Abstract
Distinct genes encode 6 human receptors for IgG (hFcγRs), 3 of which have 2 or 3 polymorphic variants. The specificity and affinity of individual hFcγRs for the 4 human IgG subclasses is unknown. This information is critical for antibody-based immunotherapy which has been increasingly used in the clinics. We investigated the binding of polyclonal and monoclonal IgG1, IgG2, IgG3, and IgG4 to FcγRI; FcγRIIA, IIB, and IIC; FcγRIIIA and IIIB; and all known polymorphic variants. Wild-type and low-fucosylated IgG1 anti-CD20 and anti-RhD mAbs were also examined. We found that (1) IgG1 and IgG3 bind to all hFcγRs; (2) IgG2 bind not only to FcγRIIAH131, but also, with a lower affinity, to FcγRIIAR131 and FcγRIIIAV158; (3) IgG4 bind to FcγRI, FcγRIIA, IIB and IIC and FcγRIIIAV158; and (4) the inhibitory receptor FcγRIIB has a lower affinity for IgG1, IgG2, and IgG3 than all other hFcγRs. We also identified parameters that determine the specificity and affinity of hFcγRs for IgG subclasses. These results document how hFcγR specificity and affinity may account for the biological activities of antibodies. They therefore highlight the role of specific hFcγRs in the therapeutic and pathogenic effects of antibodies in disease.
Introduction
The biological activities of antibodies depend on the interaction of their Fc portion with effector systems. These are essentially complement and cells. Antibodies bind to cells that express receptors for the Fc portion of antibodies (FcRs). FcRs exist for all classes of antibodies. They are expressed by different cell types having different biological activities, which they modulate when they are aggregated by multivalent antigen-antibody complexes.1 Most cells express several FcRs, and different FcRs can generate different signals in a single cell, depending on their intracytoplasmic domains. Activating FcRs possess immunoreceptor tyrosine-based activation motifs (ITAMs).2 ITAMs are present in the intracytoplasmic domain of FcRγ, a homodimeric common subunit which associates with the ligand-binding subunit of most activating FcRs.3 They are also present in the intracytoplasmic domain of 2 single-chain activating receptors. Inhibitory FcRs are single-chain receptors that possess an immunoreceptor tyrosine-based inhibition motif (ITIM) in their intracytoplasmic domain.4 Other FcRs are inserted in the outer layer of the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor and contain no signaling motif.5 FcRs have been associated with many antibody-dependent diseases6 and are key molecules in antibody-based immunotherapy. These include the treatment, for instance, of non-Hodgkin lymphomas by mouse/human chimeric IgG1 anti-CD20 antibodies7 and the prevention of hemolytic disease of the newborn by a mixture of polyclonal IgG1 and IgG3 anti-RhD antibodies (eg, Rophylac). Therapeutic antibodies are, however, potentially harmful, as exemplified by a recent clinical trial using IgG4 anti-CD28 antibodies
Four human subclasses of IgG are produced in different amounts in response to various antigens. T-dependent protein antigens elicit primarily IgG1 and IgG3 antibodies, whereas T-independent carbohydrate antigens elicit primarily IgG2 antibodies. Chronic antigen stimulation, as in allergic desensitization, elicits IgG4 antibodies. The biological activities of each subclass of IgG are poorly known. IgG receptors (FcγRs) are strikingly numerous in humans. They comprise high-affinity and low-affinity receptors.8 Both high-affinity and low-affinity FcγRs bind IgG-immune complexes with a high avidity, but only high-affinity FcγRs bind monomeric IgG. There is one high-affinity IgG receptor in humans, hFcγRI (CD64), and 2 families of low-affinity IgG receptors, hFcγRIIA, IIB, and IIC (CD32), and hFcγRIIIA and IIIB (CD16). hFcγRI and hFcγRIIIA are FcRγ-associated activating receptors, hFcγRIIA and hFcγRIIC are single-chain activating receptors, hFcγRIIB are single-chain inhibitory receptors, and hFcγRIIIB are GPI-anchored receptors whose function is uncertain.1
The multiplicity of hFcγRs is further increased by a series of polymorphisms in their extracellular domains (reviewed in van Sorge et al9 ). Two alleles of the gene encoding hFcγRIIA generate 2 variants differing at position 131, named low-responder (H131) and high-responder (R131).10 The H131 and R131 alleles are differentially distributed in whites, Japanese, and Chinese.11 Two alleles of the gene-encoding hFcγRIIIA generate 2 variants differing at position 158 (V158 and F158).12 Two alleles of the gene-encoding hFcγRIIIB generate 2 variants differing at 4 positions, NA1 (R36 N65 D82 V106) and NA2 (S36 S65 N82 I106), with different glycosylation patterns.13 A point mutation (A78D) of the NA2 allele generates another hFcγRIIIB variant named SH.14 Besides, duplications of the gene-encoding hFcγRIIIB may generate a variable number of gene copies in different individuals. A single individual may therefore express all 3 hFcγRIIIB variants.15 hFcγR polymorphisms have been linked to autoimmune and infectious diseases. hFcγRIIAR131 has been associated with an increased prevalence of nephropathy,16 bacterial infections,17 and possibly systemic lupus erythematosus (SLE).18,19 hFcγRIIIAF158 has been linked to SLE20 and to rheumatoid arthritis (RA).21 hFcγRIIIBNA1 has been linked to Wegener granulomatosis22 and systemic vasculitis23 and hFcγRIIIBNA2 to SLE in Japanese people.24
The subclass specificity of hFcγRs has been investigated since the 1980s, that is, at a time when the complexity of hFcγRs was not suspected. Some studies were performed before hFcγRs were cloned. Others were performed using cell lines expressing several hFcγRs. Several techniques with different sensitivities were used. Finally, total human serum IgG or human myeloma IgG1 were used in most studies. IgG2, IgG3, and IgG4 were rarely considered. Marked variations,25,26 up to 1 log, were reported in the binding affinities of human IgG for the same hFcγR.27,28 Compilations of these data were nevertheless repeatedly published in reviews.6,9,29-32 hFcγR polymorphisms were rarely considered in these reviews. They, however, affect the binding of IgG to hFcγRs. hFcγRIIAH131, but not hFcγRIIAR131, was reported to bind human IgG2,33 but the affinity of this interaction was not determined. The effect of this polymorphism on the affinity of hFcγRIIA for human IgG1, IgG3, and IgG4 was not investigated. A higher efficiency of IgG1 anti-CD20 therapy was observed in hFcγRIIIA V/V158 patients than in F/F158 patients, and peripheral blood lymphocytes from V/V158 donors killed more efficiently anti-CD20–coated target cells than PBLs from F/F158 donors in antibody-dependent cell-mediated cytotoxicity (ADCC) assays.34,35 Indeed, hFcγRIIIAV158 has a higher affinity for monoclonal hIgG1 than hFcγRIIIAF158.36 The affinity of hFcγRIIIAF158 and hFcγRIIIAV158 for other IgG subclasses was not investigated. The NA1 and NA2 alleles of hFcγRIIIB were described as having similar affinities for total human IgG37 or IgG1, but discordant results were reported for IgG3.38 The binding properties of hFcγRIIIBSH have not been studied.
In view of these incomplete, heterogeneous, and sometimes discrepant set of data, we undertook a systematic investigation of the binding specificity of every hFcγR for all 4 subclasses of human IgG. These were assayed, both as monomers and as complexes, on a collection of CHO transfectants expressing comparable levels of FLAG-tagged receptors. All hFcγRs, including all polymorphic variants, were studied. Soluble glycosylated ectodomains of all hFcγRs were also produced, and their affinity for IgG subclasses was measured by surface plasmon resonance (SPR). We found that IgG1 and IgG3 bind to all hFcγRs, that IgG2 binds to 3 hFcγRs, and that IgG4 binds to 6. We found that the inhibitory receptor FcγRIIB has a lower affinity for IgG1, IgG2, and IgG3 than all other hFcγRs. We also establish a hierarchy of affinities of all hFcγRs and their variants for polyclonal IgG of the 4 subclasses. Our data account for the previously reported association of inflammatory diseases with hFcγR polymorphisms and should unravel novel candidate strategies in optimization of therapeutic antibodies.
Methods
Cells and cDNAs
CHO-K1 and HEK-293T cells were purchased from ATCC (Manassas, VA), and J558L transfectants producing anti-NIP (VH mouse/CH human) chimeric IgG1, IgG2, IgG3, IgG4, and IgE from the European Collection of Cell Cultures (Salisbury, United Kingdom), and cultured as recommended. Culture reagents were obtained from Invitrogen Life Technologies (Carlsbad, CA).
Human FcγRIIB (FcγRIIB1 isoform) cDNA and mouse FcRγ-chain cDNA were previously cloned in the laboratory. Human FcγRI and FcγRIIA(R131) cDNAs were from J. Van de Winkel (University Medical Center Utrecht, Utrecht, The Netherlands). Human FcγRIIIB (NA1, NA2, and SH)13,14 cDNAs were provided by S. Santoso (Institute for Clinical Immunology and Transfusion Medicine, Giessen, Germany). Blood cells from healthy donors were used as a template for mRNA extraction, cDNA synthesis, and cloning of cDNAs encoding FcγRIIA(H131) and hFcγRIIIA(V158).12 The latter served as template to generate cDNA encoding hFcγRIIIA(F158) by site directed mutagenesis using the Quickchange Multi Kit (Stratagene, La Jolla, CA). Human FcγRIIC (FcγRIIC1 isoform) cDNA was generated by a 2-step PCR using FcγRIIB1 cDNA and FcγRIIA(R131) cDNA as templates. hFcγRIIIA(EC domain)-hFcγRIIB (TM + IC domains) chimeric cDNA was generated similarly using FcγRIIIA(V158) cDNA and FcγRIIB1 cDNA as templates. Sequencing of all cDNAs did not reveal variation compared with published data. A cDNA sequence coding for a FLAG tag was inserted immediately 3′ of the signal sequence cleavage site in all FcR cDNAs by PCR using overlapping primers. Resulting constructs were cloned into pNT NEO (pBR322 containing a SRα promoter, neomycin). cDNAs corresponding to EC domains of all FcRs were cloned into p3xFLAG-CMV-14 (Sigma-Aldrich, St Louis, MO), to generate soluble FcR ectodomains linked to a 3xFLAG tag at their C-terminus. CHO-K1 stable FLAG-tagged FcR transfectants were obtained by selection in 1 mg/mL Geneticin and/or 0.5 mg/mL Zeocin (Invitrogen, Paisley, United Kingdom), and were sorted to equivalent surface expression by flow cytometry on a MoFlo (Dako, Ely, United Kingdom) or FACSAria (Becton Dickinson, Franklin Lakes, NJ).
Antibodies and reagents
FITC-mIgG1 and anti-hCD32 (clone FLI8.26) were purchased from PharMingen (San Diego, CA); anti-hCD64 (clone 10.1) from Biolegend (San Diego, CA); HRP- or FITC-labeled anti-FLAG (M2) from Sigma-Aldrich; PE-F(ab′)2 anti–human Fab-specific from Jackson Immunoresearch (West Grove, PA); neutravidin-PE from Molecular Probes (Eugene, OR); polyclonal human IgG1, IgG2, IgG3, and IgG4 from The Binding Site (Birmingham, United Kingdom); myeloma human IgG2 from Sigma-Aldrich; myeloma human IgG4, polyclonal human IgE, and polyclonal human IgA from Biodesign (Saco, ME); human IgE (PHP142) and anti-hCD16 (clone 3G8) from Serotec (Oxford, United Kingdom); and human IgEκ from Calbiochem (San Diego, CA). Anti-NIP IgGs and anti-NIP IgE were purified from cell culture supernatants as described.39 Monoclonal mouse anti–human FcγRIIB/C (clone GB3) was provided by U. Jacob (SuppreMol, Munich, Germany). Monoclonal human IgG1 anti-RhD40 antibodies, monoclonal murine-human chimeric anti–HLA-DR (based on the murine IgG2a anti-HLA-DR Lym-1 mAb41 ), and anti-CD2042 antibodies were provided by LFB (Lille, France): the first 106 amino acids in the light and heavy chains of the chimeric mAbs were of murine origin, the following were of human origin, these constant regions being identical to the ones of the fully human IgG1 anti-RhD mAb. Anti-RhD, anti-HLA-DR, and anti-CD20 mAbs were produced in YB2/0 cells, which have decreased levels of α-1,6-fucosyltransferase (generating low-fucose containing mAbs) or produced in α-1,6-fucosyltransferase-competent CHO cells. Anti-RhD, anti–HLA-DR, and anti-CD20 mAbs produced both in CHO and YB2/0 cells demonstrated less than 3% sialylated IgG1 by high performance capillary electrophoresis laser-induced fluorescence (HPCE-LIF; not shown). PNGase F was from New England Biolabs (Ipswich, MA) and NIP12-BSA-biotin from BioCat (Heidelberg, Germany).
Immunoglobulin binding assays
Monomeric.
Aggregates in stock solutions of human IgG were removed by an 18-hour ultracentrifugation at 100 000g followed by OD280 nm concentration measurement, or using Ultrafree-CL PTMK Ultracel-PL 300-kDa cutoff spin columns (Millipore, Billerica, MA; as an internal control of the experiment, 100% of IgM was retained on the column); 2 × 105 cells were incubated with monomeric Ig at indicated concentrations for 1 hour at 4°C. Cell-bound Ig was detected using 5 μg/mL PE-labeled F(ab′)2 fragments of goat anti–human Fab-specific for 30 minutes at 4°C.
F(ab′)2-aggregated human IgG.
Human IgG were incubated with PE-labeled F(ab′)2 fragments of goat anti–human Fab-specific for 30 minutes at 37°C and added to 2 × 105 cells for 1 hour at 4°C.
Immune complex.
Human immune complexes (ICs) were preformed by incubating 10 μg/mL NIP12-BSA-biotin with 30 μg/mL anti-NIP mAbs for 1 hour at 37°C in PBS 0.05% BSA 2 μM EDTA pH7.4; 2 × 105 cells were incubated with IC for 1 hour at 4°C. IC bound to cells were detected using neutravidin-PE at 2 μg/mL for 30 minutes at 4°C.
Heat-aggregated IgG.
A total of 100 μg/mL human IgG were incubated in borate-buffered saline pH 8.0 for 30 minutes at 63°C, diluted to indicated concentrations in PBS 0.05% BSA, 2 μM EDTA, pH 7.4, and added to 2 × 105 cells for 1 hour at 4°C. Cell-bound IgG was detected using 5 μg/mL PE-labeled F(ab′)2 fragments of goat anti–human Fab-specific for 30 minutes at 4°C.
Production of soluble FcR ectodomains-3xFLAG fusion protein
cDNA constructs coding for soluble FcR ectodomains tagged with a 3xFLAG peptide were transfected by a standard calcium chloride technique into HEK-293T cells. Fusion proteins from 96-hour supernatants were purified on anti-FLAG agarose beads and eluted using 3xFLAG peptide (Sigma-Aldrich). Purity of PNGase F-treated (following supplier's recommendations) or untreated proteins was assessed after SDS-PAGE and transfer onto Immobilon-P membranes (Millipore) by anti-FLAG M2-HRP (Sigma-Aldrich) blotting, revealed using ECL reagents (GE Healthcare, Little Chalfont, United Kingdom). As we failed to produce functional ectodomains of FcγRI, we purchased recombinant soluble C-terminal polyhistidine-tagged FcγRI ectodomains from R&D Systems (Minneapolis, MN).
Surface plasmon resonance analysis
A BIAcore 2000 SPR biosensor (GE Healthcare) was used to assay the interaction of soluble ectodomains of FcR with monoclonal Ig. An N-hydroxysuccinimide ester was formed on a CM5 sensor chip surface according to a procedure recommended by the manufacturer. Ectodomains were immobilized at acidic pH, resulting in the following densities: FcγRIIAH131: 1522 RU, FcγRIIAR131:1766 RU, FcγRIIB/C: 1493 RU, FcγRIIIAF176: 2023 RU, FcγRIIIAV176: 1952 RU, FcγRIIIBNA1: 1883-2056 RU, FcγRIIIBNA2: 2046-2161 RU, FcγRIIIBSH: 2152-2267 RU and FcγRI: 2650 RU. A range of Ig concentrations was injected into flow cells at a flow rate of 20μL/min, with a contact and dissociation time of 300 and 900 seconds, respectively. After each assay cycle, the sensor chip surface was regenerated using 10 mM NaOH. Binding response was recorded as resonance units (RU; 1 RU ≈ 1 pg/mm2) continuously, with background binding automatically subtracted. Due to the polyclonal nature of the Ig preparations used, the kinetic constants (kon, koff, t1/2) were not determined, and the KA was calculated by studying the concentration-dependence of the steady-state signal reached at the end of the injection (Req) using BIA evaluation version 4.2 software (GE Healthcare) and Scrubber version 2 software (BioLogic Software, Campbell, Australia). The steady-state response was plotted against the concentration of IgG and fitted using Origin software (OriginLabs, Northampton, MA) for FcγRI/II and for FcγRIII in Figures S4 and S5 (available on the Blood website; see the Supplemental Materials link at the top of the online article), respectively. Varying the densities of immobilized FcγR (1200-2700 RU) did not significantly affect steady-state affinities.
Results
Binding specificity of hFcγRs for IgG subclasses
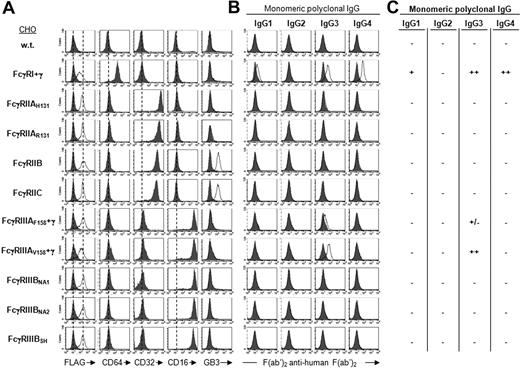
To analyze the binding of human IgG subclasses to hFcγRs, a series of CHO transfectants expressing FLAG-tagged hFcγRs were constructed. These transfectants expressed FcγRI, FcγRIIA, FcγRIIB, FcγRIIC, FcγRIIIA, or FcγRIIIB. Multisubunit hFcγRs (ie, FcγRI and FcγRIIIA) were coexpressed with FcRγ. All known polymorphic variants (ie, FcγRIIAH131 and R131, FcγRIIIAF158 and V158, and FcγRIIIBNA1, NA2, and SH) were included (Figure S1). Anti-FLAG bound comparably to all transfectants, indicating that all hFcγRs were expressed at similar levels (Figure 1A). Anti-CD64 bound to FcγRI only. Anti-CD32 bound to FcγRIIAH131, FcγRIIAR131, FcγRIIB, and FcγRIIC only. Anti-CD16 bound to FcγRIIIAF158, FcγRIIIAV158, FcγRIIIBNA1, FcγRIIIBNA2, and FcγRIIIBSH only. mAb GB3 bound to FcγRIIB and FcγRIIC, but not to FcγRIIA. The binding of human IgG to transfectants was assessed by indirect immunofluorescence. Polyclonal IgG preparations (100 000g ultracentrifuged) were used to assess the binding of monomeric IgG. IgG complexed with F(ab′)2 anti–human F(ab′)2 were used to assess the binding of IgG immune complexes (IgG-F′2 IC). Binding of monomers and immune complexes was tested in the same experiment.
Binding specificity of hFcγRs for monomeric IgG. (A) Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of anti-FcγRI (CD64), anti-FcγRII (CD32), anti-FcγRIII (CD16), and anti-FcγRIIB/C (GB3) to FLAG-tagged hFcγRs on CHO transfectants. (B) Histograms show the binding of polyclonal human IgG subclasses to hFcγR-expressing transfectants using 10 μg/mL ultracentrifugated IgG and 15 μg/mL PE-F(ab′)2 anti–human F(ab′)2. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 3 independent experiments gave identical results. (C) Summary of monomeric IgG binding ability.
Binding specificity of hFcγRs for monomeric IgG. (A) Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of anti-FcγRI (CD64), anti-FcγRII (CD32), anti-FcγRIII (CD16), and anti-FcγRIIB/C (GB3) to FLAG-tagged hFcγRs on CHO transfectants. (B) Histograms show the binding of polyclonal human IgG subclasses to hFcγR-expressing transfectants using 10 μg/mL ultracentrifugated IgG and 15 μg/mL PE-F(ab′)2 anti–human F(ab′)2. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 3 independent experiments gave identical results. (C) Summary of monomeric IgG binding ability.
Monomeric IgG1 bound weakly to FcγRI but not detectably to any other hFcγR. Monomeric IgG2 did not bind to any hFcγR. Monomeric IgG3 bound to FcγRI and FcγRIIIA only. They bound better to FcγRIIIAV158 than to FcγRIIIAF158. Monomeric IgG4 bound to FcγRI only (Figure 1B). FcγRI is therefore a high-affinity receptor for IgG1, IgG3, and IgG4; and FcγRIIIAV158 a high-affinity receptor for IgG3 only.
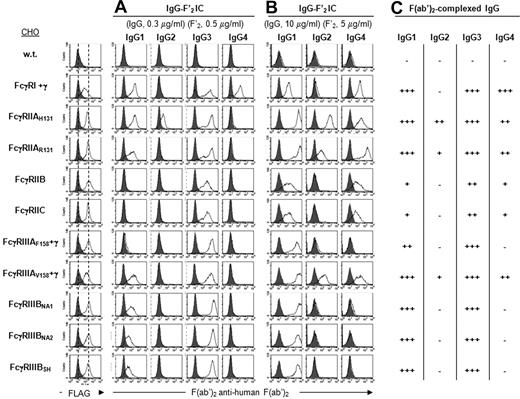
At low concentrations (0.3 μg/mL IgG), IgG1-F′2 IC bound to all hFcγRs except FcγRIIB, FcγRIIC, and FcγRIIIAF158. They indeed bound to FcγRIIIAV158 but not to FcγRIIIAF158. They bound similarly to FcγRIIAR131 and H131. They also bound similarly to FcγRIIIBNA1, NA2, or SH (Figure 2A). IgG2-F′2 IC bound only to FcγRIIAH131. IgG3-F′2 IC bound comparably to all hFcγR but less efficiently to FcγRIIB and FcγRIIC. This difference was more marked with even lower concentrations of IgG (0.1 μg/mL; data not shown). They bound similarly to FcγRIIAR131 and H131, similarly to FcγRIIIAF158 and V158, and similarly to FcγRIIIBNA1, NA2, or SH. IgG4-F′2 IC bound only to FcγRI.
Binding specificity of hFcγRs for IgG IC. Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and (A,B) the binding of IgG subclasses in complex with PE-F(ab′)2 anti–human F(ab′)2 to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 4 independent experiments gave similar results. (C) Summary of IgG-F′2 IC binding abilities to hFcγRs using data from panels A and B.
Binding specificity of hFcγRs for IgG IC. Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and (A,B) the binding of IgG subclasses in complex with PE-F(ab′)2 anti–human F(ab′)2 to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 4 independent experiments gave similar results. (C) Summary of IgG-F′2 IC binding abilities to hFcγRs using data from panels A and B.
At high concentrations (10 μg/mL), IgG1-F′2 IC bound to all hFcγR but less efficiently to FcγRIIB and FcγRIIC (Figure 2B). IgG2-F′2 IC bound to FcγRIIAH131, and to FcγRIIAR131 and FcγRIIIAV158, although less efficiently. They detectably bound to no other hFcγR, even at higher concentrations (30 μg/mL; data not shown). High concentrations of IgG3-F′2 IC were not tested, as their binding exceeded the detection range. IgG4-F′2 IC bound not only to FcγRI, but also to FcγRIIA, B, and C, and to FcγRIIIAV158. They bound better to FcγRIIAR131 than to FcγRIIAH131. They detectably bound neither to FcγRIIIAF158 nor to FcγRIIIBNA1, NA2, and SH, even at higher concentrations (30 μg/mL; data not shown).
Altogether these results demonstrate that all 4 IgG subclasses bind to hFcγR. IgG1 and IgG3, but not IgG2 and IgG4, bind to all hFcγRs (summarized in Figure 2C). Noticeably, IgG2 bind not only to FcγRIIAH131, as previously described,33 but also to FcγRIIAR131 and FcγRIIIAV158. IgG bind less efficiently to FcγRIIB and to FcγRIIC than to any other hFcγR. The H131R mutation in FcγRIIA decreases the binding of IgG2 and increases the binding of IgG4. The V158F mutation in FcγRIIIA decreases the binding of IgG1 and abrogates the binding of IgG2 and IgG4. FcγRIIIB polymorphisms do not affect the binding of IgG.
Influence of the FcRγ subunit on the binding of human IgG
Unexpectedly, monomeric IgG3 could bind to the low-affinity FcγRIIIAV158. This receptor can therefore function as a high-affinity FcR for one subclass of IgG. FcγRIIIA was previously found to have an intermediate affinity for mouse IgG2a, and this higher affinity, compared with that of other low-affinity hFcγRs, was proposed to rely on its association with FcRγ.43 To investigate whether this might apply to human IgG, we generated a chimeric FcγRIIIAV158 whose expression would not require FcRγ by replacing its transmembrane and intracellular domains by those of FcγRIIB (FcγRIIIAV158(EC)-IIB(TM + IC)). The binding of human polyclonal IgG was investigated on CHO transfectants expressing similar levels of FLAG-tagged human FcRγ-associated FcγRIIIAV158 or FcγRIIIAV158(EC)-IIB(TM + IC). Monomeric IgG1 (Figure S2), monomeric IgG2, and monomeric IgG4 (not shown) did not bind to these transfectants. Monomeric IgG3 bound similarly to both transfectants. We analyzed the behavior of heat-aggregated IgG that mimic IgG IC and that have been used in numerous FcR-binding and FcR-activation assays. Heat-aggregated IgG1, IgG2, IgG3, and IgG4 or IgG1-F′2 IC and IgG3-F′2 IC bound similarly to both transfectants. FcγRIIIA expressed independently of FcRγ therefore has a normal affinity for human IgG.
Binding affinity of hFcγRs for IgG subclasses
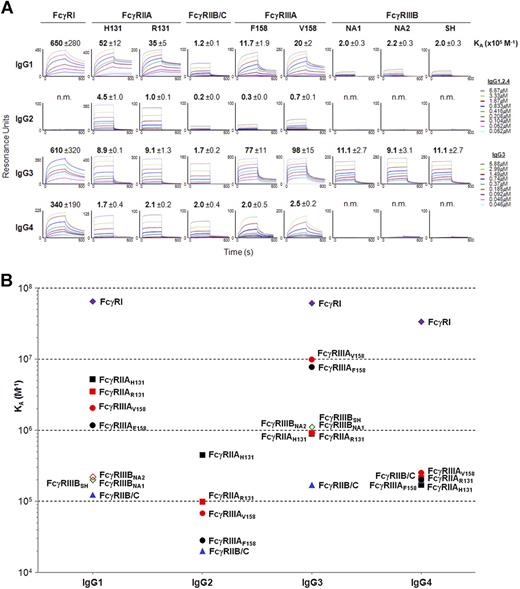
To measure the affinity of hFcγRs for the 4 IgG subclasses, FLAG-tagged extracellular domains of all FcγRII and FcγRIII and their polymorphic variants were produced in HEK-293T cells. The extracellular domains of FcγRIIB and FcγRIIC being identical, a single extracellular domain, herein referred to as FcγRIIB/C, was produced to analyze the affinity for FcγRIIB and FcγRIIC. Polyhistidine-tagged ectodomains of FcγRI were used instead of their FLAG-tagged equivalents that were not functional. These molecules were N-glycosylated as demonstrated by SDS-PAGE analysis before and after peptide:N-glycosidase F treatment (data not shown). They were covalently immobilized onto activated dextran surfaces and used for surface plasmon resonance (SPR) analysis (Figure 3A).
Binding affinity of hFcγRs for IgG subclasses. (A) Real-time surface plasmon resonance sensorgrams and affinity constants (×105 M−1) were determined from SPR analysis. The standard deviation of the affinity constant (KA) determination is indicated. n.m. indicates not measurable (ie, no detectable binding). (B) The affinity constants calculated in panel A are plotted against IgG subclasses. Data correspond to 1 experiment per interaction tested, which is representative of 2 to 6 independent experiments that gave similar results.
Binding affinity of hFcγRs for IgG subclasses. (A) Real-time surface plasmon resonance sensorgrams and affinity constants (×105 M−1) were determined from SPR analysis. The standard deviation of the affinity constant (KA) determination is indicated. n.m. indicates not measurable (ie, no detectable binding). (B) The affinity constants calculated in panel A are plotted against IgG subclasses. Data correspond to 1 experiment per interaction tested, which is representative of 2 to 6 independent experiments that gave similar results.
IgG1 bound to hFcγR ectodomains with a wide range of affinity. They bound with a KA approximately 2 × 105 M−1 to FcγRIIB/C and FcγRIIIBNA1, NA2, or SH, with a KA approximately 1.5 × 106 M−1 to FcγRIIIAV158 or F158, with a KA approximately 4 × 106 M−1 to FcγRIIAH131 or R131 and with a KA approximately 6.5 × 107 M−1 to FcγRI (Figure 3A,B).
IgG2 bound to hFcγR ectodomains with a similarly wide range of affinities. They bound to FcγRIIAR131 and FcγRIIIAV158 with a KA approximately 8 × 104 M−1. They bound with a higher affinity to FcγRIIAH131 (KA = 4.5 × 105 M−1) and with a lower affinity to FcγRIIIAF158 and FcγRIIB/C (KA approximately 2.5 × 104 M−1). They had no detectable affinity for FcγRI or FcγRIIIBNA1, NA2 or SH (Figure 3A,B).
IgG3 also bound to hFcγR with a wide range of affinities. They bound to FcγRIIAH131 or R131 and to FcγRIIIBNA1, NA2, or SH with a KA approximately 1 × 106 M−1. They bound with a higher affinity (KA approximately 5 × 106 M−1) to FcγRIIIAV158 or F158 and (KA approximately 6.1 × 107 M−1) to FcγRI, and with a markedly lower affinity (KA = 1.7 × 105 M−1) to FcγRIIB/C (Figure 3A,B).
IgG4 bound to hFcγRII/III ectodomains with a narrow range of affinities. They bound to FcγRIIAH131 or R131, FcγRIIB/C, and FcγRIIIAV158 or F158 with a KA approximately 2 × 105 M−1. They bound with a higher affinity (KA approximately 3.4 × 107 M−1) to FcγRI. They had no detectable affinity for FcγRIIIBNA1, NA2, or SH (Figure 3A,B).
Altogether SPR analysis data revealed that interactions of human IgG with low-affinity hFcγRs segregate into 2 groups: half of them have affinities of approximately 1 to a few × 105 M−1, half have affinities of 1 to a few × 106 M−1. Noticeably, FcγRIIB/C have the lowest affinities for all 4 IgG subclasses. FcγRIIA has an affinity 35-fold higher for IgG1 than FcγRIIB/C. Likewise, FcγRIIIA has an affinity 5-fold higher for IgG3 than FcγRIIIB. The R131H polymorphism affects the affinity of FcγRIIA for IgG2, but hardly its affinity for IgG1, IgG3, and IgG4. The F158V polymorphism increases moderately the affinity of FcγRIIIA for all 4 subclasses. The NA1/NA2/SH polymorphism does not affect the affinity of FcγRIIIB for any IgG subclass. As expected, FcγRI has the highest affinity for IgG1, IgG3, and IgG4 but has no affinity for IgG2. SPR analysis also unraveled measurable interactions of IgG2 with FcγRIIB/C and FcγRIIIAF158 and of IgG4 with FcγRIIIAF158, which had not been detected by immunofluorescence analysis.
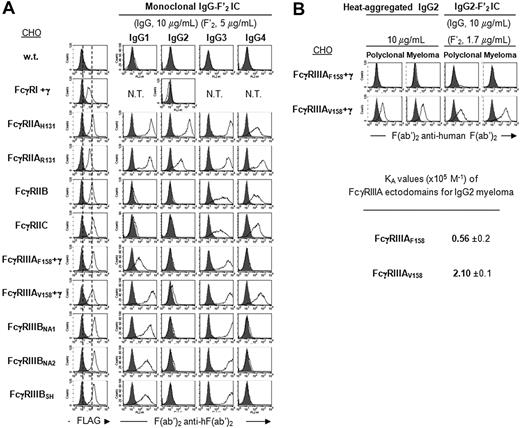
Binding specificity of hFcγRs for monoclonal IgG
To determine whether hFcγRs have the same specificity for human monoclonal and for human polyclonal IgG, we first examined the binding of mouse/human chimeric anti-NiP antibodies having a human IgG1, IgG2, IgG3, or IgG4 Fc portion39 to the same set of CHO transfectants as in Figure 1. Preformed ICs were made either with NiP-BSA (IgG-Ag IC; Figure S3A) or with F(ab′)2 anti–human F(ab′)2 (IgG-F′2 IC; Figure 4A). Higher fluorescence intensities were observed for all hFcγRs with IgG-F′2 IC than with IgG-Ag IC. IC made with monoclonal IgG1, IgG3, and IgG4 displayed the same binding specificity for all hFcγRs, including polymorphic variants, as polyclonal IgG1, IgG3, and IgG4, respectively. Monoclonal IgG2 also bound similarly as polyclonal IgG2, except IgG2-Ag IC which bound weakly to FcγRIIB/C and FcγRIIIAF158 (Figure 4A and Figure S3A). Interestingly, a weak affinity of these 2 receptors for polyclonal IgG2 was measurable by SPR analysis (Figure 3A,B). When heat-aggregated or complexed with F(ab′)2 anti-human F(ab′)2, a human myeloma IgG2 antibody, however, bound similarly and with a similar affinity as polyclonal IgG2 to FcγRIIIAV158 and F158 (Figure 4B).
Binding specificity of hFcγRs for monoclonal IgG. (A) Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of monoclonal human IgG subclasses in complex with PE-F(ab′)2 anti–human F(ab′)2 to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 2 independent experiments gave similar results. N.T. indicates not tested. (B) FcγRIIIAV158 is a low-affinity receptor for IgG2. Histograms show the binding of heat-aggregated IgG2 or IgG2-F′2 IC to FLAG-tagged hFcγRIIIA on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone. Affinity constants (×105 M−1) were determined from SPR analysis on immobilized FcγRIIIA ectodomains using the same concentrations of IgG2 as in Figure 3A. The standard deviation of the affinity constant (KA) determination is indicated.
Binding specificity of hFcγRs for monoclonal IgG. (A) Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of monoclonal human IgG subclasses in complex with PE-F(ab′)2 anti–human F(ab′)2 to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone; 2 independent experiments gave similar results. N.T. indicates not tested. (B) FcγRIIIAV158 is a low-affinity receptor for IgG2. Histograms show the binding of heat-aggregated IgG2 or IgG2-F′2 IC to FLAG-tagged hFcγRIIIA on CHO transfectants. Concentrations of human IgG and PE-F(ab′)2 anti–human F(ab′)2 are indicated. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human F(ab′)2 alone. Affinity constants (×105 M−1) were determined from SPR analysis on immobilized FcγRIIIA ectodomains using the same concentrations of IgG2 as in Figure 3A. The standard deviation of the affinity constant (KA) determination is indicated.
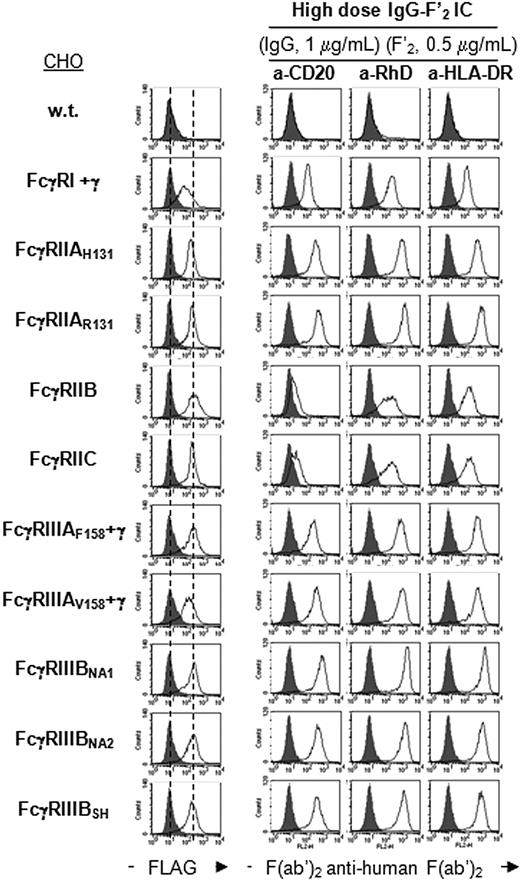
mAbs are increasingly used for the treatment of human diseases, such as lymphomas (anti-CD20 or anti–HLA-DR), allergic asthma (anti-IgE), or hemolytic disease of the newborn (anti-RhD). Most therapeutic mAbs are chimeric mouse/human or fully human IgG1 Abs. hFcγRs engaged by these mAbs remain mostly unknown. We therefore studied the binding of IgG-F′2 IC made with 1 fully human IgG1 anti-RhD mAb40 and 2 chimeric mouse/human IgG1 (anti-CD2042 and anti–HLA-DR) to the same set of transfectants as in Figure 1, and measured the affinity of anti-CD20 and anti-RhD for the extracellular domains of hFcγRs. All 3 mAbs bound to all hFcγRs (Figure S3B) with measurable affinities (Table 1, CHO columns). Higher concentrations of complexes, however, were required for FcγRIIB and FcγRIIC, which had a lower affinity for anti-CD20 and anti-RhD than other hFcγRs (Figure 5). Noticeably, up to 4-fold variations in affinity were observed for the 2 mAbs studied by SPR. FcγRIIIB polymorphisms did not detectably affect binding. FcγRIIAH131 and FcγRIIIAV158 had a higher affinity for anti-CD20 and anti-RhD than FcγRIIAR131 and FcγRIIIAF158, respectively.
KA (×105 M−1) of hFcγR ectodomains for monoclonal human IgG1
| . | Anti-CD20 . | KA fold induction . | Anti-RhD . | KA fold induction . | ||
|---|---|---|---|---|---|---|
| CHO . | YB2/0 . | CHO . | YB2/0 . | |||
| FcγRIIAH131 | 14 ± 5 | 11 ± 1 | 0.8 | 27 ± 1 | 23 ± 1 | 0.8 |
| FcγRIIAR131 | 7.3 ± 2.1 | 6.6 ± 0.6 | 0.9 | 19 ± 1 | 22 ± 2 | 1.2 |
| FcγRIIB/C | 2.8 ± 0.5 | 1.8 ± 0.1 | 0.7 | 3.2 ± 0.4 | 2.4 ± 0.7 | 0.8 |
| FcγRIIIAF158 | 11 ± 1 | 28 ± 3 | 2.5 | 27 ± 2 | 60 ± 7 | 2.2 |
| FcγRIIIAV158 | 23 ± 2 | 55 ± 7 | 2.4 | 51 ± 7 | 117 ± 14 | 2.3 |
| FcγRIIIBNA1 | 5.5 ± 1.1 | 8.7 ± 0.5 | 1.6 | 9.2 ± 0.8 | 23 ± 2 | 2.4 |
| FcγRIIIBNA2 | 6.8 ± 0.9 | 12 ± 1 | 1.7 | 10 ± 1 | 28 ± 2 | 2.7 |
| FcγRIIIBSH | 12 ± 3 | 12 ± 1 | 1.0 | 11 ± 1 | 27 ± 3 | 2.4 |
| . | Anti-CD20 . | KA fold induction . | Anti-RhD . | KA fold induction . | ||
|---|---|---|---|---|---|---|
| CHO . | YB2/0 . | CHO . | YB2/0 . | |||
| FcγRIIAH131 | 14 ± 5 | 11 ± 1 | 0.8 | 27 ± 1 | 23 ± 1 | 0.8 |
| FcγRIIAR131 | 7.3 ± 2.1 | 6.6 ± 0.6 | 0.9 | 19 ± 1 | 22 ± 2 | 1.2 |
| FcγRIIB/C | 2.8 ± 0.5 | 1.8 ± 0.1 | 0.7 | 3.2 ± 0.4 | 2.4 ± 0.7 | 0.8 |
| FcγRIIIAF158 | 11 ± 1 | 28 ± 3 | 2.5 | 27 ± 2 | 60 ± 7 | 2.2 |
| FcγRIIIAV158 | 23 ± 2 | 55 ± 7 | 2.4 | 51 ± 7 | 117 ± 14 | 2.3 |
| FcγRIIIBNA1 | 5.5 ± 1.1 | 8.7 ± 0.5 | 1.6 | 9.2 ± 0.8 | 23 ± 2 | 2.4 |
| FcγRIIIBNA2 | 6.8 ± 0.9 | 12 ± 1 | 1.7 | 10 ± 1 | 28 ± 2 | 2.7 |
| FcγRIIIBSH | 12 ± 3 | 12 ± 1 | 1.0 | 11 ± 1 | 27 ± 3 | 2.4 |
Affinity constants were determined from SPR analysis on immobilized FcγR ectodomains using the following concentrations of IgG: 0.866, 0.433, 0.216, 0.108, 0.054, and 0.026 μM. The standard deviation of the KA determination is indicated. KA fold induction = KA(YB2/0 mAb − FcγR)/KA(CHO mAb − FcγR). Values in bold indicate a KA fold induction ≥ 2.
Binding specificity of therapeutic monoclonal IgG1-F′2 IC to hFcγRs. Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of indicated monoclonal IgG1-F′2 IC to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of monoclonal IgG and PE-F(ab′)2 anti–human Ig are indicated. Monoclonal IgG1 used here were produced in CHO cells. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human Ig alone; 2 independent experiments gave similar results.
Binding specificity of therapeutic monoclonal IgG1-F′2 IC to hFcγRs. Histograms show the binding of anti-FLAG mAb (thin line) or its isotype control (solid gray), and the binding of indicated monoclonal IgG1-F′2 IC to FLAG-tagged hFcγRs on CHO transfectants. Concentrations of monoclonal IgG and PE-F(ab′)2 anti–human Ig are indicated. Monoclonal IgG1 used here were produced in CHO cells. Solid gray histograms represent the binding of PE-F(ab′)2 anti–human Ig alone; 2 independent experiments gave similar results.
Influence of mAb fucosylation on their affinity for hFcγRs
The glycosylation of antibodies critically determines their ability to bind to FcRs, and glycosylation variants have been generated, aiming at enhancing their therapeutic efficacy.44,45 We therefore measured the affinity for hFcγR of low-fucose-containing anti-CD20 and anti-RhD mAbs produced in YB2/0 cells, which have decreased levels of α-1,6-fucosyltransferase.46 These were compared with the same mAbs produced in α- 1,6-fucosyltransferase-competent CHO cells (Table 1). Fucosylation affected neither the affinity of the 2 mAbs for FcγRIIA and FcγRIIB/C, nor their higher affinity for FcγRIIIAV158 than for FcγRIIIAF158. Fucosylation decreased the affinity of both mAbs for FcγRIIIA and the affinity of anti-RhD, but not of anti-CD20, for FcγRIIIB. Fucosylation may therefore affect the affinity of IgG1 mAbs for FcγRIII, but not for FcγRII.
Discussion
We show here that (1) IgG1 and IgG3 bind to all hFcγRs; (2) IgG2, which were thought to bind to FcγRIIAH131 only, also bind to FcγRIIAR131, FcγRIIB, FcγRIIC, FcγRIIIAF158, and FcγRIIIAV158; (3) IgG4, which were thought to bind to FcγRI only, also bind to FcγRIIA, FcγRIIB, FcγRIIC, and FcγRIIIA; and (4) the inhibitory receptor FcγRIIB has a lower affinity for IgG1, IgG2, and IgG3 than other hFcγRs. Our data establish a hierarchy of affinities of hFcγRs for polyclonal IgG of all 4 subclasses that could not be established from previous studies. Our data document parameters which determine the affinity of hFcγRs, their specificity for IgG subclasses, and how hFcγR specificity and affinity determine the biological activities of antibodies. They also document the role of specific hFcγRs in disease and therapeutics.
FcγRs have been classified in 2 types, depending on their affinity for IgG. Classically, high-affinity FcγRs have a KA higher than 107 M−1, as measured mostly by Scatchard plot analysis, whereas low-affinity FcγRs have a KA lower than 107 M−1.1 Operationally, high-affinity FcγRs can bind IgG as monomers whereas low-affinity FcγRs cannot. Both types of FcγRs, however, can bind antigen-antibody immune complexes or IgG aggregates with a high avidity. Neither the KA threshold, which discriminates high-affinity from low-affinity FcγRs, nor the lower KA, which enables low-affinity FcγRs to bind IgG-immune complexes, is known. A functionally important consequence of this distinction is that low-affinity, but not high-affinity FcγRs, remain free in spite of the high concentrations of circulating IgG. Affinities previously described in the literature and affinities measured in our study are difficult to compare, as the ligands and the technical approaches are different. They are, however, in the same order of magnitude (summarized in Table S1). From our experimental setting, we were able to define the limit between high and low-affinity for a given IgG subclass at KA approximately 9 × 106 M−1. Interestingly, our data show that the affinity of a given FcγR is not absolute but relative. It depends on the subclass of IgG. Thus, FcγRIIIAV158 is a high-affinity receptor for IgG3 as defined by its ability to bind monomeric IgG3 but a low-affinity receptor for IgG1, IgG2, and IgG4, as defined by its ability to bind immune complexes made of these IgG subclasses. Likewise, murine FcγRI has a high affinity for mouse IgG2a and a low affinity for mouse IgG2b47 and IgG3. Our data also show that hFcγR polymorphism differentially affects their affinity for IgG subclasses. Thus, FcγRIIIAV158 has a high affinity for IgG3 and a low affinity for other subclasses, whereas FcγRIIIAF158 has a low affinity for all 4 subclasses. Likewise, the FcγRIIA H131R mutation decreases the affinity of the receptor for IgG2 but increases the binding of IgG4. FcγRIIIB polymorphism affects neither the affinity nor the binding of IgG IC. Contrasting with the previous report that the FcRγ subunit increases the affinity of FcγRIIIA for mouse IgG2a,43 we found that FcγRIIIA expressed in the absence of FcRγ retained the same ability as FcRγ-associated FcγRIIIA to bind human IgG, whatever the subclass. Finally, it is well known that unglycosylated IgG do not bind to FcγRs.48 Disialylated IgG represents only 5% of polyclonal IgG49 and less than 3% of the monoclonal anti-CD20, anti-RhD, and anti-HLA-DR studied here; their contribution, if any, to the affinity for human FcγRs could not be explored in this study. We show here that the fucosylation of monomeric IgG1 antibodies decreases their affinity for FcγRIIIA (and possibly FcγRIIIB), but not their affinity for FcγRIIA, FcγRIIB or FcγRIIC. These data altogether indicate that the affinity of FcγRs depends not only on the receptor type but also on the polymorphism of its extracellular domains, on the IgG subclass, and on IgG glycosylation.
FcγRs have been named after the class of Ig they can bind but not after IgG subclasses.1 Human FcγRs indeed display a class-specificity but no IgG subclass specificity. Noticeably, murine FcγRs can display a promiscuous specificity as both IgG and IgE can bind to mFcγRIIB, mFcγRIIIA50 and mFcγRIV47 and both IgA and IgM can bind to mFcα/μR.51 We confirm that no human FcγR is specific for one IgG subclass, and we show here that most hFcγRs have a measurable affinity for all IgG subclasses. Two exceptions are FcγRI, which does not bind IgG2, and FcγRIIIB, which does not bind IgG2 and IgG4, either as monomers or as ICs. hFcγRs, however, may display a selectivity for a given subclass, due to markedly different affinities. Thus, FcγRIIIA bind IgG3 with a 25-fold higher affinity than IgG4. One step further, hFcγRs may have a measurable affinity for some subclasses but not for others. Thus, FcγRI binds monomeric IgG1, IgG3, and IgG4, but not IgG2, either as monomers or as ICs, and FcγRIIIB bind immune complexes made of IgG1 and IgG3, but not of IgG2 and of IgG4. When having marked differences in affinity for different IgG subclasses, a given hFcγR may therefore functionally behave as being specific for one or several subclasses. In other words, quantitative variations may generate qualitative differences. Interestingly, our analysis of hFcγR specificity for IgG subclasses identifies specific residues which may determine hFcγR specificity. IgG2 and IgG4 ICs bind to FcγRIIIA but not to FcγRIIIB, independently of polymorphisms. If one excludes polymorphic variations, the extracellular domains of FcγRIIIA and FcγRIIIB differ by 2 amino acids only. G147 and/or Y158, but not D147 and/or H158, enable FcγRIII to bind IgG2 and IgG4 ICs. Although lacking a true subclass specificity, hFcγRs display a subclass selectivity.
Both FcγR specificity and affinity determine the biological activities of antibodies. Because high-affinity FcγRs are occupied in vivo, but not low-affinity FcγRs, these are readily available for ICs that are formed locally at a given time against a given antigen. Low-affinity FcγRs are therefore more suitable than high-affinity FcγRs for enabling antibodies to efficiently modulate cell responses during adaptive immune responses. Activating low-affinity hFcγRs are primarily FcγRIIA and FcγRIIIA. We found that FcγRIIA bind IgG1 with an approximately 5-fold higher affinity than IgG3. They also bind IgG2 and IgG4 with the same affinity but with an approximately 23-fold lower affinity than IgG1. We also found that FcγRIIIA bind IgG3 with an approximately 3-fold higher affinity than IgG1. They also bind IgG2 and IgG4, with an approximately 35-fold and an approximately 7-fold lower affinity than IgG1, respectively. These findings have the following 3 functional consequences. First, although IgG1, IgG2, and IgG4 bind each equally well to FcγRIIA and FcγRIIIA, IgG1 binds 4- to 40-fold better than IgG2 and IgG4. IgG1 can therefore engage more efficiently these 2 activating receptors than IgG2 and IgG4. Second, IgG3 bind approximately 6-fold better to FcγRIIIA than to FcγRIIA, and they bind to FcγRIIIA approximately 3-fold better than IgG1, approximately 100-fold better than IgG2, and approximately 24-fold better than IgG4. IgG3 can therefore preferentially engage FcγRIIIA and much more efficiently than IgG1, IgG2, or IgG4. This implies that IgG3 antibodies can efficiently activate FcγRIIIA-expressing NK cells, monocytes, and macrophages. Third, IgG2 and IgG4, which bind to FcγRIIA and FcγRIIIA, can trigger activation signals. Supporting this conclusion, IgG4 were reported to induce mediator release by polymorphonuclear neutrophils,52 and IgG2 and IgG4 to induce cytotoxicity in PBMC.53
We found that the inhibitory receptor FcγRIIB has a markedly lower affinity for IgG1, IgG2, and IgG3, but not for IgG4, than all other hFcγRs (except FcγRIIC). One can therefore wonder how FcγRIIB can inhibit IgG-induced cell-activation. FcγRIIB may be expressed at a higher density than activating hFcγRs on cell membranes. The relative expression of these receptors is indeed not known. It is, however, well known that FcγRIIB must be coengaged with activating hFcγRs by the same immune complex to inhibit cell activation.54 Their concomitant binding to 2 types of receptors on the same cell can markedly enhance the binding avidity of the immune complex to coengaged FcγRs. Activating hFcγRs may therefore “help” FcγRIIB to bind antibodies and to exert their inhibitory effects. As they have an identical extracellular domain as FcγRIIB, a reverse situation may apply to FcγRIIC, which is expressed on NK cells in 40% individuals.55 Under these conditions, FcγRIIC may potentiate FcγRIIIA-dependent NK cell cytotoxicity.
hFcγR affinity and specificity may account for recognized associations between disease and hFcγR polymorphism. The FcγRIIIA V158F polymorphism is associated with SLE and RA.30 FcγRIIIAF158 was described to bind IgG1 less efficiently than FcγRIIIAV158.20 We found that it also binds less efficiently IgG2 and IgG4. FcγRIIIAF158 may therefore be less efficient than FcγRIIIAV158 to eliminate ICs in these diseases and/or to activate inflammatory cells. Likewise, the FcγRIIA H131R polymorphism is associated with bacterial infections17 and nephropathy.16 We confirm that FcγRIIAR131 binds IgG2 IC less efficiently than FcγRIIAH131. IgG2 is a major isotype in anti-bacterial antibody responses. Although FcγRIIIB polymorphisms have been associated with autoimmune diseases,22,24 we found no differences in the binding abilities of FcγRIIIBNA1 and FcγRIIIBNA2. The association may depend on copy numbers of the gene encoding FcγRIIIB, as described recently.15,56
Finally, hFcγR affinity is critical for antibody-based immunotherapy. The mechanism of depletion of non-Hodgkin B-cell lymphoma by anti-CD20 therapy has been linked to FcγRs in humans and in mouse models (reviewed in Taylor and Lindorfer57 ), while the mechanism of suppression induced by anti-RhD therapy in the prevention of hemolytic disease of the newborn has been shown to be both FcγR-dependent and -independent (reviewed in Kumpel and Elson58 ). Oxidative bursts by myeloid cells, phagocytosis of RBC, and IL1-receptor antagonist production occurred following IgG-, but not F(ab′)2 fragments of anti-RhD,59 suggesting FcγR-dependent mechanisms. Surprisingly, mice rendered deficient for all FcγRs did not affect RBC clearance, suggesting a contribution of FcγR-independent mechanisms, such as epitope masking.60 Mechanisms underlying tumor killing by anti–HLA-DR IgG1 mAbs are not well identified yet, but FcγRI+ PMNs have been shown critical in ex vivo assays.61 Our finding that IgG1 anti-CD20 antibodies or IgG1 bind less efficiently to FcγRIIAR131 than to FcγRIIAH131 may explain the lower therapeutic efficacy of rituximab in FcγRIIAR131 patients.62 Our finding that IgG1 anti-RhD antibodies bind less efficiently to FcγRIIAR131 than to FcγRIIAH131 and to FcγRIIIAF176 than to FcγRIIIAV176 may explain the lower clearance of RhD+ RBC in R/R131 and in F/F158 subjects.63 Our finding that IgG4 ICs binds to FcγRIIA and FcγRIIIA may explain the “cytokine storm” that recently led to the hospitalization, due to multiple organ dysfunction, of volunteers involved in the IgG4 anti-CD28 clinical trial (TGN-41264 ). Our finding that IgG4 is the only IgG subclass that binds equally well to FcγRIIB and to other hFcγRs supports the suspected beneficial role of IgG4 antibodies during desensitization of allergic patients. Knowing the binding selectivity of specific IgG subclasses for specific FcγRs, the signaling capacity of these receptors and the function of IgG subclasses (ie, their ability to induce cell activation, phagocytosis, and cytotoxicity65 ) will enable therapeutic antibodies to be optimized for higher efficacy and fewer side effects.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are thankful to our colleagues for their generous gifts: U. Jacob (SuppreMol GmbH, Munich, Germany) for antibodies; J. Van de Winkel and J. Leusen (University Medical Center Utrecht, Utrecht, The Netherlands), and S. Santoso and U. Sachs (Institute for Clinical Immunology and Transfusion Medicine, Giessen, Germany) for cDNAs. We thank A. Louise and H. Kiefer-Biasizzo (Plate-Forme de Cytométrie, Institut Pasteur, Paris, France) for cell sorting, S. Hoos (Plateforme de Biophysique des Macromolécules et de leurs Interactions, Institut Pasteur, Paris, France) for assistance with Biacore experiments, and D. N. Gopaul (Unité d'Immunologie Structurale, Institut Pasteur, Paris, France) for providing 3-dimensional representations of FcγRs.
D.A.M. is the recipient of a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche. This work was supported by the Institut Pasteur and Inserm, by grants from Agence Nationale de la Recherche (Paris, France; 05-JCJC-0236-01; P.B.), Fondation pour la Recherche Médicale (Paris, France; Défis de la Recherche en Allergologie; M.D.), PTR/PIC Institut Pasteur–Institut Curie (Paris, France; Immunotherapy & Cancer; P.B. and M.D.), and by Funding under the Sixth Research Framework Program of the European Union, Project MUGEN (LSHG CT 2005-005203; P.B.). This study was supported in part by research funding from Laboratoire Français du fractionnement et des Biotechnologies (LFB) to P.B.
Authorship
Contribution: P.B., B.I., and D.A.M. performed experiments; N.F. and S.J. discussed results and provided reagents; P.B., P.E., and M.D. analyzed results; P.B. designed the research; and P.B. and M.D. wrote the paper.
Conflict-of-interest disclosure: S.J. is employed by LFB, and N.F. was employed by LFB at the time of the study, whose potential products are studied in the present work. P.B., B.I., P.E., D.A.M., and M.D. declare no competing financial interests.
Correspondence: Pierre Bruhns, Unité d'Allergologie Moléculaire et Cellulaire, Institut Pasteur, 25 rue du Docteur Roux, 75015 Paris, France; e-mail: bruhns@pasteur.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal