The p53 protein integrates the combination of mutations that cause a cancer with the set of environmental stresses that act on that cancer. Mutations in the p53 gene can therefore have dramatic impacts on the overall survival of cancer patients.
The p53 protein is a transcription factor that responds to a wide variety of stress signals (DNA damage, hypoxia, etc) that decrease the fidelity of the cellular replication processes. The duplication of damaged DNA results in a high mutation rate and activates the p53 protein, producing cell-cycle arrest, senescence, or cell death.1 The tumor suppressor properties of p53 protein prevent mutational mistakes and the development of cancers. Germ line mutations in the p53 gene result in cancers occurring at a young age, while somatic mutations in the p53 gene are common (approximately 50% incidence) in a wide variety of cancers.2 Thus, the p53 status of a person's germ line or a cancer's genome can impact not only the risk of developing a cancer, but also the response to therapy, which often activates a p53 response in cancers with a wild-type p53 gene. In this issue of Blood, Young and colleagues demonstrate that the mutational status of the p53 gene in germinal center B cell–like, diffuse large B-cell lymphoma (DLBCL), mutant or wild-type, has a large impact on overall survival (OS; 5-year survival and median survival) in this patient cohort. While the 24% of the cohort that contained p53 mutations had a poor OS, the location of the missense mutation in the p53 gene was also important.
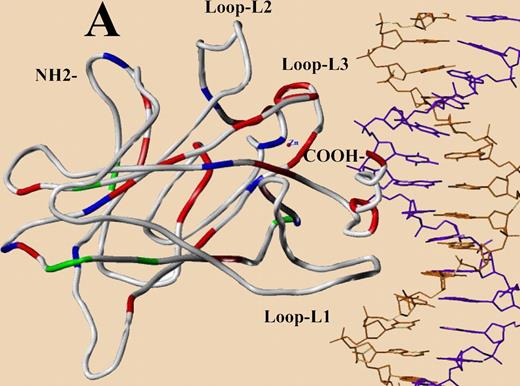
The distribution pattern of TP53 mutations in the central core domain model designed from published crystal structure (red, patients with poor survival; green, those with survival similar to WT group; blue, survival data unavailable). See the complete figure in the article beginning on page 3088.
The distribution pattern of TP53 mutations in the central core domain model designed from published crystal structure (red, patients with poor survival; green, those with survival similar to WT group; blue, survival data unavailable). See the complete figure in the article beginning on page 3088.
The human p53 gene contains 393 codons and the p53 protein this gene produces is commonly divided into 5 domains: an RNA polymerase transactivating domain, a proline rich domain, a DNA-binding domain, a tetramerization domain, and a carboxy-terminal regulatory domain. Virtually all of the p53 mutations found in cancers are localized to the DNA-binding domain. That protein domain is commonly divided into 3 loops: L1 (codons 112-141), L2 (codons 163-195), and L3 (codons 236-251), and 2 loop-sheet-helix regions (codons 119-135; 272-287) that reflect the structure of this protein. Loops 1 and 3 and the Loop-Sheet-Helix (LSH) region from codons 272 to 287 make direct contacts with the DNA adjacent to a gene that is regulated by the p53 transcription factor, while loop 2 is required for folding and stabilization of this DNA-binding domain and is not in direct contact with the DNA. P53 mutations (62 mutations) in the part of the protein that make DNA contacts, loops 1 and 3, and the LSH codons 272 to 287 had poor median survivals (1 year) and poor 5-year survivals (19%, P = .008), while the p53 mutations in portions of the DNA-binding domain of the protein not in contact with DNA (loop 2) had no significant impact on OS. This has also been observed in a variety of other cancers.3,4
In a second paper in this issue of Blood, by O'Shea and colleagues, p53 mutational status of a follicular lymphoma (FL) was also associated with a shorter progression-free survival and with a shorter overall survival. In FLs, only a small percentage of these cancers have p53 mutations (6%), and these 12 cases were correlated with older ages of patients and a higher International Prognostic Index. P53 mutations also correlated with a poorer nature of the immune response in FL. The consistent and strong results in both studies suggest that analysis of the type of p53 mutation in a lymphoma could be very useful prognostic information with utility for treatment decisions. These ideas could be validated in a clinical trial, providing rational selection criteria for patients who could benefit from more intensive up-front or postremission therapy rather than waiting for a tumor relapse.
How can these results be functionally interpreted? First, there are 2 classes of missense mutations in the DNA binding domain, one that disrupts the structure of the entire protein commonly by eliminating the binding of zinc molecules (cystines-176, -238, and -242 and histidine-179), and a second where mutations in the protein's DNA contact regions that weaken the binding of the protein to its targeted DNA but retain a similar protein structure. It is this second class that both gives rise to the adverse prognosis of DLBCL and often demonstrates allele-specific “gain-of-function mutations”5,6 that confers new properties on a mutant p53 protein. These mutant proteins apparently bind to other transcription factors (like p73) and alter their functions, making the cell more resistant to apoptosis and chemotherapy. These “gain-of-function” phenotypes have been demonstrated in cell cultures and in transgenic mice.7 The observations made in these papers suggest the possibility that allele-specific “gain-of-function” mutations can act in humans to contribute to poor overall survival. Unlike many tumor suppressor gene mutations, most of the p53 mutations found in tumors are missense mutations (92 in the DLBCL study), rather than deletions or insertions (10 in that study) that destroy the structure of the protein. That observation suggests that tumors may select for a p53 missense mutation that provides a helpful “gain-of-function” for tumor growth and survival before and during treatments. This idea will clearly need additional studies examining the possible altered transcriptional patterns brought about in a p53 mutant allele-specific fashion in DLBCL and FL. There ought to be a transcriptional “signature of genes” of that mutant p53 protein, disrupting the function of other transcription factors. The concept that tumor suppressor genes contribute to the origins and development of cancers by their loss of a function will now have to be modified to include the oncogene-like activity of a mutant p53 protein that has a “gain-of-function.”
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal