Abstract
Controlled regulation of Rho GTPase activity is an essential component mediating growth factor–stimulated migration. We have previously shown that angiomotin (Amot), a membrane-associated scaffold protein, plays a critical role during vascular patterning and endothelial migration during embryogenesis. However, the signaling pathways by which Amot controls directional migration are not known. Here we have used peptide pull-down and yeast 2-hybrid (Y2H) screening to identify proteins that interact with the C-terminal PDZ-binding motifs of Amot and its related proteins AmotL1 and 2. We report that Amot and its related proteins bind to the RhoA GTPase exchange factor (RhoGEF) protein Syx. We show that Amot forms a ternary complex together with Patj (or its paralogue Mupp1) and Syx. Using FRET analysis, we provide evidence that Amot controls targeting of RhoA activity to lamellipodia in vitro. We also report that, similar to Amot, morpholino knockdown of Syx in zebrafish results in inhibition of migration of intersegmental arteries. Taken together, our results indicate that the directional migration of capillaries in the embryo is governed by the Amot:Patj/Mupp1:Syx signaling that controls local GTPase activity.
Introduction
Controlled cell migration is essential for normal embryonic development, neurogenesis, immune function, and angiogenesis.1 During neovascularization of the mouse retina or the formation of intersegmental vessels in zebrafish, vessel migration is stimulated by local secretion of VEGF-A that is detected by the VEGF receptors of the leading endothelial cells.2,3 Upon growth factor stimulation, these so-called tip cells become polarized, in that the leading front extends filopodia whereas the rear maintains contact with the stalk cells. Activation of migration, then, involves the differential polymerization of filamentous actin (F-actin) at the front of a cell, leading to protrusion of the membrane surface and “forward” movement.4
Genetic analyses have shown that cell polarity is controlled by several proteins that are conserved through evolution. This is exemplified by the small GTPase Cdc42 that is recruited to the leading edge of migrating cells where local GTPase activity regulates polarization and cell orientation. The state of activity of these small GTPases is determined by 2 classes of proteins: G-nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs).5 Whereas GAPs are responsible for the inactivation of small GTPases, GEFs are the activating components and stimulate specific small GTPases. RhoGEFs contain a C-terminal PDZ-binding motif, and numerous interactions between RhoGEFs and PDZ domain proteins have recently been reported.6 It has been proposed that the association of the PDZ-binding motif with scaffolding proteins could be a general mechanism of confining GTPase activity to distinct subcellular compartments.6 One example is the RhoGEF Syx/GEF720/PLEKHG5/TECH (for sake of simplicity, now depicted as Syx), which is targeted to the plasma membrane in a synectin-dependent manner.7-9
We have previously shown that angiomotin (Amot) is required in the control of endothelial cell polarization and migration.10,11 Amot belongs to a protein family comprising 2 additional members, AmotL1 (initially identified as JEAP) and AmotL2 (also referred to as LCCP or MASCOT), that are characterized by a glutamine-rich domain, a conserved coil-coil domain, and a C-terminal PDZ-binding domain.12-15
The functional importance of the PDZ-binding domain is indicated by the migratory defect displayed by cells expressing a C-terminal mutant form of Amot.11 Transgenic mice expressing mutant Amot under the EC-specific Tie promoter lose their response to growth factors, which leads to insufficient vascularization and death around embryonic day 9.5 (E9.5).11 Recently, it has been shown that the GAP protein Rich1 binds to Amot, and they together interact with a protein complex containing Pals1, Patj/Mupp1, and Par-3, which are crucial for cell polarity.16 Consistent with these findings, Amot-deficient endothelial cells exhibit loss of polarity, as analyzed by Golgi orientation, and have impaired directional migration in response to chemotactic factors.17
In this report, we set out to identify proteins that bind to the PDZ-binding motif of Amot and promote migration of endothelial cells. Since all 3 members of the Amot family are expressed in endothelial cells, we analyzed the protein complexes associated with the respective PDZ-binding motifs. We report that Amot forms a ternary complex with Syx via the interaction with the Patj/Mupp1 scaffold proteins. We further provide evidence that the Amot:Patj:Syx complex regulates focal RhoA activity in the leading front of migrating cells and provide evidence that this signaling complex is essential for endothelial migration during zebrafish angiogenesis in vivo.
Methods
Plasmids
Using the pEYFP-Patj construct as a template (kindly provided by Dr Margolis, Ann Arbor, MI), we generated mutants by polymerase chain reaction (PCR) that encode the various deletion mutants of human Patj (accession no. NM_176877.218 ). The PCR products were subcloned into pENTR plasmids (Gateway; Invitrogen, Carlsbad, CA) and then shuttled into GAL4 DNA-binding domain vectors (pDEST22; Invitrogen) or directly cloned into the bait plasmid pDB Leu (Invitrogen). The C-termini of Amot (aa's 1020-1081), AmotL1 (aa's 892-956), and AmotL2 (aa's 716-780) as well as the last 62 aa's of human Syx were cloned into pENTR and subsequently sequenced. For yeast-2 hybrid (Y2H) assays Amot, AmotL1, AmotL2, and Syx inserts were shuttled into pDEST22 and cotransformed together with the bait constructs described here. Details of constructs and primers are available from M.S. or T.W. The p80 and p130 Amot expression constructs have been described previously.10,19 AmotL1 and AmotL2 were subcloned into pcDNA3 (Invitrogen).12,13 The myc-tagged Pals1 and Patj constructs were kindly provided by Dr Margolis and the Syx constructs have been described by Liu and Horowitz.19
Cell lines and reagents
HEK 293T cells, HEK 293 cells, and spontaneously immortalized mouse aortic endothelial (MAE) cells were grown in DMEM medium supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin. The culturing conditions for bovine capillary endothelial (BCE) hTERT+ and polyoma middle T-endothelial cells (PmT-ECs)17 were described by Ernkvist et al.19
Cells were transfected using Lipofectamine 2000 according to the protocol of the manufacturer (Invitrogen).
Antibodies
The antibody against Amot was generated in rabbit using the 24 C-terminal amino acids of human angiomotin.11 For Western blot, the following antibodies were used: anti-Mupp1 (BD Biosciences, Franklin Lakes, NJ), anti–c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA), anti-6xHis (Clontech, Mountain View, CA), anti-Flag (Sigma-Aldrich, St Louis, MO), and anti-GST (Santa Cruz Biotechnology). Horseradish peroxidase–conjugated secondary antibodies were purchased from GE Healthcare (Piscataway, NJ). Antibodies used for immunofluorescence were as follows: phalloidin (Invitrogen), ZO-1 (Zymed, Invitrogen), 6xHis, and the generated antibodies for the Amot isoforms. Secondary antibodies were purchased from Dako (Carpinteria, CA), Vector Laboratories (Burlingame, CA), and Invitrogen.
Yeast cotransformation assays
For Y2H assays, yeast cells (strain MaV203; Invitrogen) were simultaneously transformed with bait and prey plasmids encoding various deletion mutants of human Patj and the C-termini of human Amot, AmotL1, AmotL2, and Syx, respectively. Interaction of bait and prey molecules was tested by growth of yeast cells on selective medium containing 50 to 75 mM 3AT (3-amino 1,2,4,-triazole), but lacking leucine, tryptophan, and histidine (-L-T-W), and by LacZ assays with filter-immobilized cells.
Peptide pull-down assay and mass spectrometric analysis
Peptides corresponding to carboxyl terminals of mouse Amot, AmotL1, and AmotL2 were synthesized (Innovagen, Lund, Sweden) and cross-linked to NHS-activated Sepharose 4 Fast Flow beads according to the manufacturer's instructions (GE Healthcare). Pull-down experiments were performed using MAE cell lysates and interacting proteins were identified by mass spectrometry after in-gel digestion. Briefly, beads containing peptides were added to cell lysates and incubated for 2 hours at 4°C. Beads were washed in lysis buffer (50 mM Hepes, pH 7.5, 1 mM EGTA, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100), and proteins bound to beads were separated on NuPage 4% to 12% Bis-tris acrylamide gels in Mops buffer according to the manufacturer's instructions (Invitrogen). Protein bands were stained with SimplyBlue SafeStain kit (Invitrogen), cut from the gel, and digested with trypsin (Promega, Madison, WI) according to Shevchenko et al.21
Mass spectrometric analyses were performed using a MALDI-TOF instrument (Ultraflex; Bruker Daltonics, Bremen, Germany) with reflector and positive modes, an ions acceleration of 25 keV, a 5-Hz laser frequency, and a delay extraction of 110 ns. Six hundred shots were accumulated for each spectrum. Spectra were recalibrated using internal trypsin monoprotonated monoisotopic masses 842.509, 1045.564, 2211.104, and 2283.180. Raw data were processed using Flex Analysis and Biotool software (Bruker Daltonics). Protein identification was achieved using a Mascot search engine against the MSDB database22 (Matrix Science, London, United Kingdom). Parameters fixed for each search were as follows: taxonomy, mouse: error tolerance, 50 ppm: miss-cleavage, not allowed: peptides charge, monoprotonated. Fixed carbamidomethylated cysteine and potential oxidized methionine were selected as modifications.
Immunoprecipitation
Cells were extracted with lysis buffer and centrifuged. The supernatant was precleared with Sepharose A beads (GE Healthcare), followed by incubation with 1 μg antibody for 2 hours and thereafter with Sepharose A beads for 2 hours, all at 4°C. Pellets were washed 4 times with lysis buffer and resuspended in sample buffer and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE).
Western blot
Cell lysates and immunoprecipitation yields were analyzed by SDS-PAGE and proteins were transferred to nitrocellulose membranes. Nonspecific binding was blocked for 1 hour in 10% dried milk in PBS containing 0.1% Tween (PBS-T). Membranes were incubated in 5% dried milk in PBS-T with primary antibody and thereafter with the HRP-conjugated secondary antibody. Afterward, the nitrocellulose membrane was visualized using a detection system from Santa Cruz Biotechnology and by exposing the membrane to x-ray film (GE Healthcare).
Immunofluorescence
Cultured cells were plated in chamber slides and allowed to grow and adhere overnight. The cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature and permeabilized with 0.1% Triton X-100. Nonspecific reactivity was blocked with 5% horse serum in PBS before addition of primary antibody in blocking buffer. Antibody binding was detected with fluorescent-labeled secondary antibodies. F-actin was visualized with Texas red phalloidin (Molecular Probes, Invitrogen, Eugene, OR). The slides were mounted with mounting media from Vector Laboratories and viewed on a Zeiss Axioplan 2 microscope, and images collected using an AxioCam HRm Camera and Axiovision 4.5 software (Carl Zeiss, Heidelberg, Germany).
FRET experiments
wt and Amot−/− PmT-ECs (0.5-0.8 million) were grown in fibronectin-coated (25 ng/mL) 35-mm plates with optical-quality 1-mm-thick glass bottoms (FluoroDish; World Precision Instruments, Sarasota, FL). Cells were transfected with the RhoA FRET probe pRaichu-1237X23 gift of M. Matsuda (Kyoto University, Kyoto, Japan) using the Nucleofector apparatus and kit (Amaxa, Gaithersburg, MD) according to the manufacturer's instructions. Plates were placed 24 hours after transfection on the stage of an inverted Olympus IX71 microscope (Tokyo, Japan) equipped with a Sutter DG4 white light source (Novato, CA) for excitation of fluorescence and an Olympus 60×/1.45 NA objective. The microscope stage was enclosed in a Plexiglas chamber maintained at 37°C and 5% CO2. Images were acquired by a cooled software-controlled (QED in Vivo software; Media Cybernetics, Bethesda, MD) CCD camera (Cooke SensiCam; Romulus, MI). The dual-wavelength emission of the probe was excited via a FF458-Ex01-25 filter (Omega Optical, Brattleboro, VT), and split with a CFP (HQ 465/30m)/YFP (HQ 560/55m) mirror. Series of 20 images, each with an exposure time of 0.5 seconds, were taken at 20-second intervals. Cell-free areas of the plate were imaged for each series and were subtracted from the raw images by dedicated software (ImagePro; Media Cybernetics). The ratio of the YFP/CFP emissions (FRET efficiency) was calculated by the same software. The FRET efficiency is displayed as a pseudocolor thermal map on which blue corresponds to low efficiency and red to high efficiency.
Zebrafish angiogenesis
Adult zebrafish were grown in the fish facility with a 14-hour light/10-hour dark cycle. Zebrafish embryos harboring the Tg (fli:EGFP)y1 transgene (used by Lawson and Weinstein2 ) were injected at the 1- to 4-cell stage with morpholinos and raised at 28°C in standard E3 water supplemented with 0.003% PTU (phenyl-2-thiourea; Lawson and Weinstein2 ). BLAST searches on Ensembl24 and NCBI25 were performed to identify zebrafish syx-a (ENSDARG00000025902/ XP_691320) on chromosome 11 and syx-b (ENSDARG00000025288/ XP_001336734). For syx-a, 2 morpholinos were used that target translation initiation by binding near the ATG start; syx-a MO1: GCAGTCCACAGCATTGCACGACATC and syx-a MO2: AGATCCAGG CCCATGCTGATCCCGG. For syx-b, 2 splice interfering morpholinos syx-b MO1: AGACCACACAGGAACAGCTGCACAG and syx-b MO2: GCTGAAACCGGGATGCAAACTAACC targeting the Dbl/Rho GEF domain were made.
The zebrafish amot gene was inactivated using morpholinos described previously.17 All morpholinos (GeneTools LLC, Philomath, OR) were injected at 125 μM or 250 μM diluted in 0.3× Danieau solution with 0.05% phenol red included as a tracer. Phenotypic consequences of morpholino gene knockdown were assessed from 30 to 60 high-power fields using a Leica MZ16 stereomicroscope equipped with epifluorescence. For phenotypic scoring, an individual injected embryo was judged as being morphant if it had 3 or more blunted or missing ISV defects. Using this counting assay, we have found a background “defect” rate of 1.4 plus or minus 0.5 defects/embryo.
Results
Essential role of the p80-Amot PDZ-binding motif for endothelial migration in vivo
To assess the importance of the functional domains of Amot, we used a zebrafish angiogenesis model system. The zebrafish embryo is a valuable model for real-time studies of vasculogenesis and angiogenesis.26 Blood vessels may be visualized using fish transgenic for green fluorescent protein (GFP) driven by the endothelial-specific Fli promoter.2,27 We used a strategy by which we knocked down endogenous Amot in zebrafish using morpholino antisense oligos and sought to rescue the vascular defects by coinjecting either wt human Amot mRNA or mRNA encoding Amot minus the C-terminal YLI aa (ΔYLI). Inhibition of Amot expression in zebrafish inhibits the migration of intersegmental vessels sprouting from the dorsal aorta.17 The knockdown of Amot could be rescued by coinjecting mRNA encoding the human p80 Amot (Figure 1A,B). However, coinjection of mRNA encoding the ΔYLI mRNA could not rescue the vascular defects resulting from Amot knockdown (Figure 1A,B).
The PDZ-binding motif is essential for restoring endothelial migration in Amot knockdown in zebrafish. Zebrafish embryos (transgenic for fli:GFP visualizing endothelial cells of the developing vasculature) were injected with combinations of a morpholino against Amot and mRNA transcripts from human p80Amot or human p80 with the last 3 amino acids in the PDZ-binding motif deleted (ΔYLI). (A) Fluorescent images of zebrafish 32 hours after fertilization. Embryos were injected with morpholinos and mRNA as indicated in the figure. (B) Bar diagram showing statistics of the percentage of embryos with vascular defects. MM indicates mismatched control morpholino; MO2, morpholino targeting exon 2 of Amot; Amot, human p80Amot mRNA; and ΔYLI, human p80 mRNA with the PDZ-binding motif deleted. ***P < .001.
The PDZ-binding motif is essential for restoring endothelial migration in Amot knockdown in zebrafish. Zebrafish embryos (transgenic for fli:GFP visualizing endothelial cells of the developing vasculature) were injected with combinations of a morpholino against Amot and mRNA transcripts from human p80Amot or human p80 with the last 3 amino acids in the PDZ-binding motif deleted (ΔYLI). (A) Fluorescent images of zebrafish 32 hours after fertilization. Embryos were injected with morpholinos and mRNA as indicated in the figure. (B) Bar diagram showing statistics of the percentage of embryos with vascular defects. MM indicates mismatched control morpholino; MO2, morpholino targeting exon 2 of Amot; Amot, human p80Amot mRNA; and ΔYLI, human p80 mRNA with the PDZ-binding motif deleted. ***P < .001.
In conclusion, our data indicate the PDZ-binding motif of Amot is essential for directional migration of sprouting vessels in vivo.
Identification of proteins binding to the PDZ-binding motifs of Amot, Amot L1, and Amot L2
It has previously been shown that Amot binds to a complex of proteins involved in cell polarization and to the GAP Rich1.16,28 However, the link between the signaling pathways associated with the PDZ-binding domain and its involvement in endothelial migration remains unclear. A possible explanation is that previously reports have studied Amot activities in epithelial cells and proteins involved in regulating endothelial migration may have been missed. Therefore, we used a peptide pull-down approach to identify proteins binding to the PDZ-binding domain of Amot in lysates from endothelial cells. The last 4 amino acids of AmotL1 (EVLI) and AmotL229 contain a putative class II PDZ-binding motif that recognizes aliphatic sequences patterns of the x-Ψ-x-Ψ type (Ψ, any aliphatic residue). Angiomotin has a similar class II PDZ-binding motif (EYLI).
To identify proteins that specifically interact with the PDZ-binding motif, peptides covering the last 19 amino acids of the C-terminal domains of Amot, AmotL1, and AmotL2 were synthesized and peptides harboring a deletion of the last 3 amino acids were used as negative controls (Figure 2A). The PDZ-binding motifs were chemically coupled to Sepharose beads and incubated with lysates from MAE cells. MAE cells do not express endogenous Amot but retransfection of this gene promotes migration and cells become responsive to angiostatin or angiostatin-mimicking antibodies.10,30,31 In addition, we have shown that the C-terminal YLI sequence is essential for Amot migration-promoting activities in vitro in this cell line.11 Bound proteins were eluted after extensive washing and resolved by SDS-PAGE (Figure 2B). Proteins that specifically bound to the PDZ-binding motif containing peptides were extracted for mass spectrometry analysis. A summary of the proteins identified by this approach is shown in Figure 2C and in peptide coverage is presented in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). It shows that the PDZ-binding motifs of the Amot family binds Mupp1, Lin7, and Pals1, in accordance to previous findings.16,28 Interestingly, we also detected association of the RhoA GTPase exchange factor protein Syx with all members of the motin family (Figure 2C; Figure S1). This RhoGEF protein is expressed in endothelial and neuronal cells.7-9 In endothelial cells, Syx is targeted to the plasma membrane where it regulates RhoA activity and stimulates migration.9 To assess the potential overlap of cellular localization of Syx and Amot in endothelial cells, we performed immunofluorescent staining in MAE cells stably expressing p80-Amot, p130-Amot, or empty vector. p80-Amot localized to the leading edge of the lamellipodia, as previously shown, whereas p130-Amot was found to be expressed in a punctuated pattern that overlapped with actin as shown previously.10,19 Syx expression overlapped with p80 Amot in lamellipodia, whereas no overlap could be detected with p130 Amot (Figure S2A). We also analyzed the localization of Amot and Syx in bovine capillary endothelial cells (BCEs) that express both isoforms of Amot as well as Syx endogenously. In subconfluent cells, Syx localized to the edges of the lamellipodia (Figure S2B). In contrast, in confluent cells, Amot localized to cell-cell contacts and Syx localized in a dispersed vesicular pattern in the cytoplasm, suggesting that Syx is not targeted to cell-cell contacts (Figure S2B).
Identification of proteins associated with the PDZ-binding motifs of Amot, AmotL1, and AmotL2. (A) Peptides corresponding to the last 19 C-terminal amino acids of Amot, AmotL1, and AmotL2 including the PDZ-binding domains were used for pull-down experiments of mouse aortic endothelial lysate. Peptides lacking the 3 last amino acids were used as negative controls. (B) Proteins that bound to peptides were separated by SDS-PAGE and visualized by silver staining and analyzed by mass spectrometry. Data regarding peptide coverage can be found in Table S1. (C) The table summarizes the proteins pulled down with the full-length peptides but not with the truncated peptides. Patj was also found to associate with Amot by yeast 2-hybrid screening as described in “Yeast cotransformation assays.” (D) Myc-Pals1, myc-Patj, or His-Syx were cotransfected in HEK293 cells together with human p80 Amot or p80 ΔYLI (negative control lacking the PDZ-binding motif). Protein-protein interaction was verified by coimmunoprecipitation analysis. Endogenous Mupp1 was immunoprecipitated with transfected human p80Amot (top left panel). The vertical lines indicate repositioned gel lanes.
Identification of proteins associated with the PDZ-binding motifs of Amot, AmotL1, and AmotL2. (A) Peptides corresponding to the last 19 C-terminal amino acids of Amot, AmotL1, and AmotL2 including the PDZ-binding domains were used for pull-down experiments of mouse aortic endothelial lysate. Peptides lacking the 3 last amino acids were used as negative controls. (B) Proteins that bound to peptides were separated by SDS-PAGE and visualized by silver staining and analyzed by mass spectrometry. Data regarding peptide coverage can be found in Table S1. (C) The table summarizes the proteins pulled down with the full-length peptides but not with the truncated peptides. Patj was also found to associate with Amot by yeast 2-hybrid screening as described in “Yeast cotransformation assays.” (D) Myc-Pals1, myc-Patj, or His-Syx were cotransfected in HEK293 cells together with human p80 Amot or p80 ΔYLI (negative control lacking the PDZ-binding motif). Protein-protein interaction was verified by coimmunoprecipitation analysis. Endogenous Mupp1 was immunoprecipitated with transfected human p80Amot (top left panel). The vertical lines indicate repositioned gel lanes.
Apart from Syx, using the peptide pull-down approach, we could detect 3 proteins (Pals2, Filamin A, and PTN13) that apparently exclusively bound to Amot (Figure 2C).
In parallel with the peptide pull-down approach, we performed Y2H screening of a mouse embryo library using the last 240 C-terminal residues of Amot. Positive clones were challenged against the same bait containing a deletion of the last 3 amino acids of the PDZ-binding domain. Using this approach, we could add Patj to the list of proteins binding to the PDZ-binding motif of Amot (Figure 2C, data not shown).
The observed interactions were verified by coimmunoprecipitation analysis. Syx, Pals1, Patj, and Mupp1 were brought down together with p80-Amot, but not with the ΔYLI protein (Figure 2D), confirming that they bind via association to the PDZ-binding domain of Amot. In contrast, Pals2 and Lin7 were not pulled down with Amot, suggesting that they do not bind directly to Amot but rather form a ternary complex (data not shown).
Identification of proteins associated with the Syx PDZ-binding motif
Since Syx contains a PDZ-binding domain but not a PDZ domain, the binding between Amot and Syx is most likely indirect. To identify the proteins potentially involved in linking Amot to Syx, we used a similar strategy to what was described in Figure 2. Peptide pull-down using MAE lysate was performed using a peptide encompassing the C-terminal PDZ-binding motif of Syx (Figure 3A). Mass spectrometry analysis revealed that the PDZ-binding domain of Syx associates with Mupp1, Pals1, and Lin7 (Figure 3A). In addition, we found that the cytoskeletal proteins myosin 1c, tubulin, and actin were coimmunoprecipitated with the Syx-binding domain (Figure 3A). The binding of the polarity complex was verified by coimmunoprecipitation analysis (Figure 3B). In addition, we could also detect interaction between Syx and Patj (Figure 3B). None of the analyzed proteins bound Syx ΔEV, which is a Syx isoform lacking 2 aa's of the C-terminal PDZ-binding motif, also called Syx2 in the literature.20 These data suggest that Syx form a ternary complex with Amot via the binding of the scaffold proteins Patj/Mupp1.
Identification of proteins associated with the PDZ-binding motif of Syx. (A) A peptide corresponding to the last 19 amino acids of Syx was used for pull-down experiments in MAE cells. Proteins that bound to peptides were separated by SDS-PAGE and visualized by silver staining and analyzed by mass spectrometry. The identity of bands indicated by asterisks is shown in the figure as well as in Table S1. (B) Pals1, Patj, and Mupp1 binding to Amot was verified by coimmunoprecipitation analysis in HEK 293 cells as indicated. The Syx ΔEV (lacking 2 aa's in the PDZ-binding motif) was used as negative control.
Identification of proteins associated with the PDZ-binding motif of Syx. (A) A peptide corresponding to the last 19 amino acids of Syx was used for pull-down experiments in MAE cells. Proteins that bound to peptides were separated by SDS-PAGE and visualized by silver staining and analyzed by mass spectrometry. The identity of bands indicated by asterisks is shown in the figure as well as in Table S1. (B) Pals1, Patj, and Mupp1 binding to Amot was verified by coimmunoprecipitation analysis in HEK 293 cells as indicated. The Syx ΔEV (lacking 2 aa's in the PDZ-binding motif) was used as negative control.
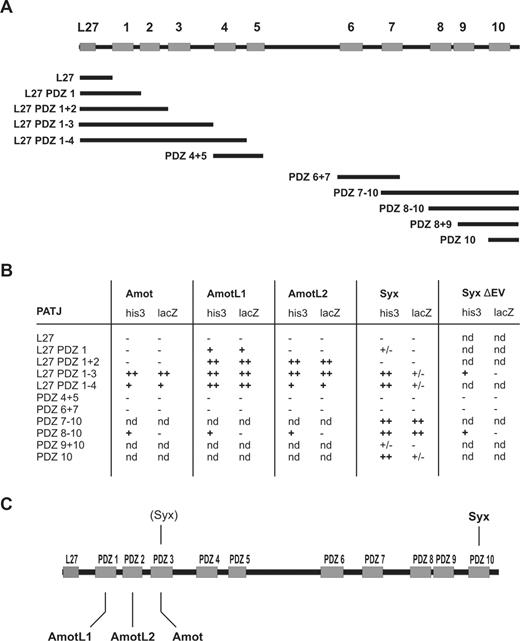
Characterization of interaction with PATJ PDZ-binding motifs.
Mupp1 and Patj are highly homologous paralogue proteins that contain multiple PDZ domains (13 and 10, respectively). Both serve as scaffolding proteins that help to organize higher-order protein complexes. Patj associates with Pals1 via an L27 domain and the transmembrane proteins CrB1 and CrB3. This protein complex has been shown to be evolutionary conserved and implicated in the regulation of polarization of epithelial cells.32 In addition, recent work has shown that Patj also localizes to lamellipodia and may control directional migration.33 It is therefore likely that Amot binds directly to Mupp1/Patj via the PDZ domains and organizes a complex of signaling molecules. To identify the specific interaction sites of Amot and Patj, we generated several constructs covering the different PDZ domains of Patj (Figure 4A).
Angiomotin family members directly interact with Patj via different PDZ domains. (A) Domain architecture of Patj constructs. Full-length Patj (1801 aa's) consists of one N-terminal L27 (blue) and 10 PDZ domains (orange). (B) Summary of the Y2H assays. S cerevisiae MaV203 cells were cotransformed with bait constructs containing cDNAs encoding different Patj deletion mutants and prey plasmids that encode the C-termini of Amot, AmotL1, AmotL2, and Syx as well as Syx lacking the last 2 amino acids. To test for interaction in the Y2H assays, corresponding strains were cultivated in synthetic media lacking leucine, tryptophan, and histidine, supplemented with 50 to 75 mM 3AT (his3). In addition, β-galactosidase reporter gene activity (lacZ) was determined on replica filters using X-gal as substrate. − indicates no growth in selection media or no β-galactosidase activity; +/−, background growth or background β-galactosidase activity; +, growth in selection media or weak β-galactosidase activity; +/+, very fast growth in selection media or high β-galactosidase activity; and nd, not determined. (C) Figure summarizing the binding specificities of Amot, AmotL1, AmotL2, and Syx to individual PDZ domains of Patj. The binding of Pals1 to L27 was shown by Roh et al.34
Angiomotin family members directly interact with Patj via different PDZ domains. (A) Domain architecture of Patj constructs. Full-length Patj (1801 aa's) consists of one N-terminal L27 (blue) and 10 PDZ domains (orange). (B) Summary of the Y2H assays. S cerevisiae MaV203 cells were cotransformed with bait constructs containing cDNAs encoding different Patj deletion mutants and prey plasmids that encode the C-termini of Amot, AmotL1, AmotL2, and Syx as well as Syx lacking the last 2 amino acids. To test for interaction in the Y2H assays, corresponding strains were cultivated in synthetic media lacking leucine, tryptophan, and histidine, supplemented with 50 to 75 mM 3AT (his3). In addition, β-galactosidase reporter gene activity (lacZ) was determined on replica filters using X-gal as substrate. − indicates no growth in selection media or no β-galactosidase activity; +/−, background growth or background β-galactosidase activity; +, growth in selection media or weak β-galactosidase activity; +/+, very fast growth in selection media or high β-galactosidase activity; and nd, not determined. (C) Figure summarizing the binding specificities of Amot, AmotL1, AmotL2, and Syx to individual PDZ domains of Patj. The binding of Pals1 to L27 was shown by Roh et al.34
Interaction of the Amot proteins with Patj domains was analyzed by Y2H cotransformation assays. The results of this in vivo approach indicate that Amot binds to the PDZ3 (and probably PDZ4) domain of Patj (Figure 4B left column). The Amot family proteins do not appear to compete for binding sites on Patj since AmotL1 binds to PDZ1 and AmotL2 to PDZ2 (Figure 4B second and third columns, respectively). The interaction sites between Syx and Patj were analyzed using cDNA encoding the last 62 aa's of Syx together with the individual Patj constructs described in Figure 4A. The L27 domain of Patj does not interact with Syx PDZ-binding motif as expected. However, some binding could be detected with PDZ3 but the dominant binding domain is PDZ10 (Figure 4B fourth column). The PDZ mutant of Syx (Syx ΔEV) does not interact with Patj (Figure 4B right column). These findings raise the possibility that Patj is able to interact simultaneously with Syx and all members of the Amot family. The binding data from Y2H are summarized in Figure 4C.
Loss of polarized RhoA GTPase activity in Amot knockout cells
Cell polarization has been shown to play an essential role in the directional movement of cells in a chemotactic gradient.35 We have previously shown that Amot is critical for polarization as measured by Golgi apparatus reorientation in migrating cells.17 Since Amot binds to the Patj/Mupp1:Pals1 polarity complex and the RhoA GEF Syx, it is feasible that Amot indirectly controls the localization of RhoA. To study the subcellular localization of RhoA activity in living cells, we transfected wt or Amot-deficient endothelial cells with a RhoA FRET (fluorescent resonance energy transfer) probe and examined by FRET analysis. Analysis of spontaneously migrating wt endothelial cells revealed that RhoA activity was concentrated to the edge of extending cellular protrusions and lamellipodia (Figure 5 left panel). This is similar to what has been previously reported in randomly migrating fibroblasts.36 In Amot-deficient cells, however, the focal activity was lost and RhoA activity was dispersed throughout the peripheral membrane (Figure 5 right panel).
Loss of focal RhoA activity in Amot-deficient endothelial cells. FRET images of endothelial cells expressing the Raichu-RhoA YFP-CFP probe. FRET efficiency is displayed as a thermal map corresponding to the scale shown in lower panels. Randomly migrating Amot wt cells exhibited high RhoA activity in the leading front of lamellipodia and cellular protrusions. In contrast, the RhoA activity in Amot-deficient cells was not focal but was uniformly localized to the outer membrane. Scale bar represents 10 μm.
Loss of focal RhoA activity in Amot-deficient endothelial cells. FRET images of endothelial cells expressing the Raichu-RhoA YFP-CFP probe. FRET efficiency is displayed as a thermal map corresponding to the scale shown in lower panels. Randomly migrating Amot wt cells exhibited high RhoA activity in the leading front of lamellipodia and cellular protrusions. In contrast, the RhoA activity in Amot-deficient cells was not focal but was uniformly localized to the outer membrane. Scale bar represents 10 μm.
Morpholino knockdown of Syx in zebrafish
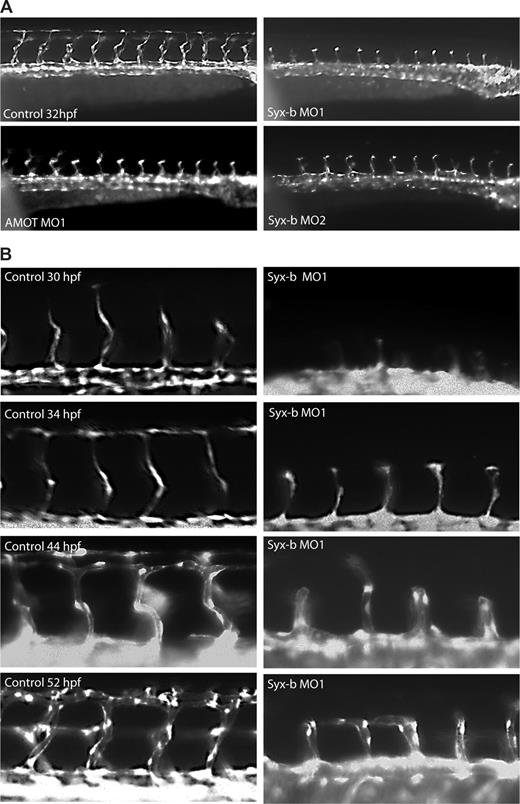
Analysis of Syx homologues in zebrafish revealed that there are 2 Syx genes located on chromosomes 11 (syx-a) and 23 (syx-b). Zebrafish syx-a and syx-b share 60% and 59% amino acid identity, respectively, with mouse Syx1 and 55% amino acid identity with each other. The C-termini containing the PDZ-binding motifs were shown to be 100% conserved between mouse and zebrafish, suggesting that this motif associates with similar signaling pathways in both species. As presented in Figure 1, Amot knockdown in zebrafish results in impaired migration of intersegmental vessels (ISVs) in a PDZ-binding motif-dependent manner. To assess syx function in zebrafish angiogenesis, we injected morpholinos that specifically target either syx-a or syx-b. We found that knockdown of either gene, similar to Amot knockdown, resulted in inhibition of ISV migration (Figure 6A). However, the effects of the syx-b morpholinos were more dramatic in that the phenotypic penetrance was stronger and more vessels were affected per embryo (Figure 7). The efficiency of Syx-b knockdown was quantified by reverse-transcription (RT)–PCR as shown in Figure S3. The effect of knocking down syx-b was relatively stable since arrested vessels did not migrate and form dorsal anastomosing vessels (DLAVs) even after 55 hours (data not shown). However, the sprouts that had arrested at the myoseptum formed a lumen after 40 hours and later actually fused with neighboring vessels (Figure 6B). In addition, knockdown of either syx-a or syx-b led to dilation of the primordial midbrain and hindbrain channels, as observed previously in amot morphants17 (data not shown). These phenotypes were replicated with a second nonoverlapping morpholino (Figure 6A and “Zebrafish angiogenesis”). This indicates that the effect of syx knockdown is primarily on migration, whereas lumen formation and fusion of sprouts appear to be intact.
Syx-b and amot morphants show a common ISV phenotype. (A) Knockdown of zebrafish syx-b using 2 nonoverlapping morpholinos, MO1 and MO2, resulted in identical ISV migration defects. The sprouts in syx-b MO arrest at the horizontal myoseptum. (B) In syx-b morphants, outgrowth of the ISV tip cells was delayed. With time, defective ISVs appear to form lumens that are capable of anastomosing with neighboring vessels. The truncated ISV phenotype was observed in embryos after 52 hours after fertilization (hpf). Due to the severity of the phenotype, the dorsal longitudinal anastomosing vessel (DLAV) does not form. In all panels, anterior is to the left and dorsal is to the top.
Syx-b and amot morphants show a common ISV phenotype. (A) Knockdown of zebrafish syx-b using 2 nonoverlapping morpholinos, MO1 and MO2, resulted in identical ISV migration defects. The sprouts in syx-b MO arrest at the horizontal myoseptum. (B) In syx-b morphants, outgrowth of the ISV tip cells was delayed. With time, defective ISVs appear to form lumens that are capable of anastomosing with neighboring vessels. The truncated ISV phenotype was observed in embryos after 52 hours after fertilization (hpf). Due to the severity of the phenotype, the dorsal longitudinal anastomosing vessel (DLAV) does not form. In all panels, anterior is to the left and dorsal is to the top.
Genetic interactions between syx genes and between syx-b and amot. (A) Phenotypic penetrance of amot, syx-a, and syx-b morphants. (B) Phenotypic severity of amot, syx-a, and syx-b morphants measured in terms of ISV defects per embryo. (C) Phenotypic penetrance of syx-b; amot double morphants and syx-a; syx-b double morphants. Phenotypic severity of syx-b; amot double morphants and syx-a; syx-b double morphants measured in terms of ISV defects per embryo. (D) Phenotypic severity of amot, syx-a, and syx-b morphants as well as for amot syx-b and syx-a/syx-b double morphants. Error bars indicate the standard deviation. *P < .05; ***P < .001.
Genetic interactions between syx genes and between syx-b and amot. (A) Phenotypic penetrance of amot, syx-a, and syx-b morphants. (B) Phenotypic severity of amot, syx-a, and syx-b morphants measured in terms of ISV defects per embryo. (C) Phenotypic penetrance of syx-b; amot double morphants and syx-a; syx-b double morphants. Phenotypic severity of syx-b; amot double morphants and syx-a; syx-b double morphants measured in terms of ISV defects per embryo. (D) Phenotypic severity of amot, syx-a, and syx-b morphants as well as for amot syx-b and syx-a/syx-b double morphants. Error bars indicate the standard deviation. *P < .05; ***P < .001.
The observation that both syx-a and syx-b gave similar ISV defects led us to ask whether these 2 genes were functionally redundant during zebrafish angiogenesis. Therefore, we simultaneously knocked down both syx-a and syx-b using suboptimal morpholino concentrations in doubly morphant embryos. As shown in Figure 7C,D, embryos doubly morphant for syx-a and syx-b show additive effects on phenotypic penetrance (86% for syx-a MO; syx-b MO vs 16% for syx-a MO and 70% for syx-b MO) and phenotypic severity (17.0 ± 6.9 ISV defects/embryo for syx-a MO; syx-b MO vs 4.7 ± 2.0 for syx-a MO and 12.2 ± 3.3 for syx-b MO). These results led us to conclude that both syx-a and syx-b are required for ISV sprouting, that their functions overlap, and that syx-b may be more functionally significant in sprouting angiogenesis.
Given their molecular interactions and common phenotypes, amot and syx genes may function in a common signaling complex. To test this idea, we again assayed for genetic interactions between amot and syx-b, using double morpholino knockdowns. We focused our attention on syx-b, since in our assays it appears to be the more functionally important paralogue. To genetically sensitize the amot signaling pathway during ISV sprouting, we used sub-optimal concentrations of the morpholinos (Figure 7C,D). In this amot knockdown experiment, we observed ISV defects in 26% of injected embryos with a mean of 5.3 plus or minus 1.9 defects per embryo. However, when both amot and syx-b are simultaneously knocked down, we observed a phenotypic penetrance of 81% with a mean of 23.7 plus or minus 7.9 ISV defects/embryo, significantly more (P < .001) than either individual knockdown alone. The phenotypic consequences of double knockdown cannot be explained by additive effects and suggest a synergistic interaction between amot and syx-b. Therefore, the morpholino-mediated genetic interaction analysis supports amot and syx-b functioning in a common signaling pathway during ISV sprouting.
Discussion
Here we show that Amot, by binding to the Patj:Pals1 polarity complex, associates with the RhoGEF Syx. We provide evidence that this complex controls RhoA activity at the leading edge of migrating cells and that knockdown of either Amot or Syx results in inhibition of migration of intersegmental vessels during zebrafish angiogenesis.
The importance of the PDZ-binding motif of Amot was illustrated by the findings that human p80 Amot could rescue knockdown of Amot in zebrafish, whereas mRNA encoding a deletion mutant lacking 3 amino acids of the C-terminus could not. These findings argued that the C-terminal motif is a key functional domain in Amot signaling. We have previously shown that the C-terminal motifs of all the members of the Amot family contain consensus sequences consistent with binding PDZ proteins.
We have focused our work to understanding the regulation of migration in endothelial cells. With our peptide pull-down approach, we found that Amot associates with the Pals:Patj/Mupp1 complex in endothelial cells and could determine the precise PDZ-binding sites of the individual Amot family members to Patj by yeast 2-hybrid analysis. We also could show that Syx is associated with this polarity complex. Syx contains a RhoGEF domain, a pleckstrin homology domain, and a C-terminal PDZ-binding motif. Bioinformatics analysis on all RhoGEFs encoded in the human genome and their mouse homologues shows that 26 of 70 RhoGEFs of the human Dbl homology family contain a PDZ-binding motif. It has been proposed that the role of these PDZ-containing RhoGEFs is to associate with scaffold proteins for the precise spatio-temporal localization of GTPase activity.6 This is consistent with our findings that in wt mouse endothelial cells RhoA activity in migrating cells is confined to the lamellipodia, whereas RhoA activity is uniformly dispersed throughout the peripheral membrane in Amot-deficient cells. Since Patj and Mupp1 bind to the PDZ-binding motif of Syx, we suggest the following model, illustrated in Figure S4: angiomotin binds directly Patj/Mupp1 to form a ternary complex with Pals1, Lin7, and Syx. It has been previously shown that Patj:Pals1 form an evolutionary conserved complex with the transmembrane protein CrB that localizes to the apical surface of epithelial cells. It is therefore feasible that Amot associates indirectly to CrB, which would confer localization to distinct membrane location in the cell. It should be noted that the role of CrB in the Amot:Patj:Pals1 is still an open question as we have not successfully pulled down CrB with either Amot or Syx.
Genetic analysis of intersegmental vessel patterning in the zebrafish has provided important clues regarding the signaling pathways guiding migrating endothelial cells. Outgrowth of ISVs from the dorsal aorta follows a stereotyped sequence of sprouting, tube formation, and anastomosis.27 VEGF signaling via 3 cooperating VEGF receptors appears to drive initial outgrowth and tip cell migration of the forming ISVs. Subsequent dorsal migration is guided by repulsive semaphorin signals, secreted from somites, which bind to neuropilin/PlexinD1 receptors on the ISV endothelium.37 Our previous data show that Amot knockdown results in inhibition of migration of vessels sprouting from the dorsal aorta.17 The timing of outgrowth of intersegmental vessels was not affected, but migrating cells halted at the horizontal myoseptum. Amot deficiency affected the number of filopodia and tip cell polarization. In this study, we found that knocking down either syx-a or syx-b resulted in inhibition of dorsal migration of ISV tip cells, a phenotype shared with amot morphants. Furthermore, our analysis revealed synergistic interactions between Syx-b and Amot, leading us to conclude that Syx and Amot form a signaling pathway that controls directional migration during ISV formation. These genetic interaction studies, combined with the molecular association results, provide strong evidence for Amot and Syx functioning in a common molecular complex. The findings that Syx also associates with AmotL1 and AmotL2 indicate that Syx may also participate in other signaling pathways independent of Amot.
Cell migration is thought to occur by a coordinated cycle of leading edge protrusion in the direction of migration, substrate adhesion of the protrusion, generation of tension on new adhesions to advance the cell body, and deadhesion of the trailing cell rear. An important question in understanding vessel formation is how a guidance signal is translated to directional migration. We suggest that endothelial migration is dependent on the formation of the Amot:Patj/Mupp1:Syx complex to precisely localize RhoA activity to the leading front of migrating cells. As we have previously shown that Amot interference also inhibits pathologic angiogenesis,30,31 these findings may be of clinical significance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are deeply grateful for the help of Susan Warner, Ulla Wargh, and Sajila Kisana in the Karolinska Institute zebrafish facility. We are also indebted to Tanya Tegnebratt for the finalization of the paper.
L.H. was supported by grants from the Swedish Cancer Society (Stockholm, Sweden), Cancer Society of Stockholm (Stockholm, Sweden), Karolinska Institutet (Stockholm, Sweden), and the Swedish Research Council (Stockholm, Sweden). L.H. and J.-P.B. are supported by grants from the FP7 EUCAAD consortium (Brussels, Belgium). T.W. is supported by IMF grant WE110719 (North Hollywood, CA).
Authorship
Contribution: M.E., N.L.P., S.A., P.L., I.S., M.L., M.S., A.H., K.A., T.W., J.-P.B., and A.M. performed experiments, analyzed the results, and made the figures; T.W., J.-P.B., A.M., and L.H. designed the research; and L.H. wrote the paper.
Conflict-of-interest disclosure: L.H. is coauthor of an angiomotin patent covering antiangiogenic applications. The remaining authors declare no competing financial interests.
Correspondence: Lars Holmgren, Department of Oncology and Pathology, Cancer Centrum Karolinska, CCK R8:03 Karolinska Hospital, 171 76 Stockholm, Sweden; e-mail: lars.holmgren@ki.se.