Abstract
Megakaryoblastic leukemia 1 (MKL1), identified as part of the t(1;22) translocation specific to acute megakaryoblastic leukemia, is highly expressed in differentiated muscle cells and promotes muscle differentiation by activating serum response factor (SRF). Here we show that Mkl1 expression is up-regulated during murine megakaryocytic differentiation and that enforced overexpression of MKL1 enhances megakaryocytic differentiation. When the human erythroleukemia (HEL) cell line is induced to differentiate with 12-O-tetradecanoylphorbol 13-acetate, overexpression of MKL1 results in an increased number of megakaryocytes with a concurrent increase in ploidy. MKL1 overexpression also promotes megakaryocytic differentiation of primary human CD34+ cells cultured in the presence of thrombopoietin. The effect of MKL1 is abrogated when SRF is knocked down, suggesting that MKL1 acts through SRF. Consistent with these findings in human cells, knockout of Mkl1 in mice leads to reduced platelet counts in peripheral blood, and reduced ploidy in bone marrow megakaryocytes. In conclusion, MKL1 promotes physiologic maturation of human and murine megakaryocytes.
Introduction
Acute megakaryoblastic leukemia (AMKL, or AML variant M7) is associated with at least 2 distinct cytogenetic abnormalities: trisomy of chromosome 211,2 and the reciprocal t(1;22)(p13;q13) chromosomal translocation.3-7 Both types of AMKL are characterized by an expansion of megakaryoblasts in the bone marrow (BM), myelofibrosis, and thrombocytopenia.8 The reciprocal t(1;22)(p13;q13) chromosomal translocation4-7 results in the fusion of RNA binding motif protein 15 (RBM15, also known as OTT) and MKL1 (also known as BSAC, MAL, and MRTF-A) genes on chromosomes 1 and 22, respectively. Although both reciprocal fusion transcripts are expressed in AMKL, RBM15-MKL1 encompasses all of the putative functional motifs encoded by each gene, and is thus the candidate oncoprotein of the t(1;22) translocation.7 Investigations into the role of RBM15 suggest that it may inhibit myeloid differentiation by modulating the Notch1 signaling pathway.9,10 No role for MKL1 has yet been defined in hematopoietic differentiation.
MKL1 is a member of the myocardin family of transcriptional coactivators with MKL2 (also known as MRTF-B) and myocardin. This family has several highly conserved domains, including RPEL (arginine, proline, glutamic acid, and leucine) repeats in the N terminus that bind actin11 ; a B-box and a glutamine-rich domain, which bind serum response factor (SRF)11 ; a leucine zipper-like domain that may play a role in homo- and heterodimerization11 ; and a C-terminal transactivation domain. All myocardin family proteins bind SRF and activate transcription from promoters with SRF binding sites called serum response elements.11,12 Regulation of MKL1 activity has been studied in fibroblast and muscle cell differentiation.13,14 Until it is activated, MKL1 is bound to monomeric G-actin and cycles between the cytoplasm and the nucleus. When RhoA activates actin polymerization (F-actin formation), MKL1 is released from G-actin and accumulates in the nucleus, where it functions as a coactivator to turn on SRF target genes.15,16 MKL1 can dimerize with other myocardin family members and is required for RhoA-mediated SRF induction of immediate early and muscle specific genes.13 MKL1 inhibits tumor necrosis factor-induced cell death in embryonic fibroblasts,17 which may be relevant to its role in leukemia. MKL1 is a critical mediator of the transforming growth factor-β1–induced epithelial-mesenchymal transition via Smad3.18 Mice lacking Mkl1 expression either die in utero because of cardiac abnormalities or develop to birth but have abnormal mammary gland function.19,20 Until now, hematopoiesis and in particular megakaryocytopoiesis have not been studied in these mice.
Megakaryocytes are large, polyploid cells of the hematopoietic tissues, whose final differentiation step involves the release of platelets into the bloodstream. The differentiation sequence of megakaryocytopoiesis progresses from hematopoietic stem/progenitor cells, to more committed biphenotypic megakaryocyte-erythroid precursors,21 burst-forming unit–megakaryocytes, colony-forming unit megakaryocytes (CFU-MK), and megakaryoblasts.22 Maturation continues with the cells enlarging as they undergo endomitosis, which results in polyploid megakaryocytes. After polyploidization, proplatelet formation begins, which involves complex cytoskeletal rearrangements,23 and is a prerequisite for platelet release.
To better understand the mechanism by which the RBM15-MKL1 translocation causes defects in megakaryocytic differentiation associated with AMKL, we need to know the normal roles of the component proteins. Although previous reports suggest that MKL1 is ubiquitously expressed,6 we demonstrate that the Mkl1 expression level increases significantly during megakaryocytopoiesis, suggesting that MKL1 plays a role in megakaryocyte maturation. We show that overexpression of MKL1 in human erythroleukemia (HEL) cells and in primary human CD34+ cells enhances megakaryocytopoiesis, an effect that is abrogated by knock down of SRF. Consistent with this role in megakaryocytic differentiation, we find that platelet counts in mice lacking Mkl1 are decreased. On examination of the BM of Mkl1-deficient mice, we observe a higher percentage of total CD41+ megakaryocytes but a significant decrease in mature polyploid megakaryocytes, suggesting that the reduced platelet count may be the result of a block in megakaryocyte differentiation.
Methods
Stable transfection of HEL cells with inducible MKL1
HEL cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine and penicillin/streptomycin at 37°C in 5% CO2 in a humidified incubator. To induce differentiation, 2 × 105 cells per mL were cultured in the presence of 1.5 × 10−8 M 12-O-tetradecanoylphorbol 13-acetate (TPA; Sigma-Aldrich, St Louis, MO) for 3 days or as specified. To establish HEL cells with inducible MKL1, cells were first transfected with pcDNA6/TR vector (Invitrogen, Carlsbad, CA) encoding the Tet repressor on the CMV promoter by nucleofection (Amaxa Biosystems, Gaithersburg, MD) using Solution V and program T030 as described by the manufacturer. Stable transfectants containing pcDNA6/TR were selected by growth in 5 μg/mL blasticidin. MKL1-His cDNA was then cloned into a pcDNA5/TO vector (Invitrogen) to express MKL1-His on the Tet responsive promoter. Next, MKL1-pcDNA5/TO vector was transfected into the HEL-pcDNA6/TR cells by nucleofection; 375 μg/mL hygromycin B (Invitrogen) was used to select stable cell lines. Induction of MKL1-His expression with 0.1 μg/mL doxycycline was confirmed by Western blotting using His-probe antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Clones with high overexpression of MKL1 were isolated and expanded. Among these clones, we chose to use MKL1 clone 5, which expresses high levels of MKL1 protein on doxycycline addition. From this point on, we will refer to this cell line as HEL/MKL1.
Bone marrow collection and MegaCult-C assays
Mkl1 knockout (KO) mice were produced as previously described.19 Two- to 4-month-old, age- and gender-matched wild-type (WT), heterozygous (HET), KO littermates, and C57BL/6 mice were used in these studies. After anesthesia and humane killing according to Yale University Animal Care–approved protocols, femurs and tibias were excised. For histology sections, femurs were immersed in 4% paraformaldehyde (PFA) overnight for fixation, followed by decalcification in Decalcifier I solution (Surgipath, Richmond, IL) for 6 hours. The bones were then dehydrated in 70% ethanol and sent to the Research Histology Facility at Yale Medical School for 5-μm longitudinal paraffin sections and hematoxylin and eosin staining. Spleens were fixed in 4% PFA overnight and processed as mentioned for paraffin sections. To quantify megakaryocytes in BM, hematoxylin and eosin-stained paraffin sections were used. The samples were then examined using an Olympus BX51 microscope equipped with objective lenses (40×/0.75, 20×/0.4, 10×/0.25, and 4×/0.1) and a 10× eyepiece, and an Olympus Q-Color5 camera (Olympus America, Melville, NY) and QCapture Pro software (QImaging, Surrey, BC). Total megakaryocyte number was counted in each 20× observation field. BM cell suspensions were harvested by flushing with Iscove modified Dulbecco medium/2% fetal bovine serum followed by filtering through a 100-μm nylon strainer to remove bone debris. For MegaCult-C assays, a total of 105 BM mononuclear cells were used according to the manufacturer's protocols (StemCell Technologies, Vancouver, BC); 50 ng/mL human thrombopoietin (TPO), 50 ng/mL human interleukin-11 (IL-11), 10 ng/mL murine IL-3, and 20 ng/mL human IL-6 (PeproTech, Rocky Hill, NJ) were used in these assays. The cultures were incubated at 37°C with 5% CO2 for 6 to 8 days. Colonies containing more than 3 megakaryocytes (AchE+ cells) were considered CFU-MKs. Duplicate assays were performed for each mouse.
Murine peripheral blood counts
Mice were anesthetized with methoxyflurane (Medical Developments International, Springvale, Australia) followed by retro-orbital bleeding (∼100 μL) using ethylenediaminetetraacetic acid–treated glass capillary tubes. The blood was evacuated into tubes with 5 μL of 0.5 M ethylenediaminetetraacetic acid to prevent clot formation. The complete blood count of blood sample was performed using a Hemavet 950FS (Drew Scientific, Oxford, CT) according to the manufacturer's protocol.
Serum TPO concentration measurement
Quantikine Murine TPO Immunoassay Kit (R&D Systems, Minneapolis, MN) was used to quantify TPO concentration in harvested sera according to the manufacturer's protocol.
Primary murine megakaryocyte enrichment
Primary megakaryocytes were derived from murine E12.5-E14.5 fetal livers. After lysis of mature red blood cells by BD PharmLyse (BD Biosciences, San Jose, CA), cells were cultured at 2 × 106 cells/mL in StemSpan (StemCell Technologies), 30% BIT9500 (StemCell Technologies), 50 ng/mL murine TPO (PeproTech), 2 mM l-glutamine, and penicillin/streptomycin. After 4 days, the cells were fractionated on a 3% discontinuous bovine serum albumin (BSA; Sigma-Aldrich) gradient.24 The fully mature polyploid megakaryocytes were mainly in the pellet below the 3% BSA fraction. For some studies, to enrich megakaryocyte progenitors, mouse BM cells were cultured for 1 day in the same medium and CD41+c-kit+Ter119− cells, stained with CD41–fluorescein isothiocyanate (FITC), c-kit–allophycocyanin, Ter119 phycoerythrin (PE)–Cy7 (BD Biosciences), were sorted using a MoFlo sorter (Dako North America, Carpinteria, CA). All procedures were performed in compliance with relevant laws and institutional guidelines and were approved by the Yale University Institutional Animal Care and Use Committee.
Flow cytometric analysis of DNA content and surface markers
HEL cell DNA content was determined by fixing cells with 50% ethanol at 4°C overnight, followed by digestion with 20 μg/mL RNase for 20 minutes at 37°C, and staining with 1 μg/mL propidium iodide (Sigma-Aldrich). To analyze human CD34+ cell-derived megakaryocytes, the cells were stained with standard protocols using allophycocyanin anti-CD42b, PE-Cy5 anti-CD41a, and PE anti-CD61 (all BD Biosciences). For ploidy analysis, human megakaryocytes were stained with FITC anti-CD41a antibody, fixed with 1% PFA at room temperature for 15 minutes, and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline at 4°C for 15 minutes. Nuclear DNA was then stained with 4,6-diamidino-2-phenylindole at 1 μg/mL. To determine mouse BM LSK cells and pre-MegE (erythro-megakaryocytic progenitor cells),21 freshly obtained BM cells were stained with PE-Cy5 anti-CD150 (BioLegend, San Diego, CA), FITC anti-CD41 (BD Biosciences), Alexa Fluor 647 anti-Sca-1, PE-Cy7 anti-CD117 (BD Biosciences), Alexa Fluor 488 anti-CD105 (BioLegend), biotin-labeled lineage cell detection cocktail (Miltenyi Biotec, Auburn, CA), and antibiotin-PE (Miltenyi Biotec). Pre-MegE cells were gated as lin−sca−kit+CD41−CD105−CD150+ cells.21 BM megakaryocyte ploidy was analyzed by the same method as for human CD41a+ cells. Flow cytometric analysis was performed with an LSRII flow cytometer (BD Biosciences) and FlowJo software (TreeStar, Ashland, OR). Isotype controls were used in each experiment.
Real time RT-PCR
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche Diagnostics, Indianapolis, IN). For samples with less than 5 × 104 cells, total RNA was extracted using the RNAqueous-Micro Kit (Applied Biosystems, Foster City, CA). For all RNA samples, genomic DNA was digested with RNase-free DNase I. cDNA was prepared using Superscript II Reverse Transcriptase (Invitrogen) with random primers (Invitrogen). Gene expression levels were determined using Applied Biosystems TaqMan Gene Expression Assays: murine Mkl1: Mm00461840_m1; murine Kit: Mm00445212_m1; murine Gata1: Mm01352636_m1; murine Nfe2: Mm00801891_m1, and detected by iCycler iQ (Bio-Rad, Hercules, CA). Eukaryotic18S rRNA: Hs99999901_s1 was used as an internal control for normalization. Standard curves were performed for each PCR.
RNA interference
The SRF Validated Stealth DuoPak 12936-64 (Invitrogen) containing 2 validated siRNA duplexes was used to decrease SRF transcript levels in the HEL cell line using nucleofection as described; 1.5 × 10−8 M TPA was added to the cells 48 hours after transfection. The transfection efficiency was 80% to 90% in all the cases. The efficiency of down-regulation was determined to be greater than 80% by Western blot. Stealth RNAi negative control (Invitrogen) was used in this study.
Human CD34+ cell lentiviral transduction and megakaryocytic differentiation
To make pCCL-C-MND-flag human MKL1 (pCCL-MKL1), the Flag-MKL1 coding sequence was cloned from pCDNA3-Flag- MKL112 using BamH1 restriction sites, and ligated to an EcoR1 linker (Invitrogen). The resulting fragment was ligated into the EcoR1 site of the pCCL-C-MND lentiviral vector (a kind gift from Dr Carolyn Lutzko, Children's Hospital, Los Angeles, CA). For this self-inactivating vector, transgene expression is driven by the MND retroviral promoter and green fluorescent protein (GFP) expression by the PGK promoter.
For lentivirus production, 90% confluent 293FT cells (Invitrogen) in T-175 flasks were transfected with 27.5 μg pCCL (or pCCL-MKL1), 17.5 μg Gag/Pol, 6.9 μg Rev, and 9.7 μg VSVG (the packaging plasmids were kind gifts from Dr Carolyn Lutzko) with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Virus was collected at 24, 48, and 72 hours after transfection, and each collection was 0.45-μm filtered, concentrated using Amicon filters (Millipore, Billerica, MA), and frozen in aliquots at −80°C for further use.
Peripheral blood mononuclear cells were collected from a granulocyte colony-stimulating factor mobilized healthy donor, and immunomagnetic selection for CD34+ cells was performed using the Baxter Isolex device (Baxter, Deerfield, IL). Cells were cryopreserved in 10% dimethyl sulfoxide. For megakaryocytic differentiation on day 0, CD34+ cells were thawed and cultured at 5 × 105 cells/mL in 100 ng/mL human TPO and 50 ng/mL human stem cell factor (SCF) in StemSpan Serum Free Expansion Medium (Stem Cell Technologies). On day 1, cells were infected with pCCL-MKL1 or control virus by spinfection in a volume of 750 μL in the presence of polybrene (8 μg/mL). Spinfection was performed at 900g for 2 hours at 30°C. On day 9 after culture in the presence of TPO and SCF, megakaryocytic markers CD41a, CD61, and CD42b (BD Biosciences) were measured by flow cytometry in the GFP-positive fraction. Transduction efficiencies of pCCL control and pCCL-MKL1 viruses were generally 50% to 60% and 5% to 10%, respectively. For the ploidy experiments, CD34+ cells were expanded in StemSpan medium containing 10 μM hydrocortisone (Sigma-Aldrich), 100 ng/mL human SCF, 13.3 ng/mL human BMP4, 1.33 U/mL human erythropoietin, 33.3 ng/mL human Flt3-ligand, 13.3 ng/mL human IL-3, and 50 ng/mL human TPO (PeproTech). After 3 days of expansion, the cells were washed, differentiated in StemSpan with 50 ng/mL SCF and 100 ng/mL TPO, and infected with pCCL control or MKL1 lentivirus. After 7 days of differentiation, the CD41+ cell number was generally expanded 10-fold compared with the culture without 3 days of expansion. The CD41+ cells were then analyzed for ploidy. Use of human subjects was approved by the Yale Institutional Review Board, and informed consent was obtained in accordance with the Declaration of Helsinki.
Gene expression analysis using cDNA microarrays
HEL/MKL1 and control cells were treated with TPA and doxycycline for 24 hours, and total RNA prepared from 2 biologic repeats was used on Affymetrix Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA). Normalization and statistical analysis of the expression data were carried out using the limma software package for the R programming environment.25,26 Differential expression of the combined MKL/control cells' expression data before and after exposure to 1 day of TPA was analyzed using linear model methods.27,28 The linear models allowed for assessment of general changes in gene expression between different conditions and across different biologic replicates. Each probe was tested for changes in expression based on a bifactorial design in which the presence or absence of exposure to TPA was one factor and doxycycline-inducible activation of MKL1 was the other factor. This test is similar to an analysis of variance method for each probe except that the residual SDs are moderated across genes, as a compromise between the individual gene-wise SDs and an overall pooled SD, to provide more stable inference for each gene. Statistics were computed for each probe to test for a response to TPA, for a response to induction with doxycycline, and for any differences in the pattern of response to TPA compared with doxycycline. Each probe therefore gave 3 statistics and 3 P values: one for TPA response, one for doxycycline response, and one contrasting the TPA and doxycycline responses. P values were adjusted for multiple testing using the method of Benjamini and Hochberg29 to control for false discovery rate. The same RNA samples from TPA-treated HEL/MKL1 cells with and without doxycline were analyzed using Illumina Sentrix Human Ref-8_V2 Expression Beadchips (Illumina, San Diego, CA). The Illumina data were quantile normalized using the Affy package for the R programming environment. The Illumina data were then analyzed using the limma package to detect genes that were changed in response to MKL overexpression. The 1000 probe sets that were most significantly affected by MKL on both microarray platforms were combined to yield an overlap set of 176 genes.
Statistical analysis
Statistical significance of differences between different conditions was performed using Prism (GraphPad Software, La Jolla, CA) with a 2-tailed unpaired t test.
Results
Mkl1 is up-regulated during murine megakaryocytic differentiation
We first assessed whether Mkl1 is differentially expressed during megakaryocyte differentiation. Cells from E12.5-E14.5 fetal livers were cultured with TPO and harvested for RNA daily for 4 days. On day 4, the cells (comprised predominantly of myelomonocytic cells and megakaryocytes) were fractionated on a discontinuous BSA gradient based on cell size.24 The pellet below the 3% BSA fraction was most enriched (∼ 80% purity) for fully mature polyploid megakaryocytes based on morphology (data not shown) and DNA content (Figure 1A). Real-time quantitative reverse-transcribed polymerase chain reaction (RT-PCR) analysis indicated that Mkl1 is expressed throughout megakaryocytic differentiation. However, Mkl1 is expressed at the highest levels on day 4 (Figure 1B left) and is enriched in the fraction of mature megakaryocytes (Figure 1B right).
Expression of Mkl1 during megakaryocyte differentiation. (A) Freshly isolated E12.5-E14.5 fetal liver cells were differentiated in the presence of TPO, IL-6, and IL-11. At day 4, different cell types (myelomonocytic cells vs megakaryocytes) were separated on a discontinuous BSA gradient. Analysis of cell ploidy validates the megakaryocyte fractionation. (B) Relative Mkl1 mRNA levels were assessed by quantitative RT-PCR on fetal liver subpopulations. The RNA from total fetal liver cells was used from day 0 to day 4 (left). At day 4, RNA from fractionated cells was used (right). The relative expression levels were normalized to 18S rRNA. (C) c-kit+CD41+ BM cells were sorted (day 1) and cultured with TPO for 4 more days. RNA at different time points was used to assess relative Mkl1 levels by quantitative RT-PCR. The Mkl1 expression was normalized to 18S rRNA or absolute cell number. Mean plus or minus SEM of duplicate experiments is represented.
Expression of Mkl1 during megakaryocyte differentiation. (A) Freshly isolated E12.5-E14.5 fetal liver cells were differentiated in the presence of TPO, IL-6, and IL-11. At day 4, different cell types (myelomonocytic cells vs megakaryocytes) were separated on a discontinuous BSA gradient. Analysis of cell ploidy validates the megakaryocyte fractionation. (B) Relative Mkl1 mRNA levels were assessed by quantitative RT-PCR on fetal liver subpopulations. The RNA from total fetal liver cells was used from day 0 to day 4 (left). At day 4, RNA from fractionated cells was used (right). The relative expression levels were normalized to 18S rRNA. (C) c-kit+CD41+ BM cells were sorted (day 1) and cultured with TPO for 4 more days. RNA at different time points was used to assess relative Mkl1 levels by quantitative RT-PCR. The Mkl1 expression was normalized to 18S rRNA or absolute cell number. Mean plus or minus SEM of duplicate experiments is represented.
To further clarify whether Mkl1 mRNA levels increase during megakaryocytic maturation as opposed to simply being expressed maximally in megakaryocytes versus other blood cell types, we cultured murine BM cells with TPO for 1 day, sorted c-kit+CD41+ megakaryocytic progenitors (day 1), and differentiated them into mature megakaryocytes in vitro (day 2, day 5). Because of the active biogenesis during differentiation,30 we could underestimate the change when the data are normalized to regular internal controls. Therefore, we graphed our quantitative RT-PCR in 2 ways, normalized to 18S levels (Figure 1C left) or to cell number (Figure 1C right). The data showed a 2.26-fold increased in Mkl1 mRNA levels when normalized to 18S and 26.6-fold increase when normalized to cell number on day 5 of differentiation (Figure 1C). Normalization using hypoxanthine phosphoribosyl transferase gave a similar fold change as 18S (data not shown). As further confirmation that megakaryocytes were maturing in this culture system (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), we found that Nf-e2 and Gata1 mRNA were up-regulated, and c-kit was down-regulated over time (Figure S1B). In conclusion, Mkl1 is up-regulated during megakaryocyte differentiation.
Ectopic expression of MKL1 promotes megakaryocytic differentiation
The HEL cell line, which can be differentiated along the megakaryocytic and erythroid lineages, was used as a model system to assess the effects of MKL1 on megakaryocyte differentiation. When HEL cells are treated with TPA, they differentiate into megakaryocyte-like cells that are polyploid, and in which megakaryocyte markers such as CD41a and CD61 are up-regulated, whereas erythroid markers such as glycophorin A are down-regulated.31,32
To examine whether overexpression of MKL1 protein affects megakaryocytic differentiation, we established stably transfected HEL cell clones with doxycycline-inducible MKL1 (Figure 2A). In the presence of TPA for 4 days, enforced MKL1 expression increased the average percentage of mature megakaryocytes from 42% to 94% based on analysis of Wright-Giemsa–stained cytospins (P < .05; Figures 2B, S2). The percentage of cells with 2N ploidy decreased from 45% to 20% (P < .05), 4N ploidy increased from 30% to 32%, 8N ploidy increased from 17% to 36% (P < .05), and 16N ploidy increased from 6% to 10% (Figure 2C). Megakaryocytic differentiation is accompanied by increased cell adhesion properties.33 In the HEL cell system, TPA-treated MKL1 overexpressing cells had increased adherence to the culture plates and were more spread out compared with control cells (Figure 2D). The adherent cells were 100% CD41+ and were enriched for megakaryocytes with more than 2N ploidy (data not shown). Overall, overexpression of MKL1 significantly enhanced megakaryopoiesis in HEL cells.
MKL1 promotes human megakaryocytic differentiation. (A) Validation of 3 HEL cell clones (clone numbers 5, 8, and 9) using Western blot analysis of MKL1 expression in the absence (left) and presence (right) of Dox for 2 days. MKL1-His protein is detected by anti-His antibody. (B) Based on analysis of Wright-Giemsa–stained cytospins, MKL1 increases the percentage of mature megakaryocytes in response to TPA over 4 days (P < .05). (C) Representative data showing ploidy of TPA-stimulated HEL/MKL1 cells or control cells after 4 days of differentiation with (solid line) or without (tinted) Dox. MKL1 increases the ploidy of cells exposed to TPA. Note increased 8N and 16N peaks in HEL/MKL1 cells. (D) Morphology of HEL cells treated with TPA and/or Dox as indicated. MKL1 has no effect on cell morphology in the absence of TPA (compare untreated with Dox), and promotes adhesion and spreading when cells are exposed to TPA (compare TPA with TPA + Dox). (E,F) The effects of enforced expression of MKL1 in human CD34+ cells during megakaryocytic differentiation. (E) The figures shown are gated for GFP-positive cells infected with pCCL and pCCL-MKL1 virus. Average percentage-positive cells for CD61, CD41a, and CD42b expression plus or minus SD of 3 independent experiments. *P < .05 from pCCL versus pCCL-MKL1 in the respective immunophenotype group. (F) Ploidy distribution of CD41a+ fraction in pCCL and pCCL-MKL1–infected cells plus or minus SD of 3 independent experiments. *P < .05 from pCCL versus pCCL-MKL1 in the respective ploidy group.
MKL1 promotes human megakaryocytic differentiation. (A) Validation of 3 HEL cell clones (clone numbers 5, 8, and 9) using Western blot analysis of MKL1 expression in the absence (left) and presence (right) of Dox for 2 days. MKL1-His protein is detected by anti-His antibody. (B) Based on analysis of Wright-Giemsa–stained cytospins, MKL1 increases the percentage of mature megakaryocytes in response to TPA over 4 days (P < .05). (C) Representative data showing ploidy of TPA-stimulated HEL/MKL1 cells or control cells after 4 days of differentiation with (solid line) or without (tinted) Dox. MKL1 increases the ploidy of cells exposed to TPA. Note increased 8N and 16N peaks in HEL/MKL1 cells. (D) Morphology of HEL cells treated with TPA and/or Dox as indicated. MKL1 has no effect on cell morphology in the absence of TPA (compare untreated with Dox), and promotes adhesion and spreading when cells are exposed to TPA (compare TPA with TPA + Dox). (E,F) The effects of enforced expression of MKL1 in human CD34+ cells during megakaryocytic differentiation. (E) The figures shown are gated for GFP-positive cells infected with pCCL and pCCL-MKL1 virus. Average percentage-positive cells for CD61, CD41a, and CD42b expression plus or minus SD of 3 independent experiments. *P < .05 from pCCL versus pCCL-MKL1 in the respective immunophenotype group. (F) Ploidy distribution of CD41a+ fraction in pCCL and pCCL-MKL1–infected cells plus or minus SD of 3 independent experiments. *P < .05 from pCCL versus pCCL-MKL1 in the respective ploidy group.
To determine whether MKL1 affects megakaryocytic differentiation in primary cells, we used lentiviral vectors to express MKL1 in primary human CD34+ cells. CD34+ cells were infected with pCCL (encoding GFP) or pCCL-MKL1 (encoding MKL1 and GFP) and cultured in medium containing TPO and SCF to promote megakaryocytopoiesis. Flow cytometric analysis for CD41a, CD61, and CD42b was performed after 8 days of culture. In 3 independent experiments, we observed an increase in the percentages of CD41a, CD61, and CD42b-positive cells in the GFP-positive MKL1 overexpressing cells compared with control GFP-positive cells. As shown in Figure 2E, enforced MKL1 expression increased CD41a expression from 37% to 61%, CD61 from 37% to 64%, and CD42b from 15% to 39%. Consistent with the HEL cell data, MKL1 overexpression also promotes primary human megakaryocytes to become polyploid. In the CD41a+ fraction, the percentage of cells with 2N ploidy decreased from 35% to 23%, 4N ploidy decreased from 31% to 21%, 16N ploidy increased from 9% to 19%, and 32N ploidy increased from 2% to 8% (P < .05; Figure 2F).
The effects of MKL1 in HEL cells are mediated through SRF
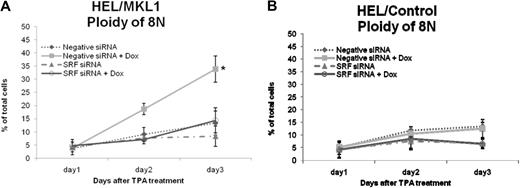
In muscle and fibroblast cells, MKL1 is associated with G-actin. In response to RhoA activation, MKL1 dissociates from G-actin and accumulates in the nucleus where it forms a complex with SRF to activate transcription of SRF target genes.11 Here we tested whether MKL1 and SRF together play a role in megakaryocytic differentiation. We transiently transfected SRF siRNA into HEL/MKL1 cells and assessed the ploidy in the presence or absence of doxycycline. We first validated that SRF protein was knocked down at least 80% by siRNA, whereas the MKL1 protein level was not affected (Figure S3). Figure 3 shows the percentage of cells with 8N ploidy under different culture conditions with doxycycline and transfection of siRNA against SRF. MKL1 overexpression increases the percentage of 8N cells from 4% on day 1 to 34% on day 3 compared with from 4% to 13% in the controls (Figure 3A, closed squares and closed diamonds, respectively). This effect of MKL1 on ploidy was abrogated when cells were transfected with siRNA against SRF; ploidy increases from 5% on day 1 to only 14% on day 3 (Figure 3A open circles), indicating that MKL1 acts via SRF to enhance megakaryocytic differentiation. Interestingly, SRF siRNA blocked the TPA induced polyploidy in HEL/MKL1 cells in the absence of doxycycline (8% compared with 13% on day 3; Figure 3A, closed triangles and closed diamonds, respectively, P = .02) and in control cells (7% compared with 13% on day 3; Figure 3B, closed triangles and closed diamonds, respectively, P = .001), suggesting that SRF plays a role in polyploidization. In conclusion, the results show that MKL1 promotes polyploidy through SRF in HEL cells.
Knockdown of SRF inhibits MKL1-induced increase in ploidy in response to TPA. HEL/MKL1 cells (A) and HEL/control cells (B) were treated with Dox and siRNA against SRF as indicated, and ploidy was measured over time (x-axis) after TPA addition. The y-axis represents the percentage of cells that had a ploidy of 8N. *P < .005, negative siRNA + Dox versus the rest of the groups.
Knockdown of SRF inhibits MKL1-induced increase in ploidy in response to TPA. HEL/MKL1 cells (A) and HEL/control cells (B) were treated with Dox and siRNA against SRF as indicated, and ploidy was measured over time (x-axis) after TPA addition. The y-axis represents the percentage of cells that had a ploidy of 8N. *P < .005, negative siRNA + Dox versus the rest of the groups.
To better elucidate the molecular mechanisms by which enforced MKL1 expression enhances megakaryocytic differentiation, we compared the RNA expression profiles of TPA-differentiated HEL/MKL1 cells with and without MKL1 overexpression using both Affymetrix and Illumina microarray platforms. After 24 hours of differentiation with TPA, MKL1 overexpression increased megakaryocytic markers, such as GATA1, GATA2, and GP5 (Table S1; Figure S4). Furthermore, MKL1 expression led to increased expression of cytoskeletal and adhesion molecule genes that are known to be SRF targets34-36 (Table S1), and are up-regulated during megakaryocytic differentiation,37-39 such as CALD1, MYLK, TPM, and MYOM1. Expression data described in this report have been deposited in the Gene Expression Omnibus under the accession series number GSE 13710.40
Reduced platelet number in Mkl1 KO mice
To determine the role of Mkl1 in vivo, we examined the hematopoietic system in Mkl1 KO mice. Complete peripheral blood counts from age-matched littermates of Mkl1 KO, HET, and WT mice were performed. Although there were no significant differences in total white blood cell and red blood cell counts, the platelet count was significantly decreased in Mkl1 KO mice (Table 1; P < .001 for both WT vs KO and HET vs KO). For mean platelet volume, the data revealed a reduction in Mkl1 KO mice (P = .001, WT vs KO; and P = .15, HET vs KO).
Blood cell counts in WT, HET, and Mkl1 KO mice
| . | WT (n = 23) . | HET (n = 47) . | MKL1 KO (n = 31) . |
|---|---|---|---|
| WBC × 103 | 6.62 ± 0.55 | 7.38 ± 0.35 | 5.59 ± 0.30 |
| RBC × 106 | 8.79 ± 0.17 | 8.86 ± 0.12 | 8.40 ± 0.16 |
| PLT × 103 | 817.9 ± 28.25 | 764.4 ± 20.96 | 522.0 ± 18.85* |
| MPV, fL | 4.83 ± 0.04 | 4.70 ± 0.04 | 4.60 ± 0.05 |
| . | WT (n = 23) . | HET (n = 47) . | MKL1 KO (n = 31) . |
|---|---|---|---|
| WBC × 103 | 6.62 ± 0.55 | 7.38 ± 0.35 | 5.59 ± 0.30 |
| RBC × 106 | 8.79 ± 0.17 | 8.86 ± 0.12 | 8.40 ± 0.16 |
| PLT × 103 | 817.9 ± 28.25 | 764.4 ± 20.96 | 522.0 ± 18.85* |
| MPV, fL | 4.83 ± 0.04 | 4.70 ± 0.04 | 4.60 ± 0.05 |
WBC indicates white blood cells; RBC, red blood cells; PLT, platelets; and MPV, mean platelet volume.
P < .001, Mkl1 KO versus WT and Mkl1 KO versus HET.
Reduced mature megakaryocyte number and ploidy in Mkl1 KO mice
To investigate whether the reduced platelet number in the Mkl1 KO mice might result from abnormal hematopoiesis, we assessed the BM in these mice. Total BM cellularity is not affected (Figure 4A) by knockout of Mkl1, and there is no significant difference in the percentage of LSK cells (Figure 4B) or pre-MegE cells (erythro-megakaryocytic progenitor cells21 ; Figure 4C). We found a slight increase in CFU-MK numbers, but this was not statistically significant (Figure 4D).
Mkl1 deficiency does not affect LSK, pre-MegE cell numbers, and CFU-MK formation. (A) Two tibias and 1 femur from each mouse were collected. After BM harvest and red blood cell lysis, total cell numbers were counted (n = 6 per genotype). (B) Comparison of percentage of LSK cells in WT, HET, and Mkl1 KO BM (n = 9 per genotype). (C) Comparison of percentage of pre-MegE cells in WT, HET, and Mkl1 KO BM (n = 9 per genotype). (D) CFU-MK potential from total BM (n = 6 per genotype).
Mkl1 deficiency does not affect LSK, pre-MegE cell numbers, and CFU-MK formation. (A) Two tibias and 1 femur from each mouse were collected. After BM harvest and red blood cell lysis, total cell numbers were counted (n = 6 per genotype). (B) Comparison of percentage of LSK cells in WT, HET, and Mkl1 KO BM (n = 9 per genotype). (C) Comparison of percentage of pre-MegE cells in WT, HET, and Mkl1 KO BM (n = 9 per genotype). (D) CFU-MK potential from total BM (n = 6 per genotype).
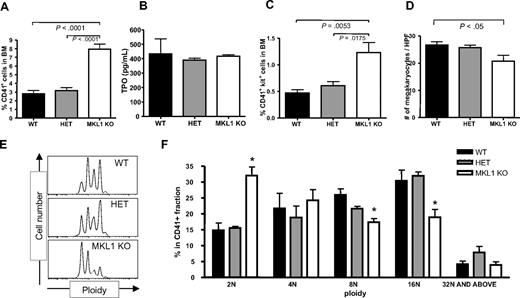
CD41 (GPIIb) is expressed throughout megakaryocyte differentiation, starting from early progenitors to mature polyploid megakaryocytes and platelets.41 Interestingly, there is a significant increase of more than 2-fold in the percentage of CD41+ cells in Mkl1 KO BM (Figure 5A) with essentially identical serum TPO levels (Figure 5B). There is also a significant increase of more than 2-fold in the percentage of CD41+ c-kit+ progenitor cells in Mkl1 KO BM, suggesting that the lack of Mkl1 leads to expansion of immature megakaryocytes (Figure 5C). To quantify the number of mature megakaryocytes, we counted large mature megakaryocytes with typical morphologic characteristics on hematoxylin and eosin-stained BM sections (Figure S5). As shown in Figure 5D, there were decreased numbers of mature megakaryocytes in Mkl1 KO BM, assessed as number per high-power field (×200 magnification) in each group.
Mkl1 deficiency leads to an increased percentage of committed CD41+c-kit+ megakaryocytic progenitors and a decreased percentage of mature high-ploidy megakaryocytes. (A) Comparison of percentages of CD41+ cells in WT, HET, and Mkl1 KO BM (n = 6 per genotype). (B) Comparison of serum TPO concentration (n = 3 per genotype). (C) Comparison of percentages of CD41+ c-kit+ cells in WT, HET, and Mkl1 KO BM (n = 6 per genotype). (D) Comparison of numbers of megakaryocytes per high-power field in the femurs of WT, HET, and Mkl1 KO mice (n = 2 per genotype). (E,F) In vivo ploidy analysis of CD41-positive BM cells taken from WT, HET, and Mkl1 KO mice (n = 6 in WT and HET; n = 8 in Mkl1 KO mice). *P < .05 from both WT versus KO and HET versus KO.
Mkl1 deficiency leads to an increased percentage of committed CD41+c-kit+ megakaryocytic progenitors and a decreased percentage of mature high-ploidy megakaryocytes. (A) Comparison of percentages of CD41+ cells in WT, HET, and Mkl1 KO BM (n = 6 per genotype). (B) Comparison of serum TPO concentration (n = 3 per genotype). (C) Comparison of percentages of CD41+ c-kit+ cells in WT, HET, and Mkl1 KO BM (n = 6 per genotype). (D) Comparison of numbers of megakaryocytes per high-power field in the femurs of WT, HET, and Mkl1 KO mice (n = 2 per genotype). (E,F) In vivo ploidy analysis of CD41-positive BM cells taken from WT, HET, and Mkl1 KO mice (n = 6 in WT and HET; n = 8 in Mkl1 KO mice). *P < .05 from both WT versus KO and HET versus KO.
To clarify the discrepancy between increased CD41+ percentage and lower absolute count of mature megakaryocytes in Mkl1 KO BM, we next examined the ploidy of megakaryocytes in these mice. Mkl1 KO CD41+ cells showed significantly decreased ploidy compared with WT cells (Figure 5E,F). The percentage of CD41+ megakaryocytes that were 2N was 32% versus 15% in KO versus WT mice, respectively (WT, n = 6; KO, n = 8; P < .001). Consistent with this decreased ploidy, there were statistically significantly more WT than KO CD41+ cells with 8N and 16N ploidy (P < .05). In conclusion, these results suggest that Mkl1 deficiency impedes megakaryocyte maturation and polyploidization with a subsequent reduction in platelet counts.
Discussion
The data presented here demonstrate a novel role for MKL1 in regulating megakaryocytopoiesis. We show that Mkl1 expression is up-regulated during megakaryocytic differentiation and that enforced expression of MKL1 in HEL cells enhances megakaryocytic differentiation with up-regulation of GATA1, GATA2, and GP5, and increased cell ploidy. We confirm that, in primary human CD34+ cells, overexpression of MKL1 promotes megakaryocytic surface marker expression and ploidy. Using siRNA against SRF, we show that, as in differentiating muscle cells, the effects of MKL1 are mediated by SRF. Of note, SRF knockdown in control HEL cells stimulated with TPA also led to a significant reduction in polyploidy, suggesting that the inhibitory effects on ploidy are not artifacts of MKL1 overexpression, but rather that endogenous MKL1 acts via SRF to enhance megakaryocyte differentiation. Myocardin-related transcription factors also play a central role in differentiation of vascular smooth muscle cells,42,43 which naturally undergo polyploidization.44 It will be of interest to determine whether MKL1 has the same role in mediating polyploidization in vascular smooth muscle cells.
In Mkl1-deficient BM, there is no significant difference in the total BM cell number or the percentage of cells in the LSK or pre-MegE subpopulations, suggesting that earlier stem and progenitor stages are not affected. However, we observe a slight increase in CFU-MK and a pronounced increase in CD41+c-kit+ and CD41+c-kit− cells. In contrast, the number of morphologically recognizable large megakaryocytes in BM is reduced. Consistent with this decrease in mature megakaryocytes, the percentage of polyploid megakaryocytes (8N and above) is decreased, suggesting that polyploidization is decreased with Mkl1 deficiency. Most likely, Mkl1 deficiency in hematopoietic progenitors blocks megakaryocyte maturation and results in accumulated progenitors at the 2N stage.
How MKL1 promotes polyploidization is not yet clear. Prior studies show that the MKL1/SRF complex is a downstream target of the RhoA pathway. In addition, RhoA is a crucial regulator of cytokinesis. Studies have shown that cytokinesis is mediated by a localized contractile ring that is rich in actin and myosin and is mediated by a discrete pool of GTP-bound, active RhoA.45 In the process of megakaryocytic polyploidization, endomitosis occurs because of failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling.46 Whether MKL1 plays a role directly in endomitosis and mediates the RhoA pathway requires further investigation. The characteristics of Mkl1 KO megakaryocytes may also be caused by a microenvironmental effect rather than only by a cell-autonomous effect, which will be pursued in future studies.
Thrombocytopenia is described in Mkl1 KO mice. Although we did not observe excess bleeding when collecting retro-orbital blood and the platelet counts are well above those believed to cause bleeding problems, our preliminary data show a slight increase in bleeding time in Mkl1 KO mice (data not shown). Whether the Mkl1 KO platelets exhibit altered morphology or function should be addressed in future studies. The decreased platelet counts in Mkl1 KO mice could be the result solely of decreased megakaryocyte ploidy. However, the relationship between ploidy and platelet production is still controversial.8 Muntean et al have shown that Cyclin D1/Cdk4 could rescue the polyploidization of Gata-1-deficient megakaryocytes, but could not restore platelet biosynthesis.47 Therefore, polyploidization may be necessary but not sufficient for platelet biogenesis. The reduced platelet count and small platelets in Mkl1 KO mice could be because of additional defects in platelet biogenesis. The decreased platelet number and size in Mkl1 KO mice are reminiscent of Wiskott-Aldrich syndrome, which is associated with microthrombocytopenia. In Wiskott-Aldrich protein-deficient mice, proplatelet formation in the BM is abnormal.48,49 Because Wiskott-Aldrich protein is an important regulator of the actin cytoskeleton,50 it will be interesting to investigate whether the reduced size and number of Mkl1 KO platelets is because of the defect in cytoskeleton rearrangement in platelet biogenesis.
One could speculate on how our findings relate to the role of the t(1;22) translocation in AMKL. The phenotype of Mkl1-deficient mice with expanded precursors and decreased megakaryocyte maturation shows a similar trend toward that of AMKL pathology. Because of scarcity of primary cells bearing the translocation, it is not yet known whether this translocation encodes an oncogenic fusion protein, or whether the leukemogenesis of this translocation results from reduced expression/function of RBM15 and/or MKL1. One possibility could be that the fusion protein affects endogenous MKL1 function by dimerization via leucine zipper-like domains, thereby inhibiting terminal differentiation, which is supported by the findings of Descot et al.51 Importantly, here we show, for the first time, that MKL1 plays a role in megakaryocytopoiesis, which provides a framework for better understanding the t(1;22) translocation that is found almost uniquely in megakaryoblastic leukemia and not in other subtypes, and have begun to elucidate the mechanisms underlying MKL1-mediated megakaryocytic differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Wing Lam, Dr Stephanie Halene, Dr Yuan Gao, Dr Yue Wu, and Sharon Lin for helpful discussion; Dr Carolyn Lutzko for providing plasmids; University of California–Davis for microarray analysis; and the Yale Keck Bioinformatics Core for microarray analysis and access to Ingenuity software.
This work was supported by the National Institutes of Health (HL63357, T32-DK07556, T32-HL07262, and N01-HV-28 186), the Yale Center of Excellence in Molecular Hematology (DK072442), Cancer Center (Core grant CA21765), the American Lebanese Syrian Associated Charities, St Jude Children's Research Hospital, and Connecticut Stem Cell Research (grant no. 06SCB18).
National Institutes of Health
Authorship
Contribution: E.-c.C. designed the main research concepts, performed experiments, and wrote the manuscript; D.S.K. supervised the study, codesigned the research concepts, and wrote the manuscript; D.T. and V.S. contributed to microarray statistical analyses and wrote the manuscript; S.M.M. provided microarray tools and analysis; Q.L., E.M.B., M.J.R., S.A.M., J.A.T., and C.Q. performed experiments and/or provided scientific and technical knowledge; N.B. provided intellectual input; and S.W.M. and Y.S. produced the Mkl1 KO mice and provided technical expertise.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Diane S. Krause, Yale University School of Medicine, Department of Laboratory Medicine, PO Box 208073, 333 Cedar St, New Haven, CT 06520-8073; e-mail: diane.krause@yale.edu.