The CD40-CD154 dyad seems to play a prominent role fostering the immune-inflammatory response triggered by endothelial cell (EC)–T-cell communication. To delineate comprehensively the involvement of CD40 (TNFRSF5) in EC activation, we combined RNAi-mediated CD40 knockdown with comparative genome-wide transcriptional profiling of ECs interacting with (CD154+) T cells. We report the initiation of a profound stress response in ECs upon CD40-CD154 engagement through early up-regulation of, among others, the major proinflammatory NF-κB and MAPK/SAPK pathways and their associated transcription factors. Moreover, we have identified novel genes regulated through the CD40-CD154 interaction, and pathways previously unrecognized to be induced by CD40 signaling in ECs. Thus, we document a significant down-regulation of endothelial APLN by CD40-CD154 interaction, TNFα/IFNγ exposure, and in immune-inflammatory pathologies, which could lead to hemodynamic dysfunction. Conversely, CD40-mediated up-regulation of the viral immune surveillance system, notably TLR3, IFIH1, RIG-I, and RNASEL, establishes a reverse link from adaptive to innate immunity in ECs. Moreover, systematic enrichment analysis substantiates endothelial CD40 involvement in the transcriptional regulation of gene networks associated with adhesion and motility, immunity, cell fate control, hemostasis, and metabolism. Our study also highlights the anti-inflammatory potential of RNAi-mediated CD40 inhibition, and the relevance of CD40 signaling for therapeutic intervention.
Introduction
It has been postulated that CD40, the tumor necrosis factor receptor superfamily member 5 (TNFRSF5), when interacting with its cognate ligand CD154 (CD40L; TNFSF5), plays a prominent role in the development of immune-inflammatory conditions such as cardiovascular disorders, autoimmune diseases, and organ rejection.1
Indeed, CD154 has a restricted pattern of expression, being up-regulated mainly in activated CD4+ T cells and platelets. In contrast, CD40 is expressed at relatively low abundance in most resting cell types, showing the highest levels in B cells and dendritic cells, where it contributes to their maturation.2,3 For example, in B cells, where it was first discovered and has been more thoroughly characterized, CD40 engagement triggers the binding of different members of the family of tumor necrosis factor receptor-associated factors (TRAFs) to its intracellular domain, mediating activation of multiple signaling pathways that regulate B-cell survival, proliferation, and differentiation; immunoglobulin isotype switching; development of the germinal center; and the humoral memory response.4
In ECs, the CD40-CD154 interaction triggers proinflammatory cytokine and chemokine production, matrix metalloproteinase, and tissue factor expression, procoagulant activity, angiogenesis, and a hallmark increase in expression of the leukocyte adhesion molecules E-selectin (SELE, CD62E), vascular cell adhesion molecule 1 (VCAM1, CD106), and intercellular adhesion molecule 1 (ICAM1, CD54).5 All these events modulate an essential step in the immune-inflammatory process: leukocyte recruitment, adhesion, rolling, and extravasation to the injured or stressed tissue. Thus, interfering CD40-CD154 interaction represents a suitable therapeutic option. This has been demonstrated using blocking antibodies against CD154 in a variety of animal models of immune-inflammatory disorders and several clinical trials, although severe thromboembolic events have aroused in some of these studies.6,–8 Alternatively, we have recently shown that genetic knockdown of CD40 expression through RNAi prevents EC adhesion molecule expression and leukocyte adhesion on ECs.9 Nevertheless, our understanding of the CD40-CD154 action is not yet complete to predict the full outcome and side effects of such therapeutic approaches because CD40 has a remarkable variety of functions in different cell types. Thus, much remains to be learned about the molecular pathways that operate in ECs stimulated by interaction with CD154 displayed on the surface of activated T lymphocytes.
In this report, we combine RNAi-mediated CD40 gene knockdown and genome-wide transcriptional profiling to comprehensively delineate the molecular signature of CD40 in ECs triggered by CD154+ T lymphocytes. This approach may be exploited to (1) study the temporal expression pattern of genes regulated by CD40 signaling, (2) identify novel genes and/or pathways not previously associated with CD40 action on ECs, and (3) establish candidate targets, involved in immune-inflammatory processes through the CD40 pathway, for diagnosis/prognosis or potential therapeutic intervention.
Accordingly, our results indicate that CD40 signaling leads to robust down-regulation of the vasoactive peptide apelin by ECs, and suggest that the altered vascular homeostasis elicited by the loss of this cytokine under proinflammatory stimuli may contribute to endothelial dysfunction. Conversely, we propose a key role for CD40 triggering antiviral surveillance in ECs when challenged with activated T cells. Furthermore, gene set enrichment analysis of the dynamically regulated genes identifies sequential genetic programs characterizing CD40-induced endothelial activation.
Methods
Cell culture and siRNA transfection
Primary human umbilical vein ECs (HUVECs) representing a pool of at least 3 different umbilical cord veins (Advancell, Barcelona, Spain) were maintained in endothelial growth medium EGM-Bullet kit (Clonetics-Cambrex, Walkersville, MD). Third-passage primary ECs isolated from normal human carotid arteries (HCtAECs) were cultured in EC Growth Medium II (Cell Applications, San Diego, CA). All endothelial cultures were grown at 37°C in a 5% CO2 atmosphere. The siRNA duplex targeting human CD40 (siRNA-2) and its corresponding mismatched siRNA control (msiRNA-2) were chemically synthesized (QIAGEN, Hilden, Germany). Specific sequences and characterization of their silencing effect in HUVECs have already been described.9 Incorporation of the siRNAs into exponentially growing HUVECs (passages 3-4) was achieved by electroporation with an ECM 830 Electropulse Generator System (BTX, San Diego, CA). For details, see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
siRNA-transfected HUVECs were stimulated by coincubation with Jurkat D1.1 (CD154+) cells10 (ATCC, Manassas, VA; CRL-10915, 1:10 ratio) at 4, 10, or 16 hours prior to analysis. At 48 hours after transfection, cocultures were extensively washed with PBS to release the attached Jurkat D1.1 cells from the HUVEC monolayer. Jurkat D1.1 stimulation of HCtAECs was performed analogously to that of HUVECs. Residual Jurkat D1.1 contamination of HUVEC and HCtAEC monolayers was estimated to be less than 0.5% by flow cytometry (data not shown).
Gene expression profiling
The RNA samples from treated HUVECs were extracted using the RNeasy RNA Isolation kit (Qiagen) and incubated with RNase-free DNase I (Ambion, Austin, TX) according to the manufacturer's protocol. RNA quantification and quality assessment were performed as described in Document S1.
RNA samples (500 ng total RNA) were reverse transcribed, amplified, labeled by in vitro transcription using the Low Input Linear Amplification Kit (5184-3523; Agilent, Wilmington, DE), and hybridized following the manufacturer's instructions. The whole-genome oligonucleotide microarrays used (G4112A; Agilent) contain 44 290 spots with 60-mer probes, 41 675 of which represent in single or multiple copies a total of 37 312 human transcripts.
Two biologic replicate experiments were performed, each comparing siRNA-2–transfected with msiRNA-2–transfected HUVECs. Each experimental pair of labeled samples was cohybridized on 2 separate microarrays with dye swapping to correct for dye bias effects. Thus, 4 microarray hybridizations were processed for each of 3 EC-D1.1 Jurkat cell coincubation time points, totaling 12 array data sets. Additional details on scanning and data analysis can be found in Document S1. Differentially expressed genes were chosen, unless otherwise stated, using as cutoff criteria a Bayesian statistic percentile (B rank) above 95% and absolute fold change (FC) greater than 1.2 for at least 1 of the 3 time points. The entire dataset for all microarray experiments has been deposited in NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and is accessible through GEO Series accession number GSE10601.11
Microarray data analysis
Significance analysis of microarrays (SAM) and gene ontology (GO) functional classification were carried out using the TMEV software (TIGR Multiple Experiment Viewer; The Institute for Genomic Research, Rockville, MD; http://www.tm4.org/mev.html).12 Gene set enrichment analysis was performed with the GSEA software developed at the Massachusetts Institute of Technology (MIT, Cambridge, MA)13 using the functional set C2, which includes 522 gene sets corresponding to specific metabolic and signaling pathways from the Molecular Signature Database MSigDB 1.0.14 Either the expression intensity values or the log2 ratios from the whole-genome unfiltered microarray data were included to assess biologic functions or processes differentially regulated. We compared differences between msiRNA-2 and siRNA-2 treatment, or between 2 given stimulation times. The permutation number was fixed at 1000, and the significance at false discovery rate q-value (FDR q-val) was less than 0.25.
Quantitative real-time RT-PCR
Confirmation of regulation was achieved by quantitative real-time reverse transcription–polymerase chain reaction (RT-qPCR) on selected genes using SYBR Green and the Lightcycler technology (Roche Molecular Biochemicals, Indianapolis, IN). Reverse transcription reactions were performed with 2 μg total RNA using the Omniscript RT kit (Qiagen). Additional PCR amplification information is given in Document S1. Gene-specific primer pairs (Table S1) were designed using Oligo 4.0 software (MBI, Cascade, CO) and selected to prevent primer-dimer formation.
Results were calculated as the normalized mRNA level ratio of each gene over the housekeeping genes cyclophilin-A (CYPA) or 18S ribosomal RNA (LOC100008588), as indicated.
Northern blot analysis
For Northern blotting, 20 μg total RNA extracted from untreated or siRNA-treated HUVECs further stimulated with the proinflammatory mediators (Jurkat D1.1 [CD154+] cells, or 100 U/mL TNFα + 1000 U/mL IFNγ) was electrophoresed on 1.2% formaldehyde-agarose gels, transferred to a Hybond XL nylon membrane (Amersham Biosciences, Arlington Heights, IL), and immobilized by cross-linking with UV light (Stratalinker; Stratagene, La Jolla, CA). APLN-specific signals were detected using probes F3-R3 (244 bp) and F4-R4 (295 bp; Figure 6A, Document S1). The membranes were stripped and rehybridized using a control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (Clontech BD Biosciences, Palo Alto, CA).
Western blot analysis
Immunodetection of CD40, apelin, and IFNβ was carried out on the corresponding cell extracts in radioimmunoprecipitation assay (RIPA) buffer as previously described.9 Total protein content of the supernatants was measured using the BCA protein assay (Pierce, Rockford, IL). Total protein (40 μg from each sample) was separated by 10% SDS–polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to Hybond membranes (Amersham Biosciences). Human CD40 expression was assessed with a mouse monoclonal antibody (H-10,1:1000; Santa Cruz Biotechnology, Santa Cruz, CA). Intracellular levels of apelin were determined with a goat polyclonal antibody (S-20, 1:200; Santa Cruz Biotechnology). Intracellular IFNβ was measured with a rabbit antihuman IFNβ polyclonal antibody (ab 9662; 1:1000; Abcam, Cambridge, MA). The corresponding anti-IgG HRP-conjugated secondary antibodies (Dako, Glostrup, Denmark) were used for enhanced chemiluminescence detection (ECL + Plus; Amersham Biosciences).
Membranes were reprobed with a rabbit polyclonal antiactin antibody (1:2000; Sigma-Aldrich, St Louis, MO) for normalization.
MAPK phosphorylation
siRNA-transfected or untransfected HUVECs (2 × 106 cells) were cocultured with Jurkat D1.1 cells for 4, 10, and 16 hours prior to analysis. To investigate phosphorylation of JNK, p38, and ERK1/2 MAPKs, cells were recovered at 48 hours after transfection, washed with ice-cold PBS, and scraped into ice-cold lysis buffer (20 mM Tris-HCl pH 8.0; 150 mM NaCl; 1% Triton X-100; 10 mM NaF; 40 mM β-glycerolphosphate; 2 mM Na3VO4) supplemented with Complete Protease Inhibitor Cocktail (Roche Molecular Biochemicals). After incubation for 15 minutes on ice, samples were centrifuged at 16 000g for 15 minutes at 4°C, and supernatants were collected and processed as described in the previous section.
MAPK-specific phospho- and total anti–human antibodies (Cell Signaling, Beverly, MA) were used for probing Western blots. The Phospho-MAPK Family Antibody Sampler kit contains the rabbit polyclonal primary antibodies Phospho-p44/42 MAPK (Thr202/Tyr204), Phospho-SAPK/JNK (Thr183/Tyr185), and Phospho-p38 MAPK (Thr180/Tyr182). The MAPK Family Antibody Sampler kit contains the rabbit p44/42 MAPK (137F5) and SAPK/JNK (56G8) monoclonal antibodies and the rabbit p38 MAPK polyclonal antibody.
Nuclear extractions and transcription factor activity assays
siRNA-transfected HUVECs were cocultured with Jurkat D1.1 cells for 4 hours prior to analysis, at 48 hours after transfection. Nuclear extracts were prepared using the Transfactor Extraction Kit (Clontech BD Biosciences) according to the manufacturer's instructions. Protein concentration was determined with a Bradford-based assay (Bio-Rad Laboratories, Marnes-la-Coquette, France).
Detection of transcription factor activity was performed with 2 enzyme-linked immunosorbent assay (ELISA)–based colorimetric assays according to the supplier's protocol: the BD Mercury Transfactor Profiling Kit Inflammation 1 (Clontech BD Biosciences) for transcription factors from the NF-κB family, and the TransAM AP-1 and MAPK family kits (ActiveMotif Europe, Rixensart, Belgium) for AP-1– and MAPK-dependent transcription factor families.
Human samples
Carotid arteries (n = 5) were obtained as vascular transplants from organ donors or postmortem autopsies. Carotid plaques were classified according to the American Heart Association (AHA; Dallas, TX) criteria with some modifications.15 The study was approved by the local ethical committee, Comité Ético de Investigación Clínica, from the Bellvitge University Hospital, in accordance with institutional guidelines and with written informed consent obtained in accordance with the Declaration of Helsinki.
Experimental renal transplantation
Syngeneic renal transplantations between Wistar Agouti rats (n = 3) and allogeneic grafts using Wistar-Agouti rats as recipients of Brown-Norway kidneys (n = 6) were performed as previously described.16 The animals were maintained in accordance with the guidelines of the Committee on Care and Use of Laboratory Animals and Good Laboratory Practice from the Bellvitge Animal Care Facility (Barcelona, Spain).
Immunohistochemistry
Routine immunohistochemistry was performed on paraffin-processed sections (Document S1) from human carotid samples using the rabbit polyclonal antibodies anti–apelin-36 (human, 1:200; Phoenix Pharmaceuticals, Belmont, CA) and anti-CD40 (C-20, 1:200; Santa Cruz Biotechnology), and from rat kidneys using the rabbit polyclonal antibodies anti–apelin-36 (rat, mouse, 1:200; Phoenix Pharmaceuticals) and anti–PECAM-1 (M-20, 1:200; Santa Cruz Biotechnology).
Results
To infer the physiologic role of CD40 in the context of T-lymphocyte–EC interaction, we sought to compare global transcriptional profiles from Jurkat D1.1 (CD154+)–treated HUVECs, in which CD40 expression was either knocked down (by siRNA-2) or unaffected (by msiRNA-2).9 Moreover, to gain insight into the dynamics of CD40-mediated signal transduction in ECs, we extended our comparative transcriptomic analysis to 4, 10, and 16 hours of CD40-CD154 interaction using this cell-cell context.
As controls to assess the efficiency of CD40 gene silencing and the quality of the RNA prior to microarray processing, we quantified RNA levels of both CD40 and ICAM1, known to be up-regulated by CD40-CD154 interaction. Quantitative RT-PCR (CD40 and ICAM1) and Western immunoblotting (CD40) confirmed efficient silencing of both the costimulatory receptor as the primary target and the cell adhesion molecule as a consequence of CD40 signal transduction inhibition (Figure S1).
Microarray data mining and functional classification of CD40 responsive genes in endothelial cells
We compared gene expression profiles from CD154+ T cell–stimulated ECs with or without RNAi-mediated CD40 silencing. We considered genes as regulated when there was both B rank more than 95% and FC 1.2 or more in at least 1 of the 3 stimulation times (4, 10, and 16 hours). The analysis identified 715 transcripts (∼ 2%) that were differentially regulated on the whole genome microarrays (3% of the HUVEC transcriptome). From the regulated genes, nearly 75% were found up-regulated, whereas 25% were found down-regulated (Table S2). Moreover, the magnitude of differential gene regulation was maximal at 4 hours. This suggests an early massive response of the EC upon CD40-CD154 signaling and confirms the general role of CD40 as an EC “activator.”17
To explore the biologic functions affected by CD40 signaling in ECs, we performed a GO overrepresentation analysis from the list of regulated genes with annotated functions. As a consequence of CD40-CD154 engagement, most of the regulated genes belong to categories directly or indirectly related to the immune-inflammatory process (Figure S2). Moreover, GO analysis confirms known key aspects of CD40-related EC activation early in the inflammatory process, such as the involvement of the NF-κB signaling, and the induction of chemotactic signals allowing cross-talk with the circulating leukocytes.1
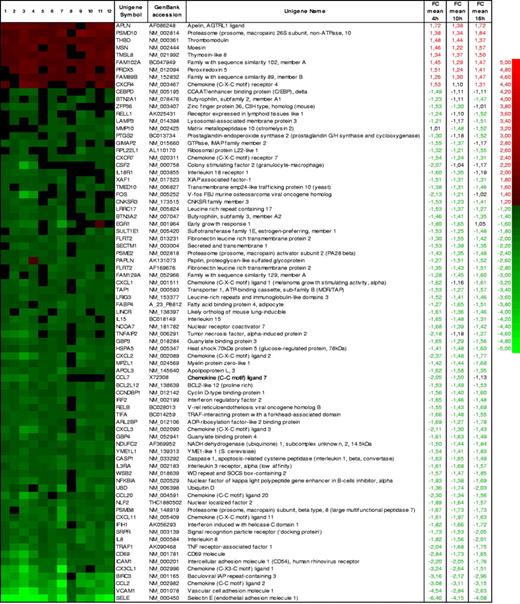
A short list of up- and down-regulated annotated genes with a B rank more than 99% and FC 1.5 or more in at least 1 of the 3 stimulation times is shown in Figure 1. The full list of differentially expressed transcripts is accessible online (Table S2). We arranged representative annotated genes into selected functional classes according to their putative activities (Figure 2). Some of these genes had been already described as targets of CD40 signaling, although we also identified many others previously unrecognized to be related to CD40 action in ECs (see Document S1, “Functional classification,” for details).
Expression profile of selected CD40-regulated genes upon EC stimulation with Jurkat D1.1 (CD154+) T cells. Heat map and the corresponding list of up-regulated (green) and down-regulated (red) genes fulfilling criteria of B rank 99 or more and mean FC 1.5 or more in at least 1 of the 3 stimulation times analyzed: 4 hours (columns 1-4), 10 hours (columns 5-8), and 16 hours (columns 9-12). Rows representing individual genes are ordered according to the mean FC across the 3 stimulation times. Unigene symbols18 and GenBank accession numbers19 were used.
Expression profile of selected CD40-regulated genes upon EC stimulation with Jurkat D1.1 (CD154+) T cells. Heat map and the corresponding list of up-regulated (green) and down-regulated (red) genes fulfilling criteria of B rank 99 or more and mean FC 1.5 or more in at least 1 of the 3 stimulation times analyzed: 4 hours (columns 1-4), 10 hours (columns 5-8), and 16 hours (columns 9-12). Rows representing individual genes are ordered according to the mean FC across the 3 stimulation times. Unigene symbols18 and GenBank accession numbers19 were used.
Representative EC gene expression patterns from CD40 signaling upon HUVEC/CD154+ T-cell interaction. Up-regulated (left flank) and down-regulated (right flank) genes, and their corresponding mean fold changes, at 4 hours (▭) and 16 hours (▬) after Jurkat D1.1 stimulation. Genes are arranged according to their related molecular and cellular functions. The underlined genes have been validated by RT-qPCR (see Figure 3 for details). Columns marked with an asterisk correspond to genes found significantly and differentially regulated between 4 and 16 hours by SAM analysis (FDR q-val = 0). Genes previously known to be induced by CD40 in ECs are highlighted in bold.
Representative EC gene expression patterns from CD40 signaling upon HUVEC/CD154+ T-cell interaction. Up-regulated (left flank) and down-regulated (right flank) genes, and their corresponding mean fold changes, at 4 hours (▭) and 16 hours (▬) after Jurkat D1.1 stimulation. Genes are arranged according to their related molecular and cellular functions. The underlined genes have been validated by RT-qPCR (see Figure 3 for details). Columns marked with an asterisk correspond to genes found significantly and differentially regulated between 4 and 16 hours by SAM analysis (FDR q-val = 0). Genes previously known to be induced by CD40 in ECs are highlighted in bold.
Validation of CD40-dependent endothelial cell activation
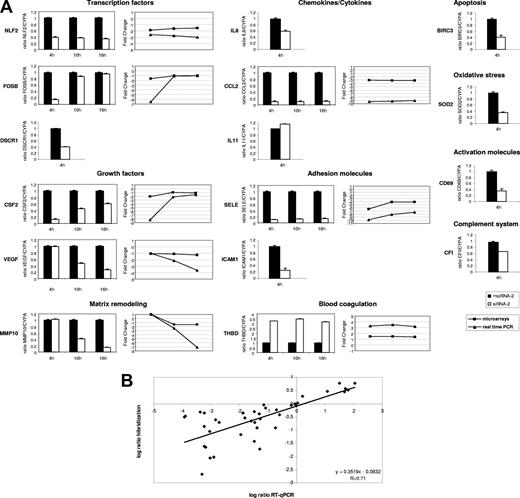
To confirm the array results, we performed RT-qPCR of a set of 15 representative genes at 4 hours after stimulation, and from 8 of them we also evaluated their behavior at 10 hours and 16 hours after stimulation (Figure 3A). Among the up-regulated genes we selected chemokines (CCL2, IL8), transcription factors (FOSB, NLF2 [and also NLF1; data not shown]), growth factors (CSF2, VEGF), the adhesion molecule SELE, the antiapoptotic BIRC3, the antioxidant SOD2, the metalloprotease MMP10, the regulator of calcineurin DSCR1, the complement factor CFI, and the T-cell activation marker CD69. The down-regulated genes included the anticoagulant THBD and the cytokine IL11. Four of these genes (SOD2, DSCR1, IF, and IL11) showed small FC values ranging 1.2 to 1.3. Our RT-qPCR validation results are in full agreement with the microarray data, both in terms of up- and down-regulated genes and in terms of early (FOSB, CSF2) and late (MMP10, VEGF) regulated ones (Figure 3A). Overall, we found correlation between the magnitude of change as measured by microarray hybridization and RT-qPCR (Figure 3B), confirming previous observations20 and suggesting that microarray-derived log2 ratio, though attenuated in high-density microarrays,21 accurately reflects gene expression changes.
Validation of microarray results by RT-qPCR. (A) Relative mRNA levels corresponding to representative genes from msiRNA-2–treated (▬) and siRNA-2–treated (▭) HUVECs stimulated with (CD154+) T cells for 4, 10, and 16 hours were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. In those genes validated at the 3 stimulation times, the comparative expression kinetics between microarrays (■) and RT-qPCR (▲) in terms of mean fold change is shown. (B) Dot-plot of the expression values in log2 ratio between microarray hybridization (ordinate) and RT-qPCR (abscissa) from all validated genes irrespective of the stimulation time used. Linear regression demonstrates a significant correlation.
Validation of microarray results by RT-qPCR. (A) Relative mRNA levels corresponding to representative genes from msiRNA-2–treated (▬) and siRNA-2–treated (▭) HUVECs stimulated with (CD154+) T cells for 4, 10, and 16 hours were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. In those genes validated at the 3 stimulation times, the comparative expression kinetics between microarrays (■) and RT-qPCR (▲) in terms of mean fold change is shown. (B) Dot-plot of the expression values in log2 ratio between microarray hybridization (ordinate) and RT-qPCR (abscissa) from all validated genes irrespective of the stimulation time used. Linear regression demonstrates a significant correlation.
T cell–stimulated endothelial CD40 activates NF-κB and MAPK family transcription factors
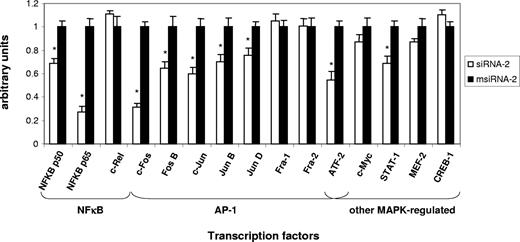
To assess the functional implication of NF-κB and additional transcription factors in the early response to CD40-mediated EC induction, we monitored their activity in nuclear extracts from Jurkat D1.1 (CD154+)–stimulated ECs pretreated with either siRNA-2 or msiRNA-2 at 4 hours after stimulation. Of 15 different transcription factors analyzed, significant differential DNA-binding activity could be monitored for the NF-κB family members NF-κB p50 and NF-κB p65, but not for c-Rel; for the AP-1 family members c-Fos, c-Jun, FosB, JunB, and JunD, but not for Fra-1 and Fra-2; and for alternative MAPK-regulated factors such as ATF-2 and STAT-1, but not for CREB-1 (Figure 4). Thus, several important transcription factor gene families such as NF-κB, AP-1, and STAT operating simultaneously appear to be essential modulators of the early immune-inflammatory process in ECs through the CD40-CD154 dyad.
Transcription factor DNA-binding assays confirm that CD40-dependent EC activation by Jurkat D1.1 (CD154+) T cells is coordinated through NF-κB, AP-1, and other MAPK-regulated activities. Nuclear extracts from HUVECs transfected with siRNA-2 (▭) or msiRNA-2 (▬) and stimulated with Jurkat D1.1 for 4 hours were analyzed through an ELISA-based assay (see “Nuclear extractions and transcription factor activity assays” for details). The histogram represents the relative DNA-binding levels of NF-κB (NF-κB p50, NF-κB p65, c-Rel), AP-1 (c-Jun, c-Fos, FosB, JunB, JunD, Fra-1, Fra-2), and several other MAPK-regulated (CREB-1, ATF-2, c-Myc, STAT-1, Mef-2) transcription factors. Data represent the average plus or minus SEM from 3 different experiments (*P < .05, vs msiRNA-2).
Transcription factor DNA-binding assays confirm that CD40-dependent EC activation by Jurkat D1.1 (CD154+) T cells is coordinated through NF-κB, AP-1, and other MAPK-regulated activities. Nuclear extracts from HUVECs transfected with siRNA-2 (▭) or msiRNA-2 (▬) and stimulated with Jurkat D1.1 for 4 hours were analyzed through an ELISA-based assay (see “Nuclear extractions and transcription factor activity assays” for details). The histogram represents the relative DNA-binding levels of NF-κB (NF-κB p50, NF-κB p65, c-Rel), AP-1 (c-Jun, c-Fos, FosB, JunB, JunD, Fra-1, Fra-2), and several other MAPK-regulated (CREB-1, ATF-2, c-Myc, STAT-1, Mef-2) transcription factors. Data represent the average plus or minus SEM from 3 different experiments (*P < .05, vs msiRNA-2).
MAPK functional validation
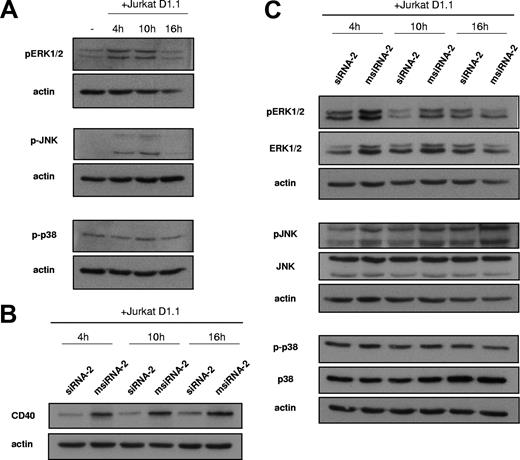
Engagement of the CD40 receptor on ECs through specific TRAF adaptor molecules leads to the induction of different proinflammatory signaling cascades.5 In agreement with previous studies, major signal transduction pathways operating in ECs by CD154 stimulation include those of the mitogen-activated protein kinase (MAPK) subfamily. ERK1/2, JNK, and p38 are responsible for the phosphorylation of many of the active transcription factors identified in the previous DNA-binding assays, including c-Fos, c-Jun, ATF-2, and STAT-1. We wished to assess the activation status of these pathways in ECs after coincubation for 4, 10, and 16 hours with Jurkat D1.1 (CD154+) cells. Thus, upon Jurkat D1.1 stimulation, ECs presented induced phosphorylation of both ERK1/2 and JNK at 4 and 10 hours, whereas at 16 hours these kinases seemed to return to basal phosphorylation levels. In contrast, p38 appeared to be constitutively activated regardless of Jurkat D1.1 stimulation (Figure 5A). To confirm that MAPK induction was indeed CD40 dependent, we assessed differential phosphorylation of ERK1/2, JNK, and p38 in protein extracts from either siRNA-2– or msiRNA-2–pretreated ECs. RNAi-mediated silencing of CD40 expression on treated cells paralleled inhibition of both total and phosphorylated ERK1/2 at 4 and 10 hours. JNK was also differentially phosphorylated, although total JNK remained constant throughout the stimulation period. In contrast, no differential regulation was apparent on either the total or the phosphorylated forms of p38 MAPK (Figure 5B-C).
CD40-dependent EC activation by Jurkat D1.1 (CD154+) T cells induces MAPK and SAPK signaling pathways. (A) HUVECs were either left untreated (lane 1) or coincubated with Jurkat D1.1 (CD154+) cells for the indicated times (lanes 2-4). Cell extracts (40 μg total protein/sample) were separated by 10% SDS-PAGE, subjected to Western blotting, and reacted with anti–phospho ERK1/2, anti–phospho JNK, or anti–phospho p38 antibodies. (B,C) HUVECs were transfected with siRNA-2 or msiRNA-2 and further stimulated with Jurkat D1.1 (CD154+) cells for the indicated times. Western blots were performed as in panel A, and probed with anti-CD40 MoAb (B), or with antibodies detecting either total or phosphorylated ERK1/2, JNK, and p38 isoforms (C). The bottom bands of the panels indicate β-actin protein levels used to normalize for equal loading of the gel lanes. All blots shown are representative from 3 different experiments.
CD40-dependent EC activation by Jurkat D1.1 (CD154+) T cells induces MAPK and SAPK signaling pathways. (A) HUVECs were either left untreated (lane 1) or coincubated with Jurkat D1.1 (CD154+) cells for the indicated times (lanes 2-4). Cell extracts (40 μg total protein/sample) were separated by 10% SDS-PAGE, subjected to Western blotting, and reacted with anti–phospho ERK1/2, anti–phospho JNK, or anti–phospho p38 antibodies. (B,C) HUVECs were transfected with siRNA-2 or msiRNA-2 and further stimulated with Jurkat D1.1 (CD154+) cells for the indicated times. Western blots were performed as in panel A, and probed with anti-CD40 MoAb (B), or with antibodies detecting either total or phosphorylated ERK1/2, JNK, and p38 isoforms (C). The bottom bands of the panels indicate β-actin protein levels used to normalize for equal loading of the gel lanes. All blots shown are representative from 3 different experiments.
The CD40-CD154 dyad and other proinflammatory stimuli mediate apelin down-regulation in endothelial cells
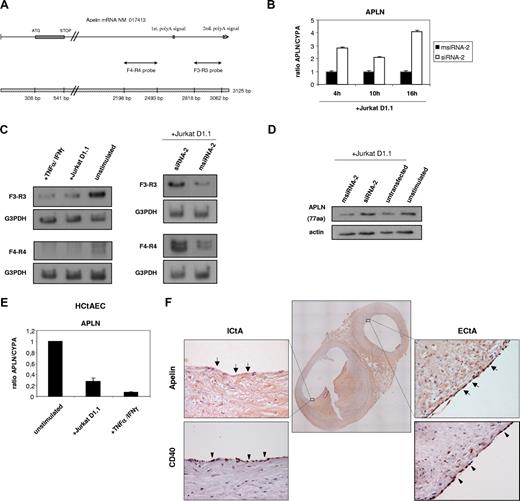
The most down-regulated gene as a consequence of CD40 signaling through EC–T-lymphocyte interaction was the AGTRL1 ligand gene APLN, coding for apelin (Figure 1). Among the multiple physiologic functions that are being attributed to this bioactive peptide, it has been suggested to have an important role in the regulation of cardiovascular function and fluid homeostasis.22 Further validation by RT-qPCR confirmed APLN down-regulation across the 3 stimulation times (Figure 6B). The APLN gene includes 2 polyadenylation signals at the 3′UTR leading to 2 alternative transcripts of 2.7 and 3.1 kb (Figure 6A). Northern analysis using 2 discriminating probes showed identical CD40-mediated down-regulation of both APLN mRNA species in ECs (Figure 6C). Interestingly, not only CD40-CD154–mediated cell-cell interactions, but also other proinflammatory soluble factors, such as TNFα plus IFNγ, were able to significantly down-regulate APLN transcript levels. Moreover, Western analysis revealed that the 77 amino acid prepropeptide apelin is repressed upon CD40 signaling in HUVECs stimulated by Jurkat D1.1 (CD154+) T cells (Figure 6D). Conversely, arterial ECs from different sources (eg, HCtAECs [Figure 6E] and human coronary artery endothelial cells [HCAECs; not shown]) demonstrated also transcriptional down-regulation of APLN upon proinflammatory stimulation. Furthermore, in agreement with previous reports,23,24 we found strong presence of apelin in ECs lining the luminal surface of normal-looking big arteries (internal and external carotid arteries and coronary arteries). Carotid arteries with type I and II atheroma lesions also presented significant staining for apelin (Figure 6F, ECtA). In contrast, carotids (Figure 6F, ICtA) and coronary arteries (not shown) with type III to VI atheromas had weak or absent staining for apelin in the intraluminal endothelium. Thus, in concordance with the decreased APLN expression observed in HCtAECs challenged by proinflammatory stimuli, there was an inverse correlation between the degree of inflammation in the neointima from carotid arteries, and the apelin-36 peptide levels within ECs lining these large conduit vessels.
The endogenous vasoactive peptide apelin is strongly down-regulated through CD154 and other proinflammatory stimuli in ECs. (A) Schematic representation of the APLN full transcript (GenBank accession no. NM 017413) showing the coding sequence and the relative positions of the 2 polyadenylation sites within the 3′ noncoding sequence of the gene. The cDNA fragments (F3-R3 probe and F4-R4 probe) used to distinguish the alternative transcripts are also shown. (B) Relative APLN mRNA levels from msiRNA-2–treated (black columns) and siRNA-2–treated (white columns) HUVECs stimulated with Jurkat D1.1 (CD154+) cells for 4, 10, and 16 hours were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (C) Northern blot analysis of total RNA isolated from HUVECs either unstimulated, or stimulated with Jurkat D1.1 (CD154+) cells or with TNFα (100 U/mL) plus IFNγ (1000 U/mL) for 4 hours (left panels). Alternatively, HUVECs were treated with siRNA-2 or msiRNA-2 and further stimulated with Jurkat D1.1 (CD154+) T cells (right panels). All Northern blots were hybridized with the APLN-specific F3-R3 or F4-R4 probes as indicated, and a G3PDH probe was used for normalization. (D) At the protein level, cell extracts (40 μg of total protein/sample) from HUVECs either untransfected and unstimulated (lane 4), transfected with msiRNA-2, transfected with siRNA-2, or untransfected, and further stimulated for 4 hours with Jurkat D1.1 (CD154+) T cells (lanes 1, 2, and 3, respectively) were fractionated by 10% SDS-PAGE and probed with a 77–amino acid preproapelin-specific antibody. The housekeeping β-actin protein levels were assessed for lane normalization. (E) Relative APLN mRNA levels from HCtAECs either unstimulated, or stimulated with Jurkat D1.1 (CD154+) cells or with TNFα (100 U/mL) plus IFNγ (1000 U/mL) for 4 hours, were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (F) Apelin-36 immunostaining in human carotid arteries. A representative section from the carotid bifurcation showing the internal (ICtA) and external (ECtA) carotid arteries (central image: ×20). In the ICtA with advanced atheroma (type V lesion) there was almost absent apelin-36 immunostaining in the intraluminal ECs ( ). In the normal-looking ECtA, strong positive intraluminal EC immunostaining for apelin-36 was evident (
). In the normal-looking ECtA, strong positive intraluminal EC immunostaining for apelin-36 was evident ( ). Both ICtA and ECtA presented significant endothelial CD40 immunoreactivity (
). Both ICtA and ECtA presented significant endothelial CD40 immunoreactivity ( ). Peripheral images, ×400. The sections were computerized as color-encoded digitized images using a Nikon Eclipse 80i microscope equipped with a CFI Plan Fluor 4×/0.13 NA and 40×/0.75 NA air objectives, a Nikon DS-2Mv camera, and the DS-U2 image processing unit and analyzing software NIKON NIS-Elements version 3.21 (all Nikon, Melville, NY).
). Peripheral images, ×400. The sections were computerized as color-encoded digitized images using a Nikon Eclipse 80i microscope equipped with a CFI Plan Fluor 4×/0.13 NA and 40×/0.75 NA air objectives, a Nikon DS-2Mv camera, and the DS-U2 image processing unit and analyzing software NIKON NIS-Elements version 3.21 (all Nikon, Melville, NY).
The endogenous vasoactive peptide apelin is strongly down-regulated through CD154 and other proinflammatory stimuli in ECs. (A) Schematic representation of the APLN full transcript (GenBank accession no. NM 017413) showing the coding sequence and the relative positions of the 2 polyadenylation sites within the 3′ noncoding sequence of the gene. The cDNA fragments (F3-R3 probe and F4-R4 probe) used to distinguish the alternative transcripts are also shown. (B) Relative APLN mRNA levels from msiRNA-2–treated (black columns) and siRNA-2–treated (white columns) HUVECs stimulated with Jurkat D1.1 (CD154+) cells for 4, 10, and 16 hours were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (C) Northern blot analysis of total RNA isolated from HUVECs either unstimulated, or stimulated with Jurkat D1.1 (CD154+) cells or with TNFα (100 U/mL) plus IFNγ (1000 U/mL) for 4 hours (left panels). Alternatively, HUVECs were treated with siRNA-2 or msiRNA-2 and further stimulated with Jurkat D1.1 (CD154+) T cells (right panels). All Northern blots were hybridized with the APLN-specific F3-R3 or F4-R4 probes as indicated, and a G3PDH probe was used for normalization. (D) At the protein level, cell extracts (40 μg of total protein/sample) from HUVECs either untransfected and unstimulated (lane 4), transfected with msiRNA-2, transfected with siRNA-2, or untransfected, and further stimulated for 4 hours with Jurkat D1.1 (CD154+) T cells (lanes 1, 2, and 3, respectively) were fractionated by 10% SDS-PAGE and probed with a 77–amino acid preproapelin-specific antibody. The housekeeping β-actin protein levels were assessed for lane normalization. (E) Relative APLN mRNA levels from HCtAECs either unstimulated, or stimulated with Jurkat D1.1 (CD154+) cells or with TNFα (100 U/mL) plus IFNγ (1000 U/mL) for 4 hours, were assessed by real-time RT-qPCR using the housekeeping cyclophilin A gene (CYPA) as a control. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (F) Apelin-36 immunostaining in human carotid arteries. A representative section from the carotid bifurcation showing the internal (ICtA) and external (ECtA) carotid arteries (central image: ×20). In the ICtA with advanced atheroma (type V lesion) there was almost absent apelin-36 immunostaining in the intraluminal ECs ( ). In the normal-looking ECtA, strong positive intraluminal EC immunostaining for apelin-36 was evident (
). In the normal-looking ECtA, strong positive intraluminal EC immunostaining for apelin-36 was evident ( ). Both ICtA and ECtA presented significant endothelial CD40 immunoreactivity (
). Both ICtA and ECtA presented significant endothelial CD40 immunoreactivity ( ). Peripheral images, ×400. The sections were computerized as color-encoded digitized images using a Nikon Eclipse 80i microscope equipped with a CFI Plan Fluor 4×/0.13 NA and 40×/0.75 NA air objectives, a Nikon DS-2Mv camera, and the DS-U2 image processing unit and analyzing software NIKON NIS-Elements version 3.21 (all Nikon, Melville, NY).
). Peripheral images, ×400. The sections were computerized as color-encoded digitized images using a Nikon Eclipse 80i microscope equipped with a CFI Plan Fluor 4×/0.13 NA and 40×/0.75 NA air objectives, a Nikon DS-2Mv camera, and the DS-U2 image processing unit and analyzing software NIKON NIS-Elements version 3.21 (all Nikon, Melville, NY).
In the etiology of kidney acute rejection, allorecognition is the main factor responsible for immune-inflammatory response activation. Thus, we investigated apelin regulation in a renal transplant model involving vascular dysfunction. We have previously shown that renal insufficiency in this model involves strong CD40 up-regulation and a marked increase of the Th1 proinflammatory cytokines TNFα and IFNγ.16,25 In addition, we found significant transcriptional down-regulation of APLN, and a marked decrease of apelin immunoreactivity in the microvascular and tubular cells from the rejecting kidneys (Figure S3). Taken together, these data indicate that vascular activation induced by proinflammatory states is associated with repression of apelin expression.
CD40 engagement through interaction with activated (CD154+) T cells induces the viral innate immune surveillance system in endothelial cells
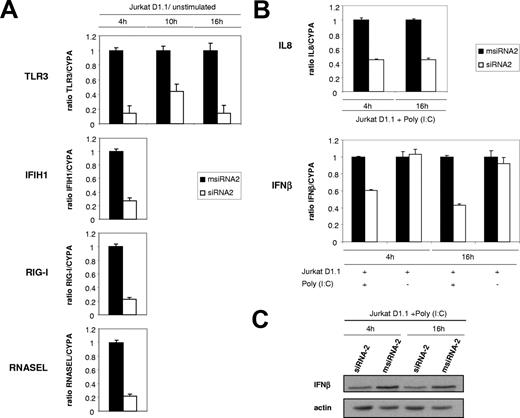
Our transcriptional profiling data suggested that T-lymphocyte–EC cross-talk through CD40 signaling alerts the EC viral innate immune surveillance system by up-regulating key sensors of viral infection: TLR3, recognizing extracellular dsRNA; the RNA helicases IFIH1 (MDA5) and RIG-I, sensing cytoplasmic dsRNA; the OAS/RNASEL axis, which mediates RNA degradation; and the IRF7 transcription factor, a common downstream regulator of the antiviral immune response. Moreover, important components of IFNγ action capable of stimulating innate cell-mediated immunity, such as both IFNγ receptor subunits (IFNGR1 and IFNGR2) and its signaling intermediate STAT1, were also found up-regulated upon CD40 engagement (Figure 2).
Again, RT-qPCR was used to validate CD40-dependent transcriptional up-regulation of the main arms of the EC innate response to dsRNA: TLR3, IFIH1, RIG-I, and RNASEL. To discriminate any potential sequence-dependent immunostimulatory effects caused by the introduction of our siRNAs into the EC, independent of CD40 action, we normalized the expression values obtained from Jurkat D1.1–stimulated HUVECs with respect to unstimulated HUVECs, both subject to the same siRNA treatment (Figure 7A).
CD40 engagement through CD154+ T cells induces the antiviral innate immune surveillance system in ECs. (A) Validation of CD40-dependent induction of key antiviral innate immune response genes by RT-qPCR. Relative mRNA levels corresponding to TLR3, IFIH1, RIG-I, and RNASEL from msiRNA-2–treated (■) and siRNA-2–treated (□) HUVECs stimulated with (CD154+) T cells for 4 hours (IFIH1, RIG-I, RNASEL), or 4, 10, and 16 hours (TLR3), were assessed by real-time RT-qPCR using CYPA as a control, and normalized from the corresponding values obtained from unstimulated, msiRNA-2–treated, or siRNA-2–treated HUVECs. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (B) Functional validation of the antiviral surveillance pathways induced by CD40 signaling in HUVECs. msiRNA-2–treated (■) and siRNA-2–treated HUVECs were stimulated with Jurkat D1.1 (CD154+) T cells for 4 or 16 hours and either analyzed or further challenged with poly (I:C) (25 μg/mL) for 6 hours and analyzed. Relative mRNA levels of the proinflammatory chemokine IL8 and the type I IFNβ were assessed by real-time RT-qPCR using the housekeeping CYPA as a control. Results are mean values plus or minus SEM from 3 independent experiments performed in triplicate. (C) The relative amount of synthesized IFNβ was also evaluated at the protein level by immunoblotting using an anti-IFNβ polyclonal antibody, in fractionated cell extracts from msiRNA-2–treated (lanes 2 and 4) and siRNA-2–treated (lanes 1 and 3) HUVECs stimulated with Jurkat D1.1 (CD154+) T cells for 4 and 16 hours, and further challenged with poly (I:C) (25 μg/mL) for 6 hours. β-actin protein levels were assessed for normalization. A representative blot from 3 independent experiments is shown.
CD40 engagement through CD154+ T cells induces the antiviral innate immune surveillance system in ECs. (A) Validation of CD40-dependent induction of key antiviral innate immune response genes by RT-qPCR. Relative mRNA levels corresponding to TLR3, IFIH1, RIG-I, and RNASEL from msiRNA-2–treated (■) and siRNA-2–treated (□) HUVECs stimulated with (CD154+) T cells for 4 hours (IFIH1, RIG-I, RNASEL), or 4, 10, and 16 hours (TLR3), were assessed by real-time RT-qPCR using CYPA as a control, and normalized from the corresponding values obtained from unstimulated, msiRNA-2–treated, or siRNA-2–treated HUVECs. Results are mean values plus or minus SEM from 2 independent experiments performed in triplicate. (B) Functional validation of the antiviral surveillance pathways induced by CD40 signaling in HUVECs. msiRNA-2–treated (■) and siRNA-2–treated HUVECs were stimulated with Jurkat D1.1 (CD154+) T cells for 4 or 16 hours and either analyzed or further challenged with poly (I:C) (25 μg/mL) for 6 hours and analyzed. Relative mRNA levels of the proinflammatory chemokine IL8 and the type I IFNβ were assessed by real-time RT-qPCR using the housekeeping CYPA as a control. Results are mean values plus or minus SEM from 3 independent experiments performed in triplicate. (C) The relative amount of synthesized IFNβ was also evaluated at the protein level by immunoblotting using an anti-IFNβ polyclonal antibody, in fractionated cell extracts from msiRNA-2–treated (lanes 2 and 4) and siRNA-2–treated (lanes 1 and 3) HUVECs stimulated with Jurkat D1.1 (CD154+) T cells for 4 and 16 hours, and further challenged with poly (I:C) (25 μg/mL) for 6 hours. β-actin protein levels were assessed for normalization. A representative blot from 3 independent experiments is shown.
Furthermore, we confirmed the functionality of the EC antiviral surveillance system by demonstrating CD40-dependent up-regulation of 2 downstream effectors of TLR3/MDA5/RIG-I signaling, IL8 and IFNβ, following dsRNA analog challenging. We measured both cytokines by RT-qPCR in ECs upon poly (I:C) treatment, at 4 and 16 hours after Jurkat D1.1 (CD154+) stimulation (Figure 7B). IFNβ was also up-regulated at the protein level (Figure 7C).
Toward a systematic integrated portrait of CD40 signaling in endothelial cells: gene set enrichment analysis
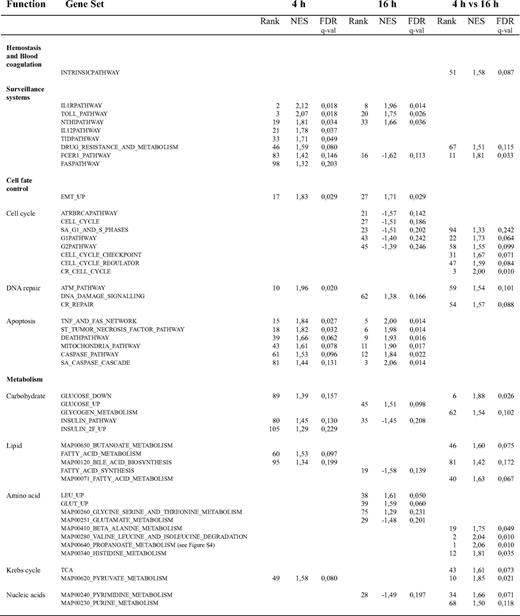
To gain additional insight into the signaling cascades and the molecular and cellular functions directly or indirectly influenced by CD40 in ECs as a consequence of their interaction with CD154-expressing T lymphocytes, we undertook a threshold-free gene set enrichment analysis (GSEA) of our RNAi-coupled transcriptional profiling data. The analysis was performed for each of the 3 stimulation times: 4, 10, and 16 hours. Moreover, to further investigate the dynamics of CD40-dependent EC activation, we also mined for gene sets differentially regulated between 4 and 16 hours of cell-associated CD154 stimulation.
Figure 8 displays a list of relevant, significantly regulated (FDR q-val < 0.25) functional gene sets: (1) at 4 hours after stimulation, (2) at 16 hours after stimulation, and (3) comparing the response between 4 and 16 hours after CD40-mediated EC activation. Overall, these results confirm the important role of the CD40 receptor as a mediator of EC activation upon interaction with activated T lymphocytes (Document S1, “GSEA”).
Representative gene sets induced upon CD40 signaling in ECs challenged by interaction with Jurkat D1.1 (CD154+) T cells. GSEA lists of selected induced gene sets (see “Microarray data analysis”) arranged according to their related molecular and cellular functions. Comparison of misRNA-2–treated versus siRNA-2–treated HUVEC transcriptional profiles at 4 hours and 16 hours after Jurkat D1.1 (CD154+) T-cell stimulation. A complementary list of CD40-dependent differentially regulated gene sets between 4 hours and 16 hours (4 h vs 16 h) is also given. For each functional gene set, its corresponding normalized enrichment score (NES) and FDR q-val is shown. Gene sets are ranked according to their FDR q-val. Positive NES values indicate up-regulated gene sets; negative NES values indicate down-regulated gene sets.
Representative gene sets induced upon CD40 signaling in ECs challenged by interaction with Jurkat D1.1 (CD154+) T cells. GSEA lists of selected induced gene sets (see “Microarray data analysis”) arranged according to their related molecular and cellular functions. Comparison of misRNA-2–treated versus siRNA-2–treated HUVEC transcriptional profiles at 4 hours and 16 hours after Jurkat D1.1 (CD154+) T-cell stimulation. A complementary list of CD40-dependent differentially regulated gene sets between 4 hours and 16 hours (4 h vs 16 h) is also given. For each functional gene set, its corresponding normalized enrichment score (NES) and FDR q-val is shown. Gene sets are ranked according to their FDR q-val. Positive NES values indicate up-regulated gene sets; negative NES values indicate down-regulated gene sets.
Discussion
In the present study, we report a key role of CD40 signaling in T cell–dependent transcriptional activation of ECs. Moreover, we further dissect the molecular pathways defining the physiologic behavior of endothelial CD40 in the immune-inflammatory process through genome-wide expression profiling at 3 different time points to obtain “snapshots” of the course of transcriptional alterations. We demonstrate that the activated T lymphocyte is a strong enough stimulus to induce EC activation by direct interaction through the CD40-CD154 dyad.
Both activated platelets and activated CD4+ T cells are the major source of CD154. However, it has been suggested that although CD154+ platelets contribute significantly to the recruitment of inflammatory cells to damaged endothelium in vivo, due to the short half-life of platelet CD154, the chronic CD154-driven inflammatory component can be sustained only by activated CD4+ T cells.26 Consequently, we undertook the study of the CD40-CD154 interaction using as experimental paradigm EC activated by T-lymphocyte coculture, to assess exclusively the relevance and consequences of CD40 engagement in ECs. This has been achieved by combining RNAi-mediated CD40 knockdown9 and comparative whole-genome microarrays. The gene silencing specificity of optimized siRNAs has been confirmed in several expression profiling studies,27,28 which allows confident investigation of gene function on genome-wide scale.29
CD40 signaling is a complex process where different pathways exert overlapping independent control of gene expression modules, as already reported in B lymphocytes30,–32 and dendritic cells.21 Our results corroborate the role of CD40 as an activator in ECs and infer a substantial perturbation of the EC transcriptome as a consequence of CD40 engagement. By gene ontology analysis, the main specific categories overrepresented, inflammatory and immune response, support the validity of our data. To further explore the biologic mechanisms behind CD40-mediated EC activation we have used GSEA.13 This knowledge-based high-content data interpretation tool has proven especially suitable to detect biologic processes affected by networks of genes and not apparent at the level of individual gene analysis.
According to our microarray results, within the first 4 hours of T cell–mediated, CD40-dependent activation of resting ECs, an engagement of the stress response program takes place in these cells, leading to the activation of multiple transcriptional programs. Our HUVEC profiling study may not fully reflect the heterogeneity of microvascular and macrovascular ECs from distinct body compartments.33,34 Nevertheless, the achievement of an EC-activated state through CD40 resembles the condition ensued when activated immune cells, inflammatory cytokines, and other acute-phase reactants interact with the vascular bed. This turns out in a prothrombotic, proadhesive, and proinflammatory phenotype typical of pathologic conditions related to the endothelium including sepsis, solid organ rejection, and atherosclerosis.35 Indeed, significant transcriptional profile similarities between ECs activated by CD154+ T cells and by inflammatory cytokines, particularly TNF-α and IL-1,36,,–39 support the proinflammatory effects of the CD40-CD154 dyad in ECs.
CD40 signaling through TRAF effectors regulates key transcription factors involved in the immune-inflammatory process such as NF-κB, AP-1, and NFAT families.40 Accordingly, we report the early induction of the NF-κB and AP-1 families, a common feature of EC activation through proinflammatory mediators. Our DNA binding results confirm CD40-mediated activation of the major proteins involved in the canonical pathway (p50, p65/RelA). Moreover, our microarray data also suggest engagement of the noncanonical pathway (through Rel B up-regulation), and perhaps of alternative pathways, such as that leading to a transcriptional module formed by a p50 homodimer interacting with the IkB-like coactivator Bcl341 (also induced by CD40). Similarly, the extensive early induction of central AP-1 components (c-Fos, FosB, c-Jun, JunB, JunD, ATF-2, ATF-3) substantiates the importance of these transcription factors for CD40-mediated transcriptional up-regulation of a diverse range of genes involved in EC proliferation, differentiation, and response to cell damage. We also found up-regulation of many other transcription factors, CD40 effectors, and regulators (Document S1, “CD40 signaling: pathway induction and transcription factor activation”). This highlights the complexity of multiple signaling pathways that cooperate in CD40-regulated gene expression in ECs. In addition, we document for the first time a strong CD40-dependent induction of the novel NLF transcription factor family (NLF1 and NLF2) in ECs, which under proinflammatory conditions has been suggested to play an important role regulating architecture and cell adhesion allowing increased vascular permeability.42
Our data imply the CD40-dependent induction of pathways regulating cell fate decisions (PI-3K/AKT, JAK/STAT), and the early and transient activation of major MAPK pathways. Erk1/2 and JNK have been functionally confirmed, analogously to that described for monocytes.43
All these events shape the transcriptional cascade elicited as a consequence of CD40-mediated EC–T-cell cross-talk, affecting fundamental EC processes such as chemotaxis, adhesion, tissue remodeling, carbohydrate and lipid metabolism, oxidative stress, and angiogenesis (Document S1).
Conversely, the fundamental contribution of CD40 as T-cell costimulatory molecule has been widely recognized.44 Moreover, it has been recently suggested than ECs themselves play a far more direct role in immunity.45 Our results indicate that CD40 signaling in ECs leads to transcriptional induction of several immunity-related molecules and actively contributes to the transformation of ECs to antigen-presenting cells (Document S1, “Immunity”). Thus, CD40-mediated leukocyte–endothelial cell interactions appear to influence immune cell function through antigen presentation and directly affect adaptive immunity, inducing transendothelial migration and/or polarization of different T-cell subsets.
In addition, several important components of the innate immune system were found induced in ECs after CD40-mediated EC–T-lymphocyte interaction (Figure 2; Document S1, “Immunity”). Remarkably, as a part of the EC stress response, we show that one of the “alert” mechanisms most relevantly induced upon CD40 signaling was the viral immune surveillance system. Thus, TLR3, recognizing the viral replication intermediate dsRNA inside intracellular organelles such as endosomes, was one of the most highly up-regulated pattern recognition receptors (PRRs) found in ECs (implied by functional annotation and GSEA, and confirmed by RT-qPCR). Moreover, additional PRRs, such as the cytosolic CARD-helicases RIG-I and MDA546,47 were also found significantly induced upon EC CD40 engagement (also validated by RT-qPCR), along with common downstream effectors such as several interferon regulatory factors (IRFs)48 (Figure 2). We verified the functionality of this induced viral recognition system by challenging CD40-activated ECs with the synthetic dsRNA analog poly (I:C), which, upon activation of their downstream signaling cascades, led to transcriptional up-regulation and production of IFNβ, and NF-κB– and AP-1–dependent proinflammatory cytokines (IL8).46
Moreover, CD40 action also induced the nonredundant type II (immune) IFN system in ECs by up-regulating both IFNγ receptor subunits and the regulatory STAT1 transcription factor49 (Figure 2), adapting the endothelium to the paracrine antiviral and antimicrobial IFNγ action from the surrounding immune cells.
Other IFN-inducible antiviral pathways that we found transcriptionally induced in ECs upon CD40 engagement include the dsRNA-dependent, 2′-5′ oligoadenylate synthetase/RNase L system mediating RNA degradation,50 and several guanylate-binding proteins (GBP1-4).51 The global induction of the antiviral sensing network in ECs when interacting with activated T lymphocytes highlights the important contribution of CD40 preserving the vasculature from the harmful consequences and the spread of systemic viral infections in the host (Figure S5). Relatedly, repression of genes encoding receptors involved in viral entry, such as FAM89B and CXCR4, a chemokine coreceptor of HIV, was also evident. Taken together, these results highlight the key role of CD40 in instructing the immune surveillance systems in ECs activated through T-lymphocyte interaction.
Thus, endothelial CD40 appears to play a fundamental role regulating both the adaptive and the innate immunity. Consequently, CD40-mediated induction of the viral immune surveillance system might also participate in the mechanisms underlying autoimmune disorders that are often associated with overproduction of type I IFNs52,53 and, perhaps, other chronic inflammatory processes.54
Furthermore, upon activation, ECs transform to a vasopressive, procoagulant state characteristic of the inflammatory process. Our results indicate that CD40 also influences such processes by regulating expression of genes involved in maintenance of vascular homeostasis, in the control of blood pressure, and in thrombin binding and inhibition (Document S1, “Hemostasis and blood coagulation”).
Interestingly, our study infers that endothelial APLN, which is expressed predominantly in normal endothelia,55,56 was significantly down-regulated not only through the CD40-CD154 dyad, but also by other proinflammatory stimuli. This previously unrecognized feature of EC activation was functionally validated both in vitro and in vivo. Atherosclerotic plaques are infiltrated by activated macrophages, T and B lymphocytes, plasma cells, and mast cells releasing inflammatory molecules, which amplify the severity of the disease. Moreover, CD40-CD154 has proven a crucial mediator not only in the initial events of atherogenesis but also during the evolution of established atheroma.57 Thus, we found an inverse correlation between the pathologic degree of stenotic plaque from human carotid arteries and the level of their endothelial apelin expression revealed by immunohistochemistry, substantiating the relevance of this vasoactive peptide in vascular immune-inflammatory processes. Several studies have indicated that apelin is an arterial and venous dilator through a NOS-dependent mechanism, causing NO release from ECs.58,59 A recent report highlights the complex and fine regulation of renal hemodynamic functions exerted by apelin through actions on preglomerular and postglomerular microvasculature and also probably on tubular functions.60 Accordingly, our results suggest that endothelial apelin down-regulation might also constitute a major factor in the hypertension and deregulation of body fluid homeostasis at the peripheral level associated with renal transplant rejection.
Given the central role of ECs in the immune-inflammatory process and the prominent position of CD40 regulating several mediators and signaling pathways that contribute to immunity and inflammation, it seems reasonable to consider the CD40-CD154 dyad as a relevant therapeutic target for anti-inflammatory action. Indeed, preclinical assessment of the efficacy of blocking the CD40-CD154 interaction is either ongoing in several autoimmune disease models, such as multiple sclerosis61 and lupus erythematosus,62 or is being considered for cardiovascular disease63,64 and to achieve immunologic tolerance in transplantation.65 Our comparative transcriptional profiling study highlights the specificity and efficiency of the siRNA anti-CD40 modulating CD40 expression and function in ECs, which supports its anti-inflammatory potential.9 Nevertheless, ongoing work using animal models of immune-inflammatory diseases with established CD40 relevance will prove the overall therapeutic effectiveness of this RNAi-based reagent. Moreover, the detailed knowledge of the molecular pathways affected by CD40 action in ECs should contribute to dissect potential targets downstream of CD40 signaling relevant to immune-inflammatory processes.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Juanjo Lozano and David Otero from the Microarray Core Facility (CRG-UPF), and Núria Bolaños from the Laboratory of Nephrology (CSUB-IDIBELL) for their expert technical assistance.
This work was supported entirely by the Ministerio de Sanidad y Consumo (Madrid, Spain) through grants 03/0516 and 05/1018 from the Fondo de Investigaciones Sanitarias (FIS), from the Red de Centros del Instituto de Salud Carlos III (Refs C03/03 and C03/07), and by an ISCIII fellowship to R.P. (BF02/9166). I.H.-F. was supported by FIS/ISCIII. J.M.A. is sponsored by the Researchers Stabilization Program from the National Health System (SNS).
Authorship
Contribution: R.P., R.O., J.K., I.H.-F., and A.L. performed the research; J.T., J.M.C., and J.M.G. contributed essential reagents and provided general support; R.P. and L.S. performed bioinformatic analyses; R.P. analyzed the data and wrote the paper; and J.M.A. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Josep M. Aran, Medical and Molecular Genetics Center, Institut de Investigació Biomèdica de Bellvitge (IDIBELL), Hospital Duran i Reynals, Gran Via s/n km 2.7, 08907 L'Hospitalet de Llobregat, Barcelona, Spain; e-mail: jaran@idibell.org.