stable transfectants expressing the FPGS gene either with or without exon 10 in both parental CCRF-CEM and MTXR10 cells. Expression levels of these exogenous genes was verified by Northern blot analysis (Figure 4C), following which FPGS activity assay and antifolate cytotoxicity experiments were undertaken. Ectopic overexpression of the normal FPGS in MTXR10 cells resulted in an 800% increase in FPGS activity relative to parental cells and consequently fully restored parental cell sensitivity to ZD1694. In contrast, the truncated form of FPGS lacking exon 10 had no effect on FPGS activity. Moreover, expressing this truncated form in parental cells failed to alter antifolate sensitivity or the catalytic activity of the endogenous FPGS.
ALL specimens exhibit FPGS exon 12 skipping
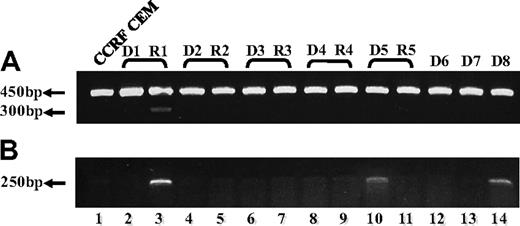
To explore the clinical relevance of our current findings of defective FPGS splicing in antifolate-resistant leukemia cell lines, we undertook an initial study aimed at screening for alterations in FPGS splicing in 13 RNA samples previously obtained from 8 ALL patients, 5 of which were matched samples at diagnosis and relapse. PCR analysis was designed to confirm the presence of all 15 exons and the absence of all introns (excluding introns 4 and 14, which are too long to be amplified using conventional PCR methodology). As depicted in Figure 5A, while using a primer set residing within exons 9 and 13, one bone marrow sample from a relapsed ALL patient (sample R1) who underwent HD-MTX–containing chemotherapy, exhibited a low-molecular-weight FPGS transcript (Figure 5A lane 3), which was absent at the time of diagnosis (sample D1, lane 2). DNA sequencing of this PCR product revealed the absence of FPGS exon 12. A second PCR was performed (Figure 5B) with the primer ex11-13 (Table 2) that can anneal only to FPGS cDNA lacking exon 12, thereby producing a 250-bp product that is diagnostic of exon 12 skipping. Consistent with the latter results, relapse sample R1 exhibited this 250-bp product that was absent at the time of diagnosis (ie, sample D1), thereby verifying FPGS exon 12 skipping at relapse (Figure 5B lanes 2 and 3, respectively). Moreover, 2 diagnosis specimens (Figure 5B; D5, D8: lanes 10 and 14, respectively) also exhibited low levels of the 250-bp product. Hence, of the 8 ALL patient specimens analyzed, 3 harbored FPGS exon 12 skipping that results in premature translation termination.
RT-PCR analysis reveals skipping of FPGS exon 12 in RNA from ALL patient specimens. PCR was performed on cDNA prepared from 5 ALL patient specimens at first diagnosis (lanes 2, 4, 6, 8, 10, 12-14) and at relapse (lanes 3, 5, 7, 9, 11), using primer ex9-10 residing in the junction between exons 9 and 10 and primer EX13-dw (A), or ex11-13 residing in the junction between exons 11 and 13 and primer EX15-dw (B). (A) All samples (including CCRF CEM, lane 1) exhibited the expected 450-bp-long product, whereas relapse sample R1 (lane 3) displayed low levels of another 300-bp product. DNA sequencing corroborated the absence of exon 12 in this 300-bp product. (B) This diagnostic PCR results in a 250-bp product only when exon 12 is absent, and was obtained for relapse sample R1 (lane 3) and to a lower extent in 2 diagnosis samples D5 and D8 (lanes 10 and 14, respectively).
RT-PCR analysis reveals skipping of FPGS exon 12 in RNA from ALL patient specimens. PCR was performed on cDNA prepared from 5 ALL patient specimens at first diagnosis (lanes 2, 4, 6, 8, 10, 12-14) and at relapse (lanes 3, 5, 7, 9, 11), using primer ex9-10 residing in the junction between exons 9 and 10 and primer EX13-dw (A), or ex11-13 residing in the junction between exons 11 and 13 and primer EX15-dw (B). (A) All samples (including CCRF CEM, lane 1) exhibited the expected 450-bp-long product, whereas relapse sample R1 (lane 3) displayed low levels of another 300-bp product. DNA sequencing corroborated the absence of exon 12 in this 300-bp product. (B) This diagnostic PCR results in a 250-bp product only when exon 12 is absent, and was obtained for relapse sample R1 (lane 3) and to a lower extent in 2 diagnosis samples D5 and D8 (lanes 10 and 14, respectively).
Discussion
The present paper provides the first evidence that aberrant splicing of FPGS mRNA, including intron retention and/or exon skipping, results in premature translation termination, loss of FPGS activity, and consequent antifolate resistance in human leukemia cell lines. At the clinical setting, although RNA specimens are not routinely derived at the time of ALL relapse, we were able to study 13 ALL specimens from 8 different patients, 5 of which were matched samples obtained both at the time of diagnosis and at relapse, along with 3 additional diagnosis samples. This preliminary study identified exon 12 skipping in FPGS transcripts both at diagnosis and relapse, thereby suggesting a possible role in the acquisition of ALL resistance to HD-MTX (3 g/m2). It is likely that this antifolate resistance factor may contribute, among other potential unknown mechanisms, to disease relapse. It should be noted that since chemotherapy of ALL is a sequential combination-based therapy, it is therefore difficult to attribute the specific contribution of a specific drug resistance modality to the patient outcome. However, MTX is currently considered a key drug in the treatment of ALL and MTX resistance may therefore affect a patient's outcome.
MTXR5P, the first leukemia cell line that was found here to harbor an FPGS splicing defect, retained introns 1, 2, 10, 11, and 12. It should be noted that the presence of the first intron alone in the FPGS transcript is sufficient to introduce a premature stop codon after a 74–amino acid–long open reading frame. The latter would certainly lead either to a truncated FPGS polypeptide that may undergo degradation and/or result in the activation of the nonsense-mediated mRNA decay (NMD) pathway. NMD is an established quality control mechanism that selectively degrades mRNAs harboring premature translation termination codons and thereby prevents their translation.25,26 Moreover, even if such a truncated FPGS polypeptide did exist, it would be devoid of FPGS activity as it lacks the C-terminal domain that contains both the binding sites for folate27 and glutamate.28 Hence, the present findings provide the first direct evidence associating antifolate resistance with aberrant FPGS splicing. Moreover, the latter further offers a novel molecular basis for the frequent emergence of tumor cells with antifolate-resistant phenotypes that display apparently normal FPGS mRNA levels commonly evaluated by RT-PCR analysis, while exhibiting loss of FPGS activity in the absence of inactivating point mutations.5,10,13,14,17 In this respect, it has been frequently shown that loss of FPGS activity occurs in the absence of a parallel decrease in mRNA levels in multiple CCRF-CEM leukemia cells with acquired resistance to various polyglutamatable antifolates,10,13,14,17,20 including MTX,10,17,29 raltirexed, edatrexate,30 or lometrexol (DDATHF).13 This finding strongly suggested the existence of alterations at the posttranscriptional level of the FPGS gene. Consistently, the mechanism that was identified in edatrexate-resistant L1210 leukemia cells was decreased rate of FPGS translation as judged from the poor translatability of the FPGS transcript in a cell-free translation system.30 In the current study, we discovered another posttranscriptional alteration involving splicing defects in FPGS mRNA leading to loss of FPGS activity. However, whereas quantitative RT-PCR analysis, particularly with nondiagnostic PCR fragments, may frequently fail to identify such splicing defects,14 Northern blot analysis proved instrumental in identification of changes in the full-length FPGS transcripts suggesting putative aberrant splicing (Figure 4A); this was corroborated with diagnostic PCR analysis. To date, there have been limited reports associating aberrant splicing with drug resistance phenomena. For instance, Perry et al31 have shown that human splicing factor SPF45 (RBM17), which is frequently overexpressed in various solid tumors, confers multidrug resistance (MDR) to anticancer drugs including doxorubicin and the antifolate pemetrexed upon ectopic overexpression of SPF45 in HeLa cells. More recently, Corsini et al32 suggested that the molecular mechanism underlying MDR in these cells is alternative splicing of FAS, a proapoptotic factor. It was further shown in this study that SPF45 induces exon 6 skipping of FAS, thereby suggesting that this alteration may result in an antiapoptotic isoform of FAS. Since various anticancer drugs exert their therapeutic cytotoxic activity by inducing apoptosis, expression of an antiapoptotic factor may confer upon cells an MDR phenotype.
Intermittent exposure of tumor cell lines to polyglutamatable antifolates, including MTX, best resembles the clinical setting when a folate antagonist is administered as a pulse (ie, bolus infusion) and the antifolate monoglutamate is cleared rapidly from the blood. In this respect, a substantial body of literature with multiple human hemato-lymphoid and carcinoma cell lines established that a predominant mechanism of resistance to polyglutamatable antifolates upon a repeated pulse exposure to high-dose drug is decreased FPGS activity and consequently defective antifolate polyglutamylation (reviewed in Assaraf5 and Zhao and Goldman6 ). In this respect, McGuire et al17 demonstrated that impaired antifolate polyglutamylation can rapidly emerge in CCRF-CEM leukemia cells following intermittent exposure to MTX via positive selection of preexisting clones with decreased FPGS activity. The major mechanism of antifolate resistance in these clonal isolates was diminished FPGS activity and subsequently impaired MTX polyglutamylation. Remarkably, even after a single pulse exposure to MTX, decreased FPGS activity, defective polyglutamylation, and consequent antifolate resistance were apparent; consistently, FPGS activity gradually decreased with the number of intermittent MTX treatment cycles. These findings of positive selection of preexisting clonal variants with decreased FPGS activity in CCRF-CEM cells are in accord with our current findings of aberrant FPGS splicing in the very same leukemia CCRF-CEM cells. We consistently find here that the population of untreated parental CCRF-CEM cells had some small, yet detectable alteration in the splicing of FPGS mRNA including exon 10 skipping (Figure 3A) as well as intron 10 retention; these alterations became dramatic in the antifolate-resistant sublines. Based on these cumulative results, a plausible scenario that can be envisioned is that pre-existing clonal variants displaying some basal, low level of aberrant splicing of FPGS mRNA may exist in the leukemia cell population prior to treatment with MTX. Under antifolate-free growth conditions, these preexisting variants are relatively infrequent in the general population as they harbor a growth disadvantage due to their inability to efficiently form and accumulate folate polyglutamates, thereby resulting in a contracted intracellular folate pool.10 However, upon repeated cycles of pulse drug exposure to relatively high concentrations (ie, clinically relevant) of polyglutamatable antifolates, these rare clones may be rapidly selected out from the general population. This is particularly true as these clones bear a growth advantage under drug selective conditions due to their inability to form antifolate polyglutamates, hence resulting in rapid efflux of antifolate monoglutamates.
MTX undergoes oxidation to its principal metabolite 7-hydroxymethotrexate (7-OHMTX) in the liver by aldehyde oxidase.33 The plasma concentration of 7-OHMTX can exceed that of MTX, as in osteosarcoma and ALL patients treated with HD-MTX, thereby becoming the predominant metabolite 10 to 12 hours after bolus MTX infusion.34 Recently we have shown that MTX and its metabolite 7-OHMTX provoke disparate mechanisms of antifolate resistance in leukemia cells14 ; hence, whereas continuous exposure to MTX leads to down-regulation of RFC expression and markedly impaired drug uptake,11,14,35 exposure to 7-OHMTX brought about a major decrease (∼ 98%) in FPGS activity with no apparent quantitative alteration at the mRNA level.14 These findings suggest the involvement of FPGS and not RFC in cases of relapse following treatment with MTX, and hence emphasize the burning need to pinpoint the molecular basis of FPGS-based antifolate resistance and its potential overcoming. In contradistinction to this report, we here show that the level of the principal transcript of FPGS is down-regulated as no specific band could be observed in these cell lines upon Northern blot analysis (Figure 4A). Instead of a discrete principal transcript band, these antifolate-resistant cells exhibited a smear extending for several hundreds of base pairs insinuating the existence of multiple FPGS transcripts differing in length, thereby reflecting the retention of introns and/or the skipping of exons. Although we do observe here defective FPGS splicing including exon 10 skipping and intron 10 retention, further studies are necessary to identify the assortment of aberrant FPGS transcripts in these antifolate-resistant leukemia cells.
To explore the possibility of the occurrence of splicing defects in the FPGS transcript in the clinical setting, we obtained 13 RNA samples from 8 different ALL patients, 5 of which were matched specimens obtained both at diagnosis and relapse. PCR amplification of a region of the FPGS transcript encompassing exons 9 to 13 established exon 12 skipping in one relapse specimen (Figure 5A lane 3); the ratio between the relatively low level of the 300-bp PCR product reflecting exon 12 skipping and the high level of the normal (ie, properly spliced) 450-bp product was found to be in a good concordance with the ratio of leukemic cells versus normal leukocytes in the bone marrow from which the RNA sample was derived (ie, patchy infiltration of leukemia cells among normal hematopoietic cells in a normocellular bone marrow). Taking into consideration the fact that complete remission (CR) is determined in the presence of less than 5% blasts in a normocellular bone marrow, patients with a smaller extent of disease might be erroneously determined as achieving a promising CR. Interestingly, 93% of all adult ALL patients achieve CR according to these criteria, whereas less than 40% eventually remain free of disease.24 Hence, a relatively low level of the aberrant PCR product reflects the poor ratio of blast cells to normal cells, rather than reflecting mere low levels of the aberrantly spliced FPGS transcript relative to the normal transcript. To corroborate these findings, a second diagnostic PCR was performed that exclusively amplified the aberrant FPGS transcript that lacks exon 12 (Figure 5B); this sensitive PCR approach allowed us to positively identify 2 additional ALL diagnosis specimens that also contained aberrant FPGS transcripts lacking exon 12. A second specimen obtained at diagnosis (D5) (Figure 5B lane 10) exhibited very low levels of the aberrant FPGS transcript that was absent at the time of relapse (Figure 5B lane 11), whereas specimen D8 (Figure 5B lane 14) had an elevated level of exon 12 skipping, suggesting a pre-existing alteration in FPGS that could contribute to drug resistance following HD-MTX–containing combination chemotherapy via positive clonal selection and expansion. Indeed, the latter patient succumbed shortly after the HD-MTX–containing chemotherapy due to a rapidly progressing disease. In the antifolate-resistant leukemia cell lines, we identified exclusion of FPGS exon 10 as well as inclusion of intron 10, each of which introduced premature stop codons that could activate the NMD pathway, and in addition, disrupt the open reading frame such that the glutamic acid binding site composed of D335, H338, and R37728 is completely absent. In the clinical ALL specimens, exon 12, however, encodes for D335 and H338; hence, exon 12 skipping eliminates glutamic acid binding as well as introduces a premature stop codon. The existence of an FPGS transcript lacking exon 12 in 3 of 8 ALL patients raises the possibility of using it as a potential molecular tool for the prediction of the response of leukemic cells to MTX. Larger studies assessing the incidence of this finding and its contribution to MTX resistance are warranted. CNS lymphoma, almost solely treated with HD-MTX, might be a good model to evaluate the clinical significance of this potential MTX-resistance mechanism. Furthermore, ALL patients presenting with such defective FPGS splicing that leads to loss of FPGS function may be treated in the future with antifolates such as plevitrexed and neutrexin that do not rely on FPGS function for their pharmacological activity.
In summary, our current findings establish that loss of FPGS function in human leukemia cells exposed to an intermittent antifolate treatment in vitro may be due to aberrant FPGS mRNA splicing including intron retention and/or exon skipping, thereby resulting in loss of cellular retention of antifolates and consequent drug resistance. The encouraging results obtained with clinical specimens from ALL patients warrant a large study aimed at exploring the frequency of occurrence of such FPGS splicing defects and their possible contribution to drug resistance phenomena.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.G.A. codesigned and wrote the present paper; M.S. codesigned and wrote the paper as well as performed the vast majority of the experiments to which C.W. contributed; and I.A. provided the ALL specimens.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yehuda G. Assaraf, The Fred Wyszkowski Cancer Research Laboratory, Department of Biology, Technion-Israel Institute of Technology, Haifa 32000, Israel; e-mail: assaraf@tx.technion.ac.il.

![Figure 4. Northern blot analysis of FPGS mRNA levels. RNA was blotted onto Zeta-Probe nylon membrane and hybridized with a [32P]-labeled FPGS probe as detailed under “Northern blot analysis.” (A) RNA from parental CCRF-CEM and Molt-4 cells (lanes 1 and 4, respectively) as well as antifolate-resistant CEM-7OH MTX, CEM MTXR10, and Molt-4-7OH MTX cells (lanes 2, 3, and 5, respectively). A discrete band with an expected size of 2400 bp can be observed for both parental cell lines (lanes 1 and 4), whereas the antifolate-resistant cells CEM-7OH MTX and MTXR10 exhibit a smear spanning several hundreds of base pairs (lanes 2 and 3, respectively). (C) RNA from parental CCRF-CEM cells and their pIRES/FPGSΔexon10 transfectant (lanes 1 and 2, respectively) as well as the antifolate-resistant cell line CEM MTXR10, its pcDNA3/hFPGS transfectant and its pIRES/FPGSΔexon10 transfectant (lanes 3 and 4, respectively). pIRES/FPGSΔexon10 is transcribed to yield an approximately 3550-bp-long operon including the FPGS gene, the IRES translation initiation sequence, and the EGFP gene. (B,D) The intensity of the FPGS transcript was normalized to the methylene blue staining of the 18S ribosomal RNA band.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-08-173799/6/m_zh80130932710004.jpeg?Expires=1769095283&Signature=FdKIRQFwpOUHJboWSKvZ3-guF7uFgbC9XOogbusQZkDYdPQU9hDji5g6VndNc91z9qnHOMy~HEfoSrQ2YVS6n9iV6WuMYXmT0m0t6dQxMURgVCtVAzXuewiesife~vPQYz0w0kcmSNDuvdWJ4WA5o9qnWPsV-Aib~meYTNgTdq6cgQ~X0zDm85JabE23FrQF2h36CF4LBCavrBwLSjkonfM6KyTc7DeqdUEjzYloA3C5yeNHBYUcyCuQaAJexLpU~Ex3KNBXtgcRTPeheFwXpAHMC1HM6JldlzwiIjH-rf4Zp67JohjBO6aqTd984JJnK9jp1yd~MNHHckZmZjJXNg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal