Although much is known about globin gene activation in erythroid cells, relatively little is known about how these genes are silenced in nonerythroid tissues. Here we show that the human α- and β-globin genes are silenced by fundamentally different mechanisms. The α-genes, which are surrounded by widely expressed genes in a gene dense region of the genome, are silenced very early in development via recruitment of the Polycomb (PcG) complex. By contrast, the β-globin genes, which lie in a relatively gene-poor chromosomal region, are not bound by this complex in nonerythroid cells. The PcG complex seems to be recruited to the α-cluster by sequences within the CpG islands associated with their promoters; the β-globin promoters do not lie within such islands. Chromatin associated with the α-globin cluster is modified by histone methylation (H3K27me3), and silencing in vivo is mediated by the localized activity of histone deacetylases (HDACs). The repressive (PcG/HDAC) machinery is removed as hematopoietic progenitors differentiate to form erythroid cells. The α- and β-globin genes thus illustrate important, contrasting mechanisms by which cell-specific hematopoietic genes (and tissue-specific genes in general) may be silenced.
Introduction
To date, research on the regulation of hematopoietic gene expression has concentrated on understanding how genes are activated, but it is equally important to discover how they are appropriately switched off and maintained in a silenced state in nonexpressing cells during differentiation and development. The well-characterized human α- and β-globin loci, which are transcriptionally silent in hematopoietic stem cells, early progenitors, and nonerythroid cells but active in erythroid cells, provide an excellent model system to address this issue. In this study we have concentrated on understanding how the human α-globin genes are silenced and contrasted this with what is known about silencing of the β-globin gene.
The human α-globin genes (16p13.3) are expressed in a highly tissue- and developmental-stage specific manner, being activated in the terminal stages of erythroid differentiation and in no other cell types.1 However, these genes and their major remote regulatory element (HS-40) lie in a gene-dense, GC-rich region of the genome, surrounded by widely expressed genes.2 When silenced, the entire region (> 500 kilobase pairs [kb]) including the α-globin cluster, its regulatory elements, and the flanking genes, remains early replicating,3 acetylated at a low level4 and never colocalizes with pericentromeric heterochromatin,5 suggesting this is a region of constitutively accessible chromatin. Like the widely expressed genes surrounding them, the promoters of the human α-globin genes lie within large CpG islands (Figure 1, islands H,I). By contrast, the coordinately regulated β-globin genes (11p15.5), which are also expressed specifically in erythroid cells, lie in a relatively gene-poor, GC-poor region of the genome and when silenced are incorporated into late replicating, hypoacetylated chromatin, which localizes with pericentromeric heterochromatin (reviewed by Higgs et al6 ), features associated with silent, inaccessible chromatin. The promoters within the β-globin cluster are not associated with CpG islands and become methylated in nonexpressing tissues.7 Therefore, whereas the β-globin genes seem to be silenced via the well-described formation of facultative heterochromatin, the α-cluster highlights an increasingly recognized general question of how tissue-specific genes lying in gene dense regions of open chromatin may be silenced, whereas neighboring genes remain fully active.
Overview of the human α-globin (HBA) locus (16p13.3). Genes of the α-locus are shown in black and other genes are in gray. Pseudogenes are in white. Genes shown above the line are transcribed toward the centromere and those below the line are transcribed toward the telomere (black oval). The gray bar above indicates the domain of erythroid-specific histone hyperacetylation.4 Below are indicated CpG islands (black bars), and previously documented erythroid-specific (black arrows) and ubiquitous (gray arrows) DNaseI hypersensitive sites. * indicates HS analyzed in this study. At bottom is shown the position of real-time PCR amplicons used to analyze quantitative ChIP experiments (gray bars). The probes spanning the duplicated adult stage-specific HBA genes (boxed) do not distinguish the HBA2 and HBA1 genes.
Overview of the human α-globin (HBA) locus (16p13.3). Genes of the α-locus are shown in black and other genes are in gray. Pseudogenes are in white. Genes shown above the line are transcribed toward the centromere and those below the line are transcribed toward the telomere (black oval). The gray bar above indicates the domain of erythroid-specific histone hyperacetylation.4 Below are indicated CpG islands (black bars), and previously documented erythroid-specific (black arrows) and ubiquitous (gray arrows) DNaseI hypersensitive sites. * indicates HS analyzed in this study. At bottom is shown the position of real-time PCR amplicons used to analyze quantitative ChIP experiments (gray bars). The probes spanning the duplicated adult stage-specific HBA genes (boxed) do not distinguish the HBA2 and HBA1 genes.
Here we show that the inactive state of the α-genes in nonerythroid cells is dependent on targeting by the multiprotein repressive PRC2 polycomb (PcG) complex, which specifically binds the α-globin genes but not the surrounding housekeeping genes. Furthermore, directed by previous biochemical analyses, we show that PcG silencing is mediated, in vivo, via histone deacetylases (HDACs). This repressive state is present in human embryonic stem (ES) cells and remains at the locus throughout differentiation in nonerythroid lineages but is removed during erythropoiesis, as the α-globin genes become activated.
Methods
Cell culture and drug treatments
Primary cells and cell lines were isolated and cultured as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Where indicated, cells were cultured with either 0.2 μmol/L TSA (Sigma-Aldrich, St Louis, MO)) or carrier (methanol) for the times stated. Where indicated, Epstein-Barr virus (EBV)–lymphoblastoid cells were pretreated for 45 minutes with either 1 μg/mL actinomycin D or 2 μg/mL cycloheximide. These treatments are in excess of those previously shown to block induction of gene expression by HDAC inhibition in a T-cell leukemia cell line.8 After pretreatment, 0.2 μmol/L TSA (or carrier) was added as indicated, and cultures were incubated for a further 8 hours before harvesting for RNA.
siRNA interference
For EZH2 knockdown, HeLa cells were transfected with duplex siRNA against EZH2 (target sequence, 5′AAG ACT CTG AAT GCA GTT GCT)9 or with control (nonsilencing) duplex siRNA (QIAGEN, Valencia, CA) at 150 nM. Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Every 2 days, cells were trypsinized, samples were removed for harvesting RNA and protein and for immunofluorescent analysis, and the remainder were reseeded (at a density of 106 cells per 100-mm dish). After being allowed to settle for 1 hour, cells were retransfected with the same siRNA as described above. At the final timepoint (6 days), cells were also harvested for chromatin immunoprecipitation (ChIP). Protein extracts and Western blots were performed as described previously10 using antibodies against EZH2 (AR-0163; Lake Placid Biologicals, Lake Placid, NY) or RNA polymerase II (H-224, sc-9001; Santa Cruz Biotechnology, Santa Cruz, CA).
ChIP assays
ChIP and ChIP on chip assays were performed essentially as described previously,11 and details are provided in Document S1.
Results
The inactive α-globin genes are marked by H3K27me3 and bound by the PRC2 complex in nonerythroid tissues
Because the human α-globin genes are embedded within a transcriptionally active, gene-dense region of the genome, it is essential that, in nonerythroid cells, they are silenced as individual genes while the surrounding genes continue to be expressed normally. It is well established that there is a large array of enzyme-catalyzed covalent modifications of histone tails that constitutes an epigenetic code which is an important determinant of gene activity (reviewed by Jenuwein and Allis12 ). It has been shown that during transcriptional up-regulation in erythroid cells, the α-globin cluster acquires activating histone modifications, specifically hyperacetylation of histone H3 and H4 (acH3 and acH4) and methylation of lysine 4 of histone H3 (H3K4me).4,13 We decided to investigate whether the inactive state of the α-globin genes in nonerythroid cells was associated with the acquisition of repressive histone modifications. We carried out ChIP experiments in nonerythroid tissues to establish profiles of H3K9me3 (trimethylation of lysine 9 of histone H3, a repressive mark usually associated with pericentric and facultative heterochromatin14,15 ) and H3K27me3 (trimethylation of lysine 27 of histone H3, a modification associated with transcriptional repression by the PcG complex.
Quantitative ChIP experiments were initially performed in nonerythroid EBV-transformed lymphocytes and analyzed by quantitative real-time polymerase chain reaction (PCR) using Taqman probes (Figure S1A). Although H3K9me3 was uniformly low at all points across the domain analyzed, we observed clear enrichment for H3K27me3 at the silenced α-globin genes. ChIP on Chip analysis in primary T lymphocytes (Figure 2A) revealed a broad domain of H3K27me3 enrichment across the α-globin locus, with discrete and prominent peaks of enrichment associated with each of the major adult αA-globin genes (HBA2 and HBA1), as well as the upstream minor αD-globin gene (HBM), and an intervening pseudogene (HBA1ps). There were lower levels of enrichment at the embryonic stage-specific ζ-globin gene (HBZ). The α-globin gene cluster is thus associated with a discrete region of H3K27me3 modification (delineated by vertical lines in Figure 2A), with only background enrichment detected for approximately 80 kb on either side, in the regions containing the widely expressed flanking genes. In addition, we observed a discrete enrichment of H3K27me3 at CpG island C (associated with the DIST1/RHBDF1 gene), which lies approximately 100 kb upstream of the HBA2 gene (Figure 2A). It is noteworthy that we did not see any repressive chromatin marks at any of the previously characterized, conserved, noncoding regulatory elements (summarized in Figure 1).
The PRC2 polycomb complex is recruited to the inactive HBA genes. ChIP experiments were carried out in primary T lymphocytes or primary proerythroblasts using antibodies against H3K27me3 (A), SUZ12 (B), or RNA pol II (C). For (A) and (B), immunoprecipitated and input material were labeled with Cy3 or Cy5, respectively, and applied to a custom tiled microarray covering the terminal region of human chromosome 16. Shown is a 250-kb genomic segment containing the α-globin locus (delineated by gray dashed lines) with the fold enrichments at each probe plotted against chromosomal position. The locus is annotated as described in Figure 1. For panel C, immunoprecipitated material was analyzed by real-time PCR at amplicons 53 (within the middle of the DIST1 gene [see Figure 1]) and an amplicon within the 18S rRNA gene (both representing background enrichment), an amplicon at the promoter of the β-actin gene (ACTB) used as a positive control, and an amplicon spanning exons 1-2 of the adult HBA genes (HBA ex1-2; see Figure 1). Data indicate the percentage of input material precipitated at each amplicon. Shown are the mean values obtained from replicate ChIPs with error bars indicating the range of values.
The PRC2 polycomb complex is recruited to the inactive HBA genes. ChIP experiments were carried out in primary T lymphocytes or primary proerythroblasts using antibodies against H3K27me3 (A), SUZ12 (B), or RNA pol II (C). For (A) and (B), immunoprecipitated and input material were labeled with Cy3 or Cy5, respectively, and applied to a custom tiled microarray covering the terminal region of human chromosome 16. Shown is a 250-kb genomic segment containing the α-globin locus (delineated by gray dashed lines) with the fold enrichments at each probe plotted against chromosomal position. The locus is annotated as described in Figure 1. For panel C, immunoprecipitated material was analyzed by real-time PCR at amplicons 53 (within the middle of the DIST1 gene [see Figure 1]) and an amplicon within the 18S rRNA gene (both representing background enrichment), an amplicon at the promoter of the β-actin gene (ACTB) used as a positive control, and an amplicon spanning exons 1-2 of the adult HBA genes (HBA ex1-2; see Figure 1). Data indicate the percentage of input material precipitated at each amplicon. Shown are the mean values obtained from replicate ChIPs with error bars indicating the range of values.
Further ChIP experiments showed that, as well as in lymphocytes, the H3K27me3 modification was present at the α-globin loci in other nonexpressing cell types tested (primary neutrophils and HeLa epithelial carcinoma cells; Figure S1B). In contrast to these nonerythroid tissues, the H3K27me3 modification was not present at the α-globin loci in K562 cells (an erythroid-like cell line which expresses α-globin mRNA; Figure S1B). In primary erythroid progenitors, we observed that H3K27me3 was reduced at the active α-globin loci (although not completely cleared [discussed further below]) relative to nonexpressing T lymphocytes (Figure 2A). Binding to the upstream DIST1/RHBDF1 gene was still very strong in proerythroblasts and serves as a good positive control for the ChIP experiment. The relative reduction of H3K27me3 signal in proerythroblasts compared with T lymphocytes could have been due to a relative paucity of nucleosomes around the α-globin genes in erythroid compared with nonerythroid cells. However, ChIP on chip experiments using an antibody against unmodified histone H3 revealed that at the resolution of the sonicated chromatin used in this assay, nucleosomal density across this region was broadly similar between T lymphocytes and proerythroblasts (Figure S2). Further, activating histone modifications (acH3, acH4, H3K4me2, and H3K4me3) are highly enriched at the α-globin genes in these erythroid cells.13 Together, these observations show that the H3K27me3 chromatin modification is strongest when the α-globin genes are silenced and lower or absent when they are active.
The H3K27me3 modification is associated with repression of gene expression mediated by the PRC2 polycomb complex, which includes the core components SUZ12 (essential for the structural and functional integrity of the complex16 ) and EZH2 (which provides the histone methyltransferase activity17 ). We therefore investigated whether enrichment of the H3K27me3 modification correlated with recruitment of components of the PRC2 complex to the inactive α-globin genes. By ChIP on chip, we observed prominent recruitment of SUZ12 to the CpG islands of the inactive α-globin genes in nonerythroid primary T lymphocytes, and this was depleted to almost baseline levels at the active locus in primary proerythroblasts (Figure 2B). The recruitment of SUZ12 to the α-globin locus in T lymphocytes was very specific and accurately reflected the pattern of H3K27me3 modification in the same cell type (Figure 2A) with no other significant peaks of SUZ12 enrichment within 80 kb on either side of the α-globin cluster. SUZ12 was also specifically recruited to the promoter region of the DIST1/RHBDF1 gene (CpG island C), where we had previously observed enrichment of H3K27me3. The level of SUZ12 enrichment at CpG island C was similar in T lymphocytes and proerythroblasts, demonstrating that the depletion of SUZ12 at the α-globin cluster in proerythroblasts was a localized effect. The pattern of SUZ12 binding in T lymphocytes and proerythroblasts was reciprocal to that observed for RNA polymerase II in the same samples (Figure 2C). As expected, RNA pol II was highly abundant at the adult-stage α-globin genes in proerythroblasts, and was absent in T lymphocytes. This reciprocity is consistent with the repressive role for the PRC2 complex. The almost complete removal of SUZ12 from the α-globin genes in proerythroblasts contrasts with the only partial depletion of the H3K27me3 modification observed in the same cells (Figure 2A) and indicates that there is a lag in clearing the histone modification from the locus even after the PRC2 complex has been removed. This probably implies a slower, passive rather than active (via histone demethylases) mechanism of clearing the methylated histones from this locus during erythroid differentiation, and moreover suggests that clearing the PRC2 complex itself is more important than removing the histone modification to achieve full activation of the genes. Binding of PRC2 to the α-globin cluster in nonerythroid cells was confirmed by conventional quantitative ChIP analysis (Figure S3A). We observed clear enrichment of both SUZ12 and EZH2 proteins at the inactive α-globin genes in EBV-lymphoblasts, whereas there was no significant recruitment above background in K562 cells (α-globin expressing).
In contrast to the α-globin locus, we observed no significant enrichment in nonerythroid cells for either the H3K27me3 modification (in EBV-lymphoblasts) or the SUZ12 protein (in primary T lymphocytes) around the adult β-globin gene (probes HBB ex1, 3′HBB) or at a probe within HS5 of the β-globin locus control region (probe βLCR HS5) by quantitative ChIP (Figures S1A,S3B) or by ChIP on chip (data not shown). Overall, these data indicate that the silenced α-globin but not β-globin genes are targeted by the PRC2 polycomb complex and modified by H3K27me3 in nonerythroid cell types and suggest that, when inactive, these clusters are silenced by different mechanisms.
RNAi-mediated depletion of EZH2 activates α-globin expression in nonerythroid cells
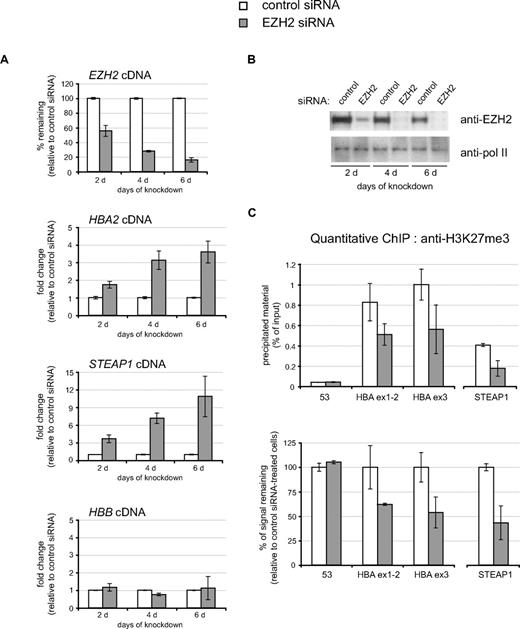
To determine the functional importance of the PRC2 complex in silencing α-globin in nonerythroid cells, we used RNAi to deplete HeLa cells of the PRC2 core component EZH2. As shown in Figure S1B(ii), the α-globin genes are modified by H3K27me3 in HeLa cells. HeLa cells were transfected with either control (nonsilencing) or EZH2-specific siRNA and analyzed every 2 days over a 6-day time course. Clear depletion of EZH2 cDNA (as measured by real-time reverse transcription PCR [real time RT-PCR]; Figure 3A) and protein (Figure 3B) was observed after only 2 days with the EZH2-specific siRNA. The extent of knockdown increased further after 4 and 6 days of treatment, and very little EZH2 protein was detected at these time points. Knock-down of EZH2 resulted in a clear depletion of nuclear levels of H3K27me3 as measured by quantitative confocal analysis of immunofluorescence-stained cells (Figure S4).
Induction of HBA expression after siRNA-mediated depletion of EZH2 in nonerythroid cells. HeLa cells were transfected with either control siRNA or EZH2-specific siRNA and were harvested every 2 days over a 6-day time course as shown. (A) Real time RT-PCR analysis of EZH2, HBA2, STEAP1, and HBB expression. Results are shown as either the percentage of EZH2 signal remaining (top panel) or the fold change (bottom panels) in the EZH2 siRNA-treated population relative to the control siRNA–treated population after normalizing for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal. (B) Protein extracts from a representative time course analyzed by Western blot using antibodies against EZH2 (top panel) or RNA polymerase II as loading control (bottom panel). (C) Chromatin was harvested after 6 days with the indicated siRNA and was analyzed by quantitative ChIP using an antibody against H3K27me3. Material was analyzed by real-time PCR at amplicons 53 (background enrichment only), HBA ex1-2 and HBA ex3 from the α-globin genes (see Figure 1) and the CpG-island of the STEAP1 gene. Data are presented as either raw enrichment (the percentage of input material precipitated at each amplicon; top panel) or the percentage of signal remaining in the EZH2 siRNA-treated cells relative to control siRNA–treated cells (bottom panel). Data in panels A and C show the mean values obtained from replicate time courses with error bars indicating the range of values.
Induction of HBA expression after siRNA-mediated depletion of EZH2 in nonerythroid cells. HeLa cells were transfected with either control siRNA or EZH2-specific siRNA and were harvested every 2 days over a 6-day time course as shown. (A) Real time RT-PCR analysis of EZH2, HBA2, STEAP1, and HBB expression. Results are shown as either the percentage of EZH2 signal remaining (top panel) or the fold change (bottom panels) in the EZH2 siRNA-treated population relative to the control siRNA–treated population after normalizing for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal. (B) Protein extracts from a representative time course analyzed by Western blot using antibodies against EZH2 (top panel) or RNA polymerase II as loading control (bottom panel). (C) Chromatin was harvested after 6 days with the indicated siRNA and was analyzed by quantitative ChIP using an antibody against H3K27me3. Material was analyzed by real-time PCR at amplicons 53 (background enrichment only), HBA ex1-2 and HBA ex3 from the α-globin genes (see Figure 1) and the CpG-island of the STEAP1 gene. Data are presented as either raw enrichment (the percentage of input material precipitated at each amplicon; top panel) or the percentage of signal remaining in the EZH2 siRNA-treated cells relative to control siRNA–treated cells (bottom panel). Data in panels A and C show the mean values obtained from replicate time courses with error bars indicating the range of values.
In response to depletion of EZH2, we detected a clear, time-dependent, increase in expression of the α2-globin gene (HBA2) as measured by real time RT-PCR (Figure 3A). This induction of HBA2 was similar to that observed in the same cDNA samples at another gene (STEAP1) previously shown to be bound by the PRC2 complex (SUZ12) and modified by H3K27me3 in HeLa cells (tracks L1 SUZ12 HeLa and L1 H3K27me3 HeLa on the UCSC Genome Browser (http://genome.ucsc.edu/)). As expected, the β-globin gene HBB, which is not bound by the PRC2 complex, did not respond to depletion of EZH2 (Figure 3A). The induction of both HBA2 and STEAP1 was associated with clear depletion of the H3K27me3 modification from each locus as measured by ChIP (Figure 3C), suggesting that the effect of EZH2 knockdown on these genes is direct. Depletion of H3K27me3 signal at these target loci (to roughly 50%-60% of the signal observed with control siRNA) was similar to the depletion of H3K27me3 signal observed globally by immunofluorescence at the same time point (Figure S4).
Polycomb-repression of α-globin is mediated by histone deacetylases
The histone methylation laid down by the PRC2 complex serves as a recruitment signal for a second polycomb complex, PRC1, whose core component (PC) recognizes H3K27me3 through its chromodomain.18 Although it is clear that targeting of a gene by the PcG machinery generally leads to transcriptional inactivation, the molecular mechanisms mediating PcG-associated repression in vivo are still poorly understood (reviewed by Sparmann and van Lohuizen19 ). The demonstration in vitro of interactions between EZH2 and DNA methyltransferases20 and between EED (another core component of the PRC2 complex) and histone deacetylases (HDACs)21 suggests that either DNA methylation and/or histone deacetylation may be involved in transcriptional repression mediated by PRC2. However, the recruitment and relative importance of these mechanisms in PcG silencing in vivo have not been fully evaluated.
The promoters of the α-globin genes are associated with large CpG islands (H and I in Figure 1), which are unmethylated in all normal tissues and cell lines previously studied.22 Here we extended these observations to investigate a wider range of normal fetal and adult nonerythoid tissues. We observed complete digestion by the methylation-sensitive restriction enzymes EagI and HpaII at sites within the α-globin CpG islands, indicating an unmethylated state in all tissues examined (Figure S5). These data indicate that even though transcriptionally inactive in these tissues, the α-globin promoters remain unmethylated during differentiation and development, demonstrating that DNA methylation is not involved in PRC2-mediated repression of the α-globin genes in nonerythroid cell types.
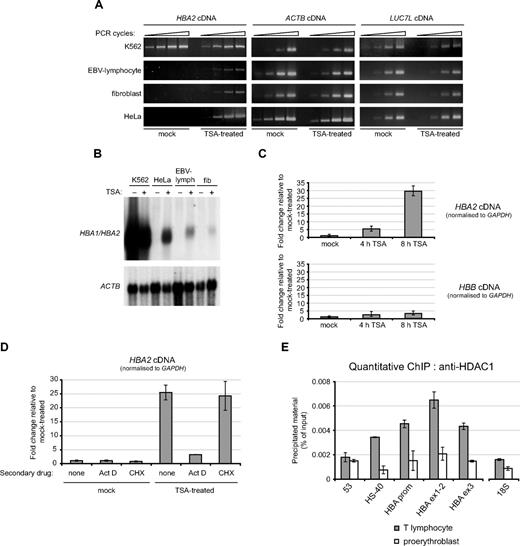
To investigate a potential role for HDACs in maintaining the transcriptionally repressed state of the α-globin genes, we initially treated a variety of nonerythroid cells (EBV-transformed lymphocytes, primary fibroblasts, and HeLa cells) with the HDAC-inhibitor trichostatin A (TSA) for 24 hours (Figure 4A). TSA induced a clear activation of α-globin mRNA expression in each of the 3 nonerythroid cell lines tested. By contrast, in the erythroid K562 cell line used as a positive control, where the α-globin genes were already active in mock-treated cultures, TSA had no significant effect on expression. Likewise, for 2 housekeeping genes (ACTB [β actin] and LUC7L, a ubiquitously expressed gene located immediately downstream of the α-globin cluster [Figure 1]), treatment with TSA had no obvious effect. Induction of α-globin mRNA expression was also observed by RT-PCR after treatment of nonerythroid cell lines with 2 other HDAC inhibitors (5 mM sodium butyrate in HeLa cells and 2 mM sodium valproate in EBV-lymphoblasts) but to a lesser degree (data not shown).
The HBA genes are repressed by histone deacetylases in nonerythroid cells. (A) Semiquantitative RT-PCR analysis of HBA2, ACTB, and LUC7L cDNA in the cell lines indicated that were either mock-treated, or treated with 0.2 μmol/L TSA for 24 hours. (B) Northern blot analysis of RNA from the cell lines indicated that were either mock-treated (−) or treated with 0.2 μmol/L TSA for 24 hours (+). The membrane was hybridized first to a probe which recognizes transcripts from both HBA1 and HBA2 genes (top panel) and subsequently to an ACTB cDNA probe as loading control (bottom panel). (C) Real-time RT-PCR analysis of HBA2 and HBB cDNA in EBV-lymphocytes that were either mock-treated or treated with 0.2 μmol/L TSA for 4 or 8 hours. Results are shown as fold change relative to mock-treated cells, after normalizing for GAPDH signal. (D) Real time RT-PCR analysis of HBA2 cDNA in EBV-lymphocyte cultures that were either mock-treated or treated with 0.2 μmol/L TSA for 8 hours, in the presence of either no secondary drug or actinomycin D or cycloheximide as indicated. Results are shown as fold change relative to mock-treated cells (with no secondary drug), after normalizing for GAPDH signal. Shown is the mean of replicate experiments, with error bars indicating the range. For each experiment, PCR was performed in triplicate. (E) ChIP experiments were carried out on primary T lymphocytes or proerythroblasts using an antibody against HDAC1, and material was analyzed at the amplicons indicated surrounding the α-globin locus (shown in Figure 1) or a control amplicon within the 18S rRNA gene. Shown are the mean values obtained from replicate experiments with error bars indicating the range.
The HBA genes are repressed by histone deacetylases in nonerythroid cells. (A) Semiquantitative RT-PCR analysis of HBA2, ACTB, and LUC7L cDNA in the cell lines indicated that were either mock-treated, or treated with 0.2 μmol/L TSA for 24 hours. (B) Northern blot analysis of RNA from the cell lines indicated that were either mock-treated (−) or treated with 0.2 μmol/L TSA for 24 hours (+). The membrane was hybridized first to a probe which recognizes transcripts from both HBA1 and HBA2 genes (top panel) and subsequently to an ACTB cDNA probe as loading control (bottom panel). (C) Real-time RT-PCR analysis of HBA2 and HBB cDNA in EBV-lymphocytes that were either mock-treated or treated with 0.2 μmol/L TSA for 4 or 8 hours. Results are shown as fold change relative to mock-treated cells, after normalizing for GAPDH signal. (D) Real time RT-PCR analysis of HBA2 cDNA in EBV-lymphocyte cultures that were either mock-treated or treated with 0.2 μmol/L TSA for 8 hours, in the presence of either no secondary drug or actinomycin D or cycloheximide as indicated. Results are shown as fold change relative to mock-treated cells (with no secondary drug), after normalizing for GAPDH signal. Shown is the mean of replicate experiments, with error bars indicating the range. For each experiment, PCR was performed in triplicate. (E) ChIP experiments were carried out on primary T lymphocytes or proerythroblasts using an antibody against HDAC1, and material was analyzed at the amplicons indicated surrounding the α-globin locus (shown in Figure 1) or a control amplicon within the 18S rRNA gene. Shown are the mean values obtained from replicate experiments with error bars indicating the range.
Although induced by HDAC inhibitors, the level of α-globin expression achieved in TSA-treated, nonerythroid cells as measured by Northern blot is still considerably lower than the expression level observed in K562 cells (which itself is < 1% of that observed in primary erythroblasts; Figure 4B) and probably represents basal (unenhanced) activity of the α-globin promoter. Shorter time-course experiments in EBV-lymphoblasts revealed that α-globin expression had been clearly induced by as early as 4 hours of TSA treatment (Figure 4C). The α-globin cDNA continued to accumulate rapidly, reaching approximately 30-fold induction relative to mock-treated culture after 8 hours of TSA treatment. The rapidity of this response is consistent with a direct effect of HDAC inhibition on the α-globin genes themselves. TSA-induced up-regulation of α-globin expression observed in EBV-lymphoblasts was unaffected by the presence of cycloheximide at protein inhibitory concentrations (Figure 4D). In contrast, pretreatment of EBV-lymphoblasts with actinomycin D (Act D; inhibitory to RNA polymerase II activity) almost completely blocked the effect of TSA on α-globin mRNA expression. Thus the induction of α-globin expression by HDAC-inhibitors involves active transcription but does not require synthesis of a mediatory protein, again consistent with HDAC-inhibition acting directly on transcription of the α-globin genes. Finally, we demonstrated directly by quantitative ChIP that the histone deacetylase HDAC1 was enriched at the α-globin genes (probes HBA prom, HBA ex1-2, and HBA ex3) and to a lesser extent at the upstream regulatory element HS-40 in primary nonerythroid (T lymphocyte) cells (Figure 4E); only background levels of enrichment of HDAC1 were observed at these points in primary proerythroblasts. In contrast to the dramatic up-regulation (∼30-fold) of α-globin expression observed in TSA-treated EBV-lymphoblasts, only a minor increase (∼3-fold) in expression of the adult β-globin gene (HBB) was observed over the same time course (Figure 4C). By Northern blot, HBB signal remained undetectable even after 24-hour treatment with TSA in HeLa, EBV-lymphoblasts, and fibroblasts (data not shown). Thus, regulation by HDACs correlates with the differential targeting by PRC2 between these 2 gene clusters.
Changes in chromatin accessibility are associated with TSA-mediated induction of α-globin expression
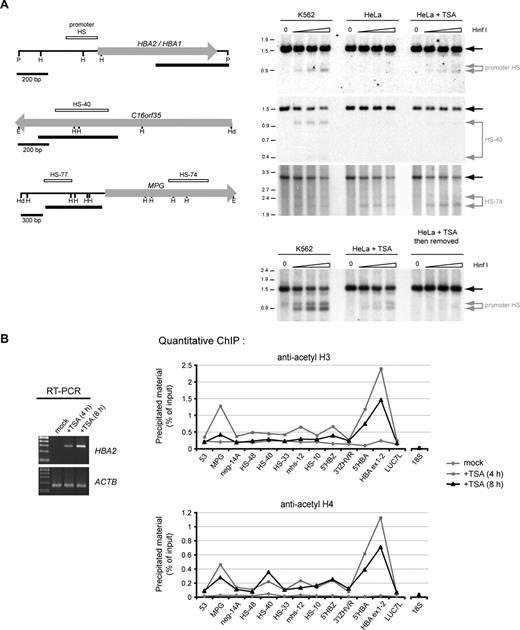
To determine the specificity of HDAC inhibition within the α-globin cluster, we next examined whether the resulting up-regulation of α-globin expression in nonerythroid cells was accompanied by changes in chromatin modification and accessibility. The α-globin promoters are usually hypersensitive to digestion by endonucleases (reflecting a relaxed chromatin structure) in expressing erythroid (fetal liver, adult bone marrow, and K562 cells) but not nonerythroid (fetal brain) tissues.23 We found that the promoter hypersensitive site (HS; assayed using the restriction enzyme HinfI)22 was clearly detected in K562 cells (used as a positive control), but as expected, was undetectable in mock-treated HeLa cells. However, treatment of this nonerythroid cell type with 0.2 μmol/L TSA for 8 hours resulted in clear formation of this HS (Figure 5A top panel). In contrast to the promoter HS, TSA had no effect on formation of the major upstream erythroid-specific HS (HS-40; Figure 1; hypersensitive to cleavage only in K562 cells) or on a ubiquitous HS (HS-74; Figure 1; hypersensitive to cleavage in all samples; Figure 5A second and third panels). The failure to activate the major upstream regulatory element (HS-40) is consistent with the observation that the level of α-globin expression in TSA-treated nonerythroid cells is still suboptimal and vastly lower than expression observed in K562 cells (Figure 4B). This expression level probably represents the basal (unenhanced) activity of the α-globin promoter. We also found that if HeLa cells in which HS formation had been induced by TSA treatment were then cultured for a further 24 hours in the absence of TSA, the HS site was no longer observed, indicating that the normal repressive chromatin structure is quickly recovered if HDAC activity is no longer inhibited (Figure 5A bottom panel). Together, these results show that HS formation at the α-globin promoter is strictly regulated by the activity of HDACs.
Chromatin changes at the HBA genes in response to HDAC-inhibition. (A) Restriction enzyme accessibility assays performed on nuclei from K562 cells, mock-treated HeLa cells, or HeLa cells treated with 0.2 μmol/L TSA for 8 hours. Nuclei were exposed to no enzyme (0) or increasing concentrations of HinfI. Shown are Southern blots to detect hypersensitive sites at the α-globin promoters, HS-40 and HS-74, as indicated. The limit digested bands are indicated by black arrows, and bands resulting from digestion by HinfI within HSs are indicated by gray arrows. The limit digested fragments analyzed on each membrane are shown at left (H, HinfI; P, PstI; E, EcoRI; Hd, HindIII). The previously mapped extent of the known hypersensitive sites (open bars) and the probes used (black bars) are indicated. Genes are shown as block arrows. In the bottom panel, the HBA promoter HS was analyzed in nuclei from K562 cells, HeLa cells after treatment with TSA, or HeLa cells which were treated with TSA and then cultured for a further 24 hours in the absence of TSA. (B) Left panel: RT-PCR analysis of HBA2 cDNA (31 cycles) and ACTB cDNA (26 cycles) in an EBV-lymphoblastoid culture which was either mock-treated, or treated with 0.2 μmol/L TSA for 4 or 8 hours. Right panels: ChIP experiments carried out on the EBV-lymphoblastoid cells analyzed by RT-PCR, using antibodies against either acetylated histone H3 or histone H4. Immunoprecipitated material was analyzed by real-time PCR at the amplicons indicated surrounding the α-globin locus (shown in Figure 1) or a control amplicon within the 18S rRNA gene. Data indicate the percentage of input material precipitated at each amplicon.
Chromatin changes at the HBA genes in response to HDAC-inhibition. (A) Restriction enzyme accessibility assays performed on nuclei from K562 cells, mock-treated HeLa cells, or HeLa cells treated with 0.2 μmol/L TSA for 8 hours. Nuclei were exposed to no enzyme (0) or increasing concentrations of HinfI. Shown are Southern blots to detect hypersensitive sites at the α-globin promoters, HS-40 and HS-74, as indicated. The limit digested bands are indicated by black arrows, and bands resulting from digestion by HinfI within HSs are indicated by gray arrows. The limit digested fragments analyzed on each membrane are shown at left (H, HinfI; P, PstI; E, EcoRI; Hd, HindIII). The previously mapped extent of the known hypersensitive sites (open bars) and the probes used (black bars) are indicated. Genes are shown as block arrows. In the bottom panel, the HBA promoter HS was analyzed in nuclei from K562 cells, HeLa cells after treatment with TSA, or HeLa cells which were treated with TSA and then cultured for a further 24 hours in the absence of TSA. (B) Left panel: RT-PCR analysis of HBA2 cDNA (31 cycles) and ACTB cDNA (26 cycles) in an EBV-lymphoblastoid culture which was either mock-treated, or treated with 0.2 μmol/L TSA for 4 or 8 hours. Right panels: ChIP experiments carried out on the EBV-lymphoblastoid cells analyzed by RT-PCR, using antibodies against either acetylated histone H3 or histone H4. Immunoprecipitated material was analyzed by real-time PCR at the amplicons indicated surrounding the α-globin locus (shown in Figure 1) or a control amplicon within the 18S rRNA gene. Data indicate the percentage of input material precipitated at each amplicon.
Concomitant with HS formation, treatment with TSA also resulted in clear increases in acetylation of both histone H3 and H4at the α-globin locus as measured by ChIP (Figure 5B). The most dramatic changes were observed at the α-globin genes themselves (Figures 1 and 5B, probes 5′HBA and HBA ex1-2), although smaller increases in acetylation were also observed elsewhere throughout the locus. Consistent with the observation that α-globin expression is up-regulated after only 4 hours of treatment with TSA (Figures 4C and 5B left panel), we found that histone hyperacetylation at the α-globin genes was also observed after only 4 hours. In fact, acetylation slightly decreases between 4 and 8 hours of treatment, indicating that the chromatin response is maximal after only 4 hours (although α-globin mRNA transcripts continue to accumulate [Figures 4C and 5B left panel]).
By quantitative ChIP, we were also able to detect clear recruitment of RNA polymerase II to the α-globin genes (probes HBA promoter and HBA ex1-2) in EBV-lymphoblasts in response to TSA (Figure S6A). In the same samples, we also observed that the SUZ12 protein was depleted from the α-globin genes in response to TSA treatment (Figure S6B), confirming that there is a direct link between these repressive pathways in regulating α-globin expression. It is noteworthy that we found that although the PRC2 complex (as indicated by SUZ12) was depleted in response to TSA, there was no reduction of H3K27me3 at the α-globin genes over the short time-course analyzed (Figure S6C), suggesting that clearing of the histone modification is less rapid than clearing of the PRC2 complex. Thus transcriptional activation of the α-globin genes in response to HDAC inhibition is associated with depletion of the PRC2 complex itself but does not require immediate removal of the H3K27me3 histone modification. This is consistent with the profiles observed in primary proerythroblasts.
The PRC2 complex is already engaged at the α-globin locus in pluripotent human embryonic stem cells
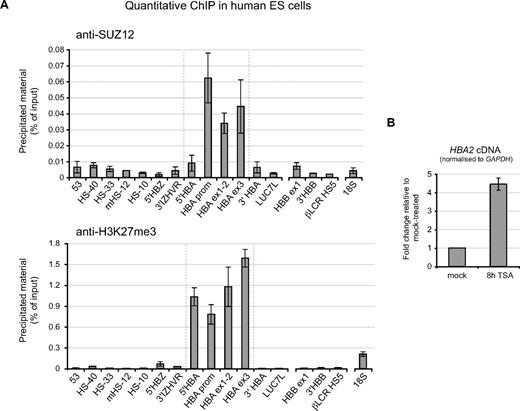
It has recently been demonstrated in both human24 and mouse25 that the PRC2 complex is targeted to a wide range of genes in pluripotent ES cells. Analysis of the PRC2 target gene sets suggested that this complex may be necessary to maintain the silence of genes that are not required for (or even inhibitory to) the pluripotent, undifferentiated state of ES cells, and which become activated during commitment and differentiation down different cellular lineages. Consistent with this model, ChIP experiments revealed that the α-globin genes, which are activated only after commitment to an erythroid fate, are bound by the PRC2 complex (as indicated by the core component SUZ12) and are strongly marked by the H3K27me3 modification in pluripotent human ES cells (Figure 6A). We also found that TSA induces up-regulation of α-globin expression in human ES cells (Figure 6B), indicating that, like the PRC2 complex, HDACs are already engaged at the α-globin genes in pluripotent cell types. Consistent with our observations in differentiated nonerythroid tissues, we found that the PRC2 complex and H3K27me3 are also not enriched at sites tested within the β-globin cluster (including the promoter of the adult HBB gene) in pluripotent ES cells (Figure 6A).
PcG-regulation at the HBA genes in human ES cells. (A) ChIP experiments carried out in human ES cells using antibodies against SUZ12 (top panel) or H3K27me3 (bottom panel). Immunoprecipitated material was analyzed at amplicons surrounding the α-globin locus (Figure 1) or at the β-globin gene promoter, 3′ of the β-globin gene, within HS5 of the β-globin locus control region (probes HBB ex1, 3′HBB, and βLCR HS5) or a control amplicon within the 18S rRNA gene. Data indicate the percentage of input material precipitated at each amplicon. (B) Real-time RT-PCR analysis of HBA2 cDNA in human ES cells which were either mock-treated, or treated with 0.2 μmol/L TSA for 8 hours. Results are shown as fold change relative to mock-treated cells after normalizing for GAPDH signal. Data in panels A and B show the mean values obtained from replicate experiments with error bars indicating the range of values.
PcG-regulation at the HBA genes in human ES cells. (A) ChIP experiments carried out in human ES cells using antibodies against SUZ12 (top panel) or H3K27me3 (bottom panel). Immunoprecipitated material was analyzed at amplicons surrounding the α-globin locus (Figure 1) or at the β-globin gene promoter, 3′ of the β-globin gene, within HS5 of the β-globin locus control region (probes HBB ex1, 3′HBB, and βLCR HS5) or a control amplicon within the 18S rRNA gene. Data indicate the percentage of input material precipitated at each amplicon. (B) Real-time RT-PCR analysis of HBA2 cDNA in human ES cells which were either mock-treated, or treated with 0.2 μmol/L TSA for 8 hours. Results are shown as fold change relative to mock-treated cells after normalizing for GAPDH signal. Data in panels A and B show the mean values obtained from replicate experiments with error bars indicating the range of values.
Discussion
Genome-wide analyses in human and mouse ES cells have revealed an important role for the PcG machinery in silencing genes, which must be repressed to maintain self-renewal and pluripotency and become activated during differentiation.24,25 These processes have also been studied in the context of specific PcG-target genes associated with muscle,26 neural,27 and germ cell28 differentiation. However, it is not clear how PcG is recruited to the particular chromosomal regions containing these genes and their cis-acting elements, and it is not certain how, once recruited, PcG complexes silence gene expression in vivo.
The globin genes provide an exemplary model for understanding how genes are switched on or off during differentiation and development. Both the α- and β-like genes are silent in hematopoietic stem cells (HSC) and specifically become activated in only 1 of the 8 mature blood lineages into which HSC differentiate, remaining completely silenced in all of the others. Many of these lineages and their progenitors can be easily purified; therefore, the discovery of PRC2- and HDAC-mediated repression of the α-globin genes in nonerythroid cells, reported here, provides a tractable model to probe how hematopoietic genes are silenced by this mechanism.
We have shown that the PcG complex and H3K27me3-modified histones are present at the α-globin genes in pluripotent human ES cells and are depleted as cells differentiate along the erythroid lineage. The activation of PcG-targeted genes during lineage-commitment and differentiation has dispelled the original dogma that the PcG-machinery constitutes a stable memory (imposed in stem cells) to maintain transcriptional silence throughout development. Instead, the work presented here and by others28 provides clear examples in which PcG-repression is reversible and shows that PcGs can be displaced by transcriptional activators at a specific target gene during commitment and differentiation along the appropriate cellular lineage. At present, the mechanisms by which the PcG machinery and H3K27me3 signal are cleared from such target loci are not understood.
While a transcriptionally repressive role for PcG is well established, the precise molecular mechanism(s) by which silencing is achieved at PcG-target genes is not known. Both DNA methylation and histone deacetylation have been implicated in Polycomb-mediated silencing, particularly from biochemical analyses. However, the recruitment and relative importance of these mechanisms in Polycomb silencing in vivo have not been comprehensively studied in normal tissues. It is noteworthy that the convergence of PRC2 with methylation and HDAC activity has been implicated in aberrant gene silencing during malignancy.29,30 Here we have shown that in nonerythroid cells, when α-globin gene expression is silenced by the PcG complex, its CpG island remains unmethylated and yet the chromatin associated with the promoter is poorly acetylated, inaccessible to nucleases, and does not bind polymerase II. However, in the presence of histone deacetylase inhibitors, the α-globin promoter rapidly becomes acetylated, depleted of SUZ12, accessible to endonucleases, binds polymerase II, and directs basal levels of α-globin mRNA transcription. Together, these findings show that PRC2-silencing of the α-globin promoter in nonerythroid cells involves the histone deacetylation pathway but does not involve DNA methylation. It is of interest that the degree of transcriptional activation upon TSA-treatment in pluripotent ES cells (Figure 6B, 4- to 5-fold) was smaller than that seen after TSA-treatment in differentiated nonerythroid cell types (Figure 4C, ∼30-fold in EBV-lymphoblastoid cell lines), suggesting that the repressive role of HDACs may be particularly important to lock in the inactive state at PcG-targeted genes in later, differentiated cell types. The mechanism of α-globin silencing reported here thus provides evidence for cooperation between the PcG and HDAC pathways in normal tissues. The more prominent response of α-globin to HDAC inhibition than PcG knockdown raises the possibility that HDACs may exert additional repressive activity at these genes, which is independent of the PcG complex. It will be of interest to examine the functional relationship between PcG and HDACs at these genes in more detail.
SUZ12 occupies the promoters of approximately 8% of all protein-coding genes in human ES cells24 but it is not clear how PcG proteins target some genes but not others. It is of interest that, unlike the human α-globin genes, we do not detect binding of the PRC2 complex and H3K27me3 modification at the orthologous mouse α-globin genes in nonexpressing cell types (data not shown). One of the most prominent differences between the genome structure of the human and mouse α-globin genes is that the large CpG islands present at the human α-globin promoters (PcG-regulated) have been eroded and are not present at the orthologous mouse genes (PcG-independent).31 Although the DNA sequence elements responsible for PcG recruitment (Polycomb response elements, PREs) have not yet been identified in mammals, our results suggest that in at least some cases, PRE elements will be contained within promoter CpG islands, and associations between PcG-binding and CpG islands have also been observed by others.32,33 This is consistent with bioinformatic analyses suggesting that binding of PRC2 is influenced to some extent by the physical characteristics of the CpG island. Among promoter-CpG islands, those that are bound by SUZ12 in human ES cells tend to be larger and with a higher overall number of CpG dinucleotides than those not bound by SUZ12 (Figure S7). Of course it is also likely that genomic factors other than DNA sequence may also influence PcG recruitment.
In summary, we have demonstrated that the PcG machinery and HDAC activity are specifically engaged at the human α-globin genes (but not the β-genes) to maintain transcriptional repression in nonerythroid cells, and that this repression is imposed in pluripotent stem cells. We propose that the PcG pathway may be particularly important in genomic contexts in which a tissue-specific gene is marked by a CpG island. In many cases, such genes are located in gene-dense regions and are surrounded by constitutively expressed genes in open chromatin domains. In these genomic contexts, where widespread repression may not be possible, we propose that repression instead often relies on the discrete recruitment of PcG proteins to specific promoters.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jim Hughes for advice and Bill Wood and Richard Gibbons for helpful discussions on the manuscript.
This work was supported by the Medical Research Council (United Kingdom).
Authorship
Contribution: D.G. and D.R.H. designed the research; D.G., M.D.G., V.S., M.R., M.H., H.A., K.L., and J.S.-S. performed the research; C.K. and I.D. provided vital new reagents; D.G., N.G., and D.R.H. analyzed and interpreted the data; and D.G. and D.R.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Doug Higgs, MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, Oxford University, Oxford, OX3 9DE, United Kingdom; e-mail: doug.higgs@imm.ox.ac.uk.

![Figure 2. The PRC2 polycomb complex is recruited to the inactive HBA genes. ChIP experiments were carried out in primary T lymphocytes or primary proerythroblasts using antibodies against H3K27me3 (A), SUZ12 (B), or RNA pol II (C). For (A) and (B), immunoprecipitated and input material were labeled with Cy3 or Cy5, respectively, and applied to a custom tiled microarray covering the terminal region of human chromosome 16. Shown is a 250-kb genomic segment containing the α-globin locus (delineated by gray dashed lines) with the fold enrichments at each probe plotted against chromosomal position. The locus is annotated as described in Figure 1. For panel C, immunoprecipitated material was analyzed by real-time PCR at amplicons 53 (within the middle of the DIST1 gene [see Figure 1]) and an amplicon within the 18S rRNA gene (both representing background enrichment), an amplicon at the promoter of the β-actin gene (ACTB) used as a positive control, and an amplicon spanning exons 1-2 of the adult HBA genes (HBA ex1-2; see Figure 1). Data indicate the percentage of input material precipitated at each amplicon. Shown are the mean values obtained from replicate ChIPs with error bars indicating the range of values.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/9/10.1182_blood-2008-06-161901/5/m_zh80200826010002.jpeg?Expires=1766012555&Signature=4Mo~-rHwiBJVytXV5~t291F00~Lbid4zU2kQ~tWTEtX7pDwvv21uPooiT~ob2yYHV1L707-YK80yq9mYpNA-9NLGg6W5YVTO4OsV~dRiuYNQi1~TD1VV-BcGNy5jCJBWfFrdd7tQvqeMTSC5ChOy48-8ltD6fJkQQtLLCJmc56ufIthaPqSyhGCtQj3p9~rYpoXVvh9Qm7Y3d60PpigpX4IVfrAzJNlLlzMXPBTm97WcfgASqyJC3P2QaV9GvrhJRhLOvaS0KUaW68C7kG3IcAegFdehxXrXVsth8b6J1x7kRDtYgQwYKxn6Q4SmfBgn8du6e4PrR657Zv7s40VaMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal