Abstract
Notch and its ligands have been implicated in the regulation and differentiation of various CD4+ T-helper cells. Regulatory T cells (Tregs), which express the transcription factor Foxp3, suppress aberrant immune responses that are typically associated with autoimmunity or excessive inflammation. Previous studies have shown that transforming growth factor beta (TGFβ1) induces Foxp3 expression and a regulatory phenotype in peripheral T cells. Here, we show that pharmacologic inhibition of Notch signaling using γ-secretase inhibitor (GSI) treatment blocks (1) TGFβ1-induced Foxp3 expression, (2) the up-regulation of Foxp3-target genes, and (3) the ability to suppress naive T-cell proliferation. In addition, the binding of Notch1, CSL, and Smad to conserved binding sites in the foxp3 promoter can be inhibited by treatment with GSI. Finally, in vivo administration of GSI results in reduced Foxp3 expression and development of symptoms consistent with autoimmune hepatitis, a disease previously found to result from dysregulation of TGFβ signaling and regulatory T cells. Together, these findings indicate that the Notch and TGFβ signaling pathways cooperatively regulate Foxp3 expression and regulatory T-cell maintenance both in vitro and in vivo.
Introduction
The Notch family is a group of evolutionarily conserved type I transmembrane receptors involved in cell fate decisions in a variety of organ systems. There are 4 mammalian Notch family members (Notch1-4) and 5 known ligands (Delta-like1,3,4, and Jagged 1,2).1 Following ligand-dependent ectodomain shedding, Notch is activated through 2 sequential enzymatic cleavage events, which are catalyzed by ADAM metalloproteases and the γ-secretase complex. Following γ-secretase–mediated cleavage, the intracellular portion of Notch (Notch1IC) translocates to the nucleus and interacts with CSL and a variety of coactivators, including Mastermind-like and p300, to activate target gene transcription. There are several targets of the γ-secretase complex in addition to Notch proteins, including Aβ, the primary constituent in Alzheimer disease plaques, and CD44.2 γ-Secretase inhibitors (GSIs), pharmacologic inhibitors of γ-secretase activity, block Notch activation by preventing the release of the intracellular domain and are currently in clinical trials for the treatment T-cell acute lymphoblastic leukemia and Alzheimer disease.3,4
Notch family members have been implicated in the differentiation of various CD4+ T-helper subsets, including TH1, TH2, and regulatory T cells (Tregs). Inhibiting Notch signaling has been shown to block TH1 and TH2 polarization by preventing Notch-mediated up-regulation of Tbx21 and GATA-3, respectively.5-7 Transgenic mice overexpressing the active form of Notch3 have increased levels of CD4+CD25+ regulatory T cells both in the thymus and in the spleen and are protected from disease onset in a mouse model of autoimmune diabetes.8 In addition, several groups have shown that overexpression of the Notch ligands Jagged1 or Delta-like1 resulted in the generation of a population of suppressive CD4+ T cells.9,10 A recent study has also shown that hematopoietic progenitors expressing Jagged2 expand peripheral regulatory T-cell populations in a Notch-dependent manner.11 To date, no studies have linked Notch signaling to the expression of foxp3, the master gene regulator of the regulatory T-cell program.12
Transforming growth factor beta 1 (TGFβ1) is a pleiotropic anti-inflammatory cytokine that also has been implicated in regulatory T-cell differentiation. Human and mouse CD4+CD25+ regulatory T cells express the transcription factor Foxp3 and can be generated from naive T cells following stimulation in the presence of TGFβ1.13-15 Although the in vivo relevance of peripheral regulatory T-cell generation is not completely clear, it is thought that TGFβ1 is required for the maintenance of the peripheral Treg pool, as mice deficient in TGFβ signaling lack peripheral, but not thymic, regulatory T cells and exhibit multiorgan lymphocyte infiltration and autoimmunity similar to Foxp3-deficient mice.16-20
Previous studies have demonstrated cross-talk between the Notch and TGFβ signaling pathways. Smads, the intracellular mediators of TGFβ signaling, bind the intracellular domains both of Notch1 and Notch4.21,22 In addition, Smad3 can form a complex with CSL and Notch1IC to cooperatively activate the transcription of the Notch target gene, Hes1.21
As TGFβ1 and Notch both are implicated in regulatory T-cell differentiation, we have conducted studies to determine whether cross-talk between the 2 signaling pathways is required for the induction of Foxp3 expression and the regulatory T-cell program in the periphery. Here, we demonstrate that in vitro GSI treatment in the presence of TGFβ1 blocks Notch-mediated up-regulation of Foxp3, Foxp3-target genes, and the ability to suppress naive T-cell proliferation. Further, in vivo GSI treatment down-regulated Foxp3 and resulted in a spontaneous lymphocyte infiltration of the liver. These data identify Foxp3 as a novel downstream target of Notch signaling, and its relevance is underscored by the fact that GSI therapy is entering the clinic, and presents a previously unrecognized side effect of long-term GSI treatment.
Methods
Cell culture
C57BL/6 and Notch1 antisense (AS) mice were housed in the animal care facility at the University of Massachusetts Amherst in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines. All animal procedures used in this study were reviewed and approved by the IACUC at the University of Massachusetts Amherst.
Notch1 AS mice were generated as previously described.23 CD4+CD25− cells were enriched to greater than 90% purity from bulk splenocytes using the IMag system and the mouse CD4 T lymphocyte enrichment set with 2.5 μg anti-CD25 biotin (BD Bioscience, San Jose, CA). In some cases, CD4+ T cells were directly isolated using mouse anti-CD4 DM particles (BD Bioscience). Cells were plated in media at a concentration of 2.25 × 106 cells/mL media in a 12-well plate coated with anti-CD3ϵ plus anti-CD28 and 2 ng/mL rhTGFβ1 (R&D Systems, Minneapolis, MN) or vehicle control. In some cultures, cells were pretreated with the γ-secretase inhibitor zIL-CHO or the Smad3 inhibitor SIS3 (Invitrogen, Frederick, MD) for 30 minutes before stimulation.7,24 In others, 50 U/mL or 100 U/mL recombinant human IL-2 (BD Bioscience) was added to the culture at the time of stimulation.
Immunoblot analysis
Immunoblot analysis was performed as previously described.7 Antibodies included anti-Notch1IC (eBioscience, San Diego, CA), anti-Foxp3 (clone eBio7979, eBioscience), anti-Gapdh (Chemicon, Temecula, CA), and anti-Hsp70 (Sigma-Aldrich, St Louis, MO).
RNA isolation and reverse-transcription–polymerase chain reaction
CD4+CD25− T cells were stimulated as described in “Cell culture” in the presence or absence of 25 μM GSI. After 72 hours, cells were harvested and total RNA was isolated using the RNAqueous kit (Ambion, Austin, TX) following the manufacturer's protocol. Total RNA samples were subjected to treatment with DNase using the TURBO DNA-free kit (Ambion), cDNA was synthesized, and transcripts were amplified by polymerase chain reaction (PCR). The following primers and temperatures (Tm's) were used: Gapdh forward, 5′-ACTTTCGATCAAGGATCAGCA-3′ and Gapdh reverse, 5′-ACGGAAGGCCATGCCAGTGAGCTT-3′; Tm = 55°C. Foxp3 forward, 5′-TTCATGCATCAGCTCTCCAC-3′ and Foxp3 reverse, 5′-CTGGACACCCATTCCAGACT-3′; Tm = 57°C. Gpr83 forward, 5′-GAAGATGCTGGTGCTTGTGGTAGTC-3′ and Gpr83 reverse, 5′-AAGTGGTGATTAGGTAGTGGAGCCC-3′; Tm = 57°C. Conditions for PCR were 94°C for 5 minutes, 94°C for 40 seconds [Tm] for 40 seconds and 72°C for 40 seconds (27 cycles), and 72°C for 10 minutes.
Flow cytometry
CD4+CD25− T cells were stimulated as described in “Cell culture” in the presence or absence of 3 μM GSI. After 72 hours, cells were harvested and stained for CD4 and CD25. Intracellular staining for Foxp3 was done using the Foxp3 staining buffer set and anti-Foxp3, clone FJK-16s (both from eBioscience). In some cases, bulk splenocytes were stained for CD4, CD25, and intracellular Foxp3 without prior cell culture. In all cases, isotype control for Foxp3 (rat IgG2a) showed no background staining. Flow cytometric data were acquired using a FACSCalibur or LSRII flow cytometer with either CellQuest or FACSDiva software, respectively (BD Bioscience). Data were analyzed using FlowJo software (TreeStar, Ashland, OR).
Suppression assay
CD4+CD25− T cells were stimulated as described in “Cell culture” in the presence or absence of 3 μM GSI (“conditioned cells”). Conditioned cells were then washed and an experimental coculture was set up at a 1:1 or 2:1 ratio, in triplicate, with freshly isolated CD4+ T cells (“responder cells”), γ-irradiated antigen presenting cells (APCs), and soluble anti-CD3ϵ for 72 hours. During the final 24 hours, cultures were pulsed with 1 μCi (0.037 MBq)/well 3H-thymidine to monitor proliferation. Total cell number per well was 1 × 106 cells. To account for the proliferation of the conditioned cells, a control coculture at a 1:1 or 2:1 ratio of conditioned cells to γ-irradiated responder cells, plus γ-irradiated APCs and soluble αCD3ϵ, was done in tandem with the experimental culture. At the end of the culture period, cells were harvested using a cell harvester and 3H incorporation was measured using a gamma counter. Proliferation of responder cells was calculated as follows: (cpmresponder cells = cpmcontrol culture − cpmexperimental culture).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) assays were performed using the Chromatin Immunoprecipitation Assay Kit (Millipore, Billerica, MA). CD4+CD25− T cells were stimulated with anti-CD3ϵ/anti-CD28 and 2 ng/mL TGFβ1; in some cases, cells were pretreated with 50 μM GSI before stimulation. After 24 hours, a ChIP assay was performed following the manufacturer's protocol. Antibodies used were anti-Notch1 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), anti–RBP-Jκ (H-50; Santa Cruz Biotechnology), anti-Smad1/2/3 (H-2; Santa Cruz Biotechnology), or anti–acetylated H3 (Upstate, Lake Placid, NY). Isotype controls were as follows: for anti-Notch1 and anti–RBP-Jκ, normal rabbit IgG (Santa Cruz Biotechnology); for anti-Smad1/2/3, normal mouse IgG (Santa Cruz Biotechnology); for anti–acetylated H3, rabbit IgG (Upstate). In the mouse Foxp3 promoter, putative CSL and Smad binding elements were amplified by PCR. Primers used were forward, 5′-CCCCCACCCTGCAATTAT-3′ and reverse, 5′-CCTGGCTTGTGGGAAACTG-3′ (primer set 1) and forward, 5′-ACAATTATGCCTCAGTTACCTTCC-3′ and reverse, 5′-TGCTCACTACAGTCCATGTATCCT-3′ (primer set 2). Enhancer region primer set D is described elsewhere.25 PCR was performed using PCR Master Mix (Promega, Madison, WI) following the manufacturer's protocol. Conditions were 94°C for 7 minutes, 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute (33 cycles), and 72°C for 10 minutes.
In vivo GSI administration and histology
Mice were fed either Harlan-Teklad mouse/rat chow (Madison, WI) formulated with a combination of 2 enantiomers of LY 411575, designated LY1 + LY2, used at a ratio of 80%:20%, respectively, to deliver 5 mg/kg per day (n = 5) or control chow (n = 4). After 3 months, mice were killed and livers were fixed overnight in 10% neutral buffered formalin. Fixed organs were then paraffin embedded, sectioned into 4-μm sections, and stained with hematoxylin and eosin.
Statistical analysis
Two-tailed t tests were performed using GraphPad Prism version 4.0 for OSX (GraphPad Software, San Diego, CA).
Results
In vitro GSI treatment blocks TGFβ1-induced Foxp3 expression and function
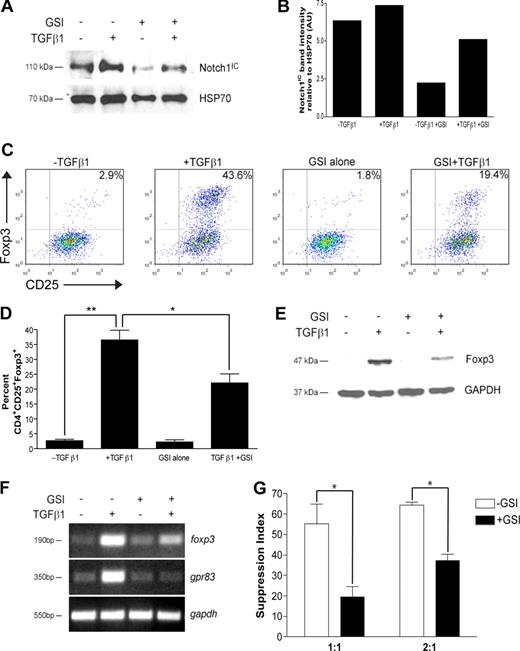
Recent data have implicated Notch signaling in the differentiation of TH1 and TH2 effector cells. To determine whether there is a CD4+ T cell–intrinsic role for Notch signaling in regulatory T-cell differentiation, CD4+CD25− cells were polarized toward a regulatory T-cell phenotype with TGFβ1 for 72 hours. In some cultures, cells were pretreated before stimulation with GSI, whose bioactivity is sustained only for the first 24 hours of culture (data not shown). To ensure GSI treatment was effective at reducing Notch1 activation, Notch1IC levels were assessed by immunoblot. Following stimulation, cells stimulated in the presence of GSI expressed a 60% or 30% reduction in Notch1IC expression in the absence or presence of TGFβ1, respectively (Figure 1A,B). Furthermore, Notch1IC expression was increased following TGFβ1 treatment, suggesting potential synergy between the TGFβ and Notch signaling pathways (Figure 1A,B).
In vitro GSI treatment blocks TGFβ1-induced Foxp3 expression and function. CD4+CD25− splenocytes were isolated and stimulated under the following conditions: no treatment, + TGFβ1, + GSI alone, or + GSI + TGFβ1. (A) Notch1 expression in cells pretreated without or with GSI was evaluated by immunoblotting using antibodies that recognized the cleaved, active form of Notch1 (Notch1IC). Antibody specific for HSP70 was used to control for loading. (B) Graphic representation of band intensities shown in panel A. (C) Effect of GSI treatment on Foxp3 expression was analyzed by flow cytometry using antibodies specific for CD4, CD25, and Foxp3. All plots are gated on live CD4+ cells. (D) Graphic representation of flow cytometry data from panel C. Data represent the means (± SD). **P = .001; *P < .05. (E) Foxp3 expression in CD4+CD25+ T cells treated without or with GSI was assessed by immunoblotting using antibodies specific for Foxp3. GAPDH was used as a loading control. (F) To assess the effects of GSI treatment on transcriptional regulation of foxp3, and its downstream target gene gpr83, total RNA was isolated from cells cultured under the conditions described in “RNA isolation and reverse-transcription–polymerase chain reaction” and analyzed by RT-PCR. Amplification of gapdh served as a loading control. (G) Cells were polarized as indicated in “Methods,” then washed, and cocultured at a 1:1 or 2:1 ratio with freshly isolated CD4+ T cells plus γ-irradiated APCs for 72 hours. *P < .05. Data are represented as mean suppression index (± SD). All results are representative of at least 2 independent experiments.
In vitro GSI treatment blocks TGFβ1-induced Foxp3 expression and function. CD4+CD25− splenocytes were isolated and stimulated under the following conditions: no treatment, + TGFβ1, + GSI alone, or + GSI + TGFβ1. (A) Notch1 expression in cells pretreated without or with GSI was evaluated by immunoblotting using antibodies that recognized the cleaved, active form of Notch1 (Notch1IC). Antibody specific for HSP70 was used to control for loading. (B) Graphic representation of band intensities shown in panel A. (C) Effect of GSI treatment on Foxp3 expression was analyzed by flow cytometry using antibodies specific for CD4, CD25, and Foxp3. All plots are gated on live CD4+ cells. (D) Graphic representation of flow cytometry data from panel C. Data represent the means (± SD). **P = .001; *P < .05. (E) Foxp3 expression in CD4+CD25+ T cells treated without or with GSI was assessed by immunoblotting using antibodies specific for Foxp3. GAPDH was used as a loading control. (F) To assess the effects of GSI treatment on transcriptional regulation of foxp3, and its downstream target gene gpr83, total RNA was isolated from cells cultured under the conditions described in “RNA isolation and reverse-transcription–polymerase chain reaction” and analyzed by RT-PCR. Amplification of gapdh served as a loading control. (G) Cells were polarized as indicated in “Methods,” then washed, and cocultured at a 1:1 or 2:1 ratio with freshly isolated CD4+ T cells plus γ-irradiated APCs for 72 hours. *P < .05. Data are represented as mean suppression index (± SD). All results are representative of at least 2 independent experiments.
Analysis by flow cytometry revealed that TGFβ1 treatment induced Foxp3 protein expression in approximately 40% of cells, a percentage similar to that which has previously been shown in the literature (Figure 1C). In the presence of GSI, however, only 20% of cells were induced to become CD4+CD25+Foxp3+, a 50% decrease compared with TGFβ1 treatment alone (Figure 1C). A 50% reduction in Foxp3 expression was maintained following either a 24-hour exposure to GSI or repeated exposure to fresh GSI, further confirming that γ-secretase activity is required during the first 24 hours of culture (Figures S1,S2, available on the Blood website; see the Supplemental Materials link at the top of the online article). The inhibition due to GSI treatment was not due to nonspecific effects on early TGFβ signaling events, including Smad2/3 phosphorylation, and could not be rescued by exogenous IL-2 (data not shown; Figure S3), which has previously been shown to be required for TGFβ1-induced Foxp3 expression.26 These data were statistically significant and reproducible over 3 experiments (Figure 1D). Western blot analysis yielded similar results, with a 60% reduction in Foxp3 protein expression in cells treated with TGFβ1 and GSI compared with TGFβ1 alone (Figure 1E). Because the GSI is bioactive in only the first 24 hours of culture, these data indicate that γ-secretase activity is required immediately following T-cell activation to induce Foxp3 expression.
As GSI treatment reduces Foxp3 expression, we hypothesized that GSI treatment would also modulate Foxp3-mediated induction of target genes and suppression of naive CD4+ T-cell proliferation. G-protein–coupled receptor 83 (Gpr83) has been identified as a bona fide downstream target of Foxp3, since retroviral transduction of Foxp3 in CD4+CD25− T cells or polarization in the presence of TGFβ1 is sufficient to induce its expression.27,28 To determine whether GSI inhibits TGFβ1-induced up-regulation of gpr83, cells were polarized as indicated in “Methods” and total RNA was isolated for analysis by reverse-transcription (RT)–PCR. Similar to the reduction in Notch1 activation, GSI treatment in the presence of TGFβ1 reduced up-regulation of foxp3 mRNA by approximately 30% (Figure 1F). In addition, although TGFβ1 is capable of inducing mRNA expression of gpr83, its up-regulation was completely abrogated in the presence of GSI (Figure 1F).
Regulatory T cells control immune responses by suppressing the proliferation of both CD4+ and CD8+ T cells. To determine whether polarization in the presence of GSI affects suppressive activity, a 3H-thymidine–based suppression assay was performed. CD4+CD25− T cells were polarized in the presence or absence of GSI for 72 hours (“conditioned cells”). Cells were then washed and cocultured at a 1:1 or 2:1 ratio with freshly isolated CD4+ T cells (“responder cells”), in the presence of γ-irradiated antigen-presenting cells (APCs) and αCD3ϵ for an additional 72 hours. Cells were pulsed with 3H-thymidine during the last 24 hours to monitor proliferation. At the end of the culture, cells were harvested and 3H-thymidine incorporation was measured using a scintillation counter. A suppression index, defined as the percentage by which the proliferation of the responder cells was reduced due to TGFβ1 treatment of the conditioned cells, was then calculated from the raw data using the equations: 1 − (cpm + TGFβ1/cpm − TGFβ1) and 1 − (cpmTGFβ1 + GSI/cpmGSI), respectively. In the absence of GSI, coculture of TGFβ1-treated conditioned cells with naive CD4+ T cells at a 1:1 ratio resulted in a high suppression index, which is indicative of a high number of Tregs in the coculture and a subsequent reduction in the proliferation of the responder cells (−GSI; Figure 1G). In contrast, coculture of cells polarized in the presence of GSI with naive CD4+ T cells at a 1:1 ratio resulted in a 64% reduction in the suppression index, which is indicative of a reduced number of Tregs in the coculture and increased proliferation of the responder cells. As expected, the suppression indices were higher when conditioned cells were cocultured with responder cells at a 2:1 ratio; however, there was a similar percentage reduction in suppression index in +GSI versus −GSI. Taken together, these data indicate that in vitro Treg polarization in the presence of GSI results in reduced Foxp3 up-regulation and function.
Foxp3 induction is impaired in cells that express reduced levels of Notch1
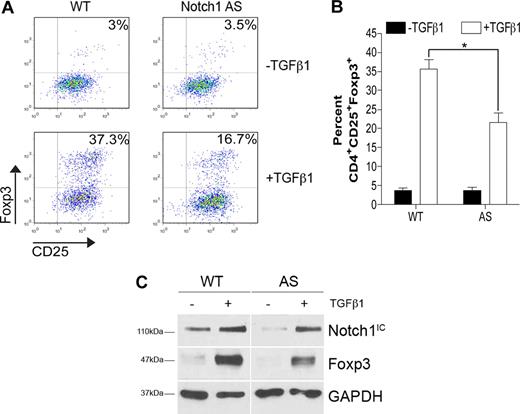
In addition to Notch proteins, there are several targets of γ-secretase–mediated proteolysis, including CD44 and E-cadherin,2 and, thus, it is important to discern whether the effects seen with GSI treatment were due to Notch inhibition. Transgenic mice expressing a Notch1 antisense transgene have previously been generated. Multiple cell types in these animals, including lymphocytes, exhibit 20% to 40% reduced levels of Notch1, with no effect on Notch2, Notch3, or Notch4.23 CD4+CD25− T cells isolated from Notch1 antisense (AS) mice or wild-type (WT) controls were polarized toward a regulatory phenotype with TGFβ1 for 72 hours. Analysis by flow cytometry revealed that TGFβ1 induced 54% fewer CD4+CD25+Foxp3+ cells when Notch1 levels were impaired (37% vs 17%) (Figure 2A). Further, Foxp3 induction in CD4+CD25− T cells from AS mice was significantly lower than that seen in cells from WT mice over 3 independent experiments (Figure 2B). Data generated by flow cytometry were also reproducible by immunoblot. When whole-cell lysates were prepared from WT or AS cells, we found that AS cells expressed a 20% reduction in Notch1IC and a subsequent 35% reduction in Foxp3 expression (Figure 2C). Taken together, these data indicate that the observed effect of in vitro GSI treatment on TGFβ1-induced Foxp3 expression is due to inhibition of Notch1 signaling.
Impaired Foxp3 induction in cells expressing reduced levels of Notch1. CD4+CD25− splenocytes from either wild-type (WT) or Notch1 antisense (AS) mice were stimulated with plate-bound αCD3ϵ plus αCD28 for 72 hours in the presence or absence of 2 ng/mL TGFβ1. (A) Cells were analyzed by flow cytometry using antibodies specific for CD4, CD25, and Foxp3. All plots are gated on live CD4+ cells. Plots are representative of 3 independent experiments. (B) Graphic representation of flow cytometry data from panel A. Data represented as the mean (± SD) of 3 samples per group. *P < .05. (C) Immunoblot detection of Foxp3, Notch1 intracellular domain (Notch1IC), and GAPDH (loading control) from whole-cell lysates prepared from cells stimulated under the same conditions as in panel A. Data are representative of 2 independent experiments.
Impaired Foxp3 induction in cells expressing reduced levels of Notch1. CD4+CD25− splenocytes from either wild-type (WT) or Notch1 antisense (AS) mice were stimulated with plate-bound αCD3ϵ plus αCD28 for 72 hours in the presence or absence of 2 ng/mL TGFβ1. (A) Cells were analyzed by flow cytometry using antibodies specific for CD4, CD25, and Foxp3. All plots are gated on live CD4+ cells. Plots are representative of 3 independent experiments. (B) Graphic representation of flow cytometry data from panel A. Data represented as the mean (± SD) of 3 samples per group. *P < .05. (C) Immunoblot detection of Foxp3, Notch1 intracellular domain (Notch1IC), and GAPDH (loading control) from whole-cell lysates prepared from cells stimulated under the same conditions as in panel A. Data are representative of 2 independent experiments.
Notch1 recruits CSL and Smad to the foxp3 promoter to directly regulate transcription
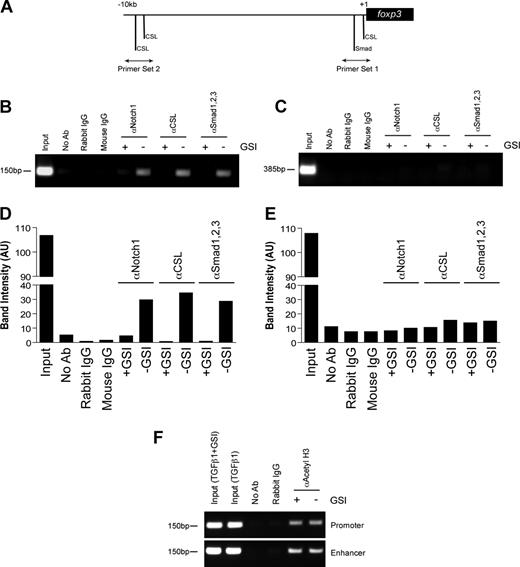
Previous studies have identified a highly conserved proximal promoter located 6.2-kb upstream of the foxp3 translational start site.29 We examined this region and identified a CSL consensus site (5′-TTTCCCA-3′) that is located −8 bp upstream of the transcriptional start site and is 100% conserved among mice, humans, rats, guinea pigs, and dogs (Figure S4). In addition, we identified a Smad binding element (5′-GTCTG-3′) located −85 bp upstream of the transcriptional start site that was also conserved among several species (Figure S4). To determine whether Notch1, CSL, and Smad1/2/3 bind the foxp3 promoter, a chromatin immunoprecipitation assay was performed. CD4+CD25− T cells were polarized as previously described, in the presence or absence of GSI. Antibodies specific for Notch1, CSL, or Smad1/2/3 were used to immunoprecipitate proteins cross-linked to DNA. After de–cross-linking, the eluted DNA was used as a template in a PCR reaction using primers specific either for the region containing the conserved CSL and Smad binding sites (primer set 1) or for a region 10-kb upstream of the transcriptional start site containing 2 putative CSL binding sites (primer set 2) (Figure 3A).
In vitro GSI treatment blocks the binding of Notch1, CSL, and Smad to the foxp3 promoter without inhibiting histone acetylation. Chromatin immunoprecipitation (ChIP) of CD4+CD25− splenocytes stimulated with plate-bound αCD3ϵ plus αCD28 and 2 ng/mL TGFβ1 in the presence or absence of GSI for 24 hours. (A) Primer sets were designed to span putative CSL and Smad binding sites within the foxp3 promoter. Rabbit αNotch1, rabbit αCSL, and mouse αSmad1/2/3 were used to immunoprecipitate protein-DNA complexes. De–cross-linked DNA was amplified by PCR using either primer set 1 (B) or primer set 2 (D). Band intensities were calculated using ImageJ software, version 1.38 (National Institutes of Health, Bethesda, MD; panels C,E). Data are representative of 3 independent experiments. (F) ChIP assay using antibody specific for acetylated histone H3 (αacetyl H3) and either primer set 1 from panel A or the region of the Foxp3 enhancer containing previously identified Smad3 and NFAT binding sites. Data are representative of 2 independent experiments. Input indicates total chromatin. No Ab (beads only), rabbit IgG, and mouse IgG were used as isotype controls.
In vitro GSI treatment blocks the binding of Notch1, CSL, and Smad to the foxp3 promoter without inhibiting histone acetylation. Chromatin immunoprecipitation (ChIP) of CD4+CD25− splenocytes stimulated with plate-bound αCD3ϵ plus αCD28 and 2 ng/mL TGFβ1 in the presence or absence of GSI for 24 hours. (A) Primer sets were designed to span putative CSL and Smad binding sites within the foxp3 promoter. Rabbit αNotch1, rabbit αCSL, and mouse αSmad1/2/3 were used to immunoprecipitate protein-DNA complexes. De–cross-linked DNA was amplified by PCR using either primer set 1 (B) or primer set 2 (D). Band intensities were calculated using ImageJ software, version 1.38 (National Institutes of Health, Bethesda, MD; panels C,E). Data are representative of 3 independent experiments. (F) ChIP assay using antibody specific for acetylated histone H3 (αacetyl H3) and either primer set 1 from panel A or the region of the Foxp3 enhancer containing previously identified Smad3 and NFAT binding sites. Data are representative of 2 independent experiments. Input indicates total chromatin. No Ab (beads only), rabbit IgG, and mouse IgG were used as isotype controls.
Stimulation in the presence of TGFβ1 induces the binding of Notch1, CSL, and Smad1/2/3 to the region of the foxp3 promoter immediately upstream of the transcriptional start site, but not to the region located 10-kb upstream (Figure 3B,C). Furthermore, GSI treatment inhibited the recruitment of all 3 proteins to the promoter, indicating Notch1 is required either to recruit both CSL and Smad to the foxp3 promoter or to stabilize their presence once bound (Figure 3B,C). Notch1 has previously been shown to bind Smad3 in an in vitro overexpression system,21 and we found that inhibition of Smad3 phosphorylation using a specific inhibitor of Smad3 phosphorylation24 reduces Foxp3 induction in a dose-dependent manner (Figure S5), leading us to hypothesize that Smad3 is the family member that is potentially binding the foxp3 promoter.
Recent studies have identified that binding of Smad3 to a specific enhancer region is required for acetylation and activity of the foxp3 promoter.25 To determine whether in vitro GSI treatment interferes with histone acetylation, a chromatin immunoprecipitation assay was performed on cells stimulated under the same culture conditions as indicated in “Methods” using an antibody specific for acetylated histone H3. We found H3 acetylation was not affected by GSI treatment (Figure 3D) and, thus, the inhibition of Notch, CSL, and Smad binding to the foxp3 promoter is not due to reduced promoter accessibility. Together, these data suggest that the TGFβ signaling pathway synergizes with Notch1 to directly regulate Foxp3 expression in a Notch-dependent manner.
In vivo GSI treatment both reduces Notch1 signaling and Foxp3 expression and induces spontaneous lymphocyte infiltration of the liver
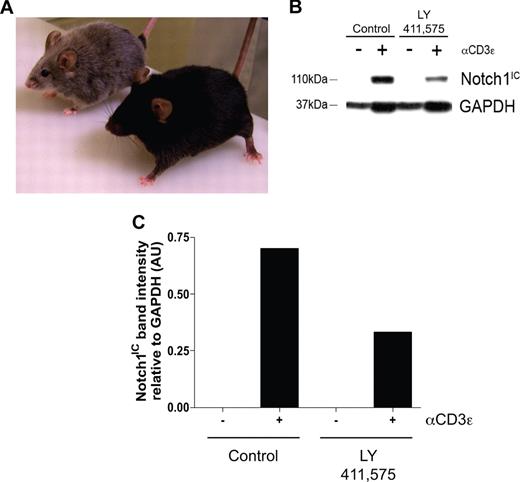
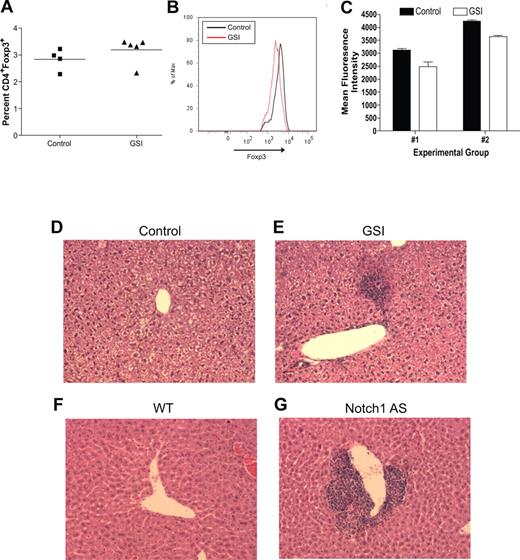
To determine the biologic relevance of our in vitro experiments, an in vivo model was established to determine what effects inhibiting Notch signaling has on maintaining the regulatory T-cell pool. Transgenic mice deficient in TGFβ signaling, by deletion of either TGFβ1 or TGFβRII, succumb to widespread, multiorgan lymphocyte infiltration caused by impaired regulatory T-cell maintenance.16-18 To determine whether a similar phenomenon occurs when Notch signaling is reduced, age- and sex-matched C57BL/6 mice were fed chow formulated with the orally active GSI, LY 411575, at a dose of 5 mg/kg per day for 3 months, a dose sufficient both to reduce Notch1 activation by greater than 40% as well as to induce progressive hair graying, which is indicative of reduced Notch activity (Figure 4A-C).30 Although there was no statistical difference in the overall percentage of CD4+Foxp3+ cells within the spleens of the GSI-treated animals, compared with controls (Figure 5A), closer examination revealed a reduction in the expression of Foxp3 within the Treg pool, as evidenced by decreased mean fluorescence intensity (Figure 5B,C). The trend in reduced Foxp3 expression was consistent over 2 experimental groups, with group 1 (control: n = 2, GSI: n = 3) displaying a 22% decrease, and group 2 (control: n = 2, GSI: n = 2) displaying a 17% decrease (Figure 5C).
Administration of LY 411575 at a dose of 5 mg/kg per day is sufficient to reduce Notch1 activation in vivo. C57BL/6 mice were fed Harlan-Teklad rodent chow formulated with LY 411575 to deliver 5 mg/kg per day for 3 months. (A) Progressive hair graying evident in GSI-fed (silver mouse, n = 5) but not control chow-fed (black mouse, n = 4) mice. (B) Bulk splenocytes were harvested from mice fed chow containing LY 411575 at the dose previously described for 3 months, and were cultured for 72 hours with or without αCD3ϵ stimulation. Whole-cell lysates were prepared, and Notch1IC and GAPDH expression was detected by immunoblot. Data are representative of n = 4 (control) and n = 5 (GSI). (C) Graphic representation of band intensities from panel B. Expression of Notch1IC was normalized to GAPDH expression.
Administration of LY 411575 at a dose of 5 mg/kg per day is sufficient to reduce Notch1 activation in vivo. C57BL/6 mice were fed Harlan-Teklad rodent chow formulated with LY 411575 to deliver 5 mg/kg per day for 3 months. (A) Progressive hair graying evident in GSI-fed (silver mouse, n = 5) but not control chow-fed (black mouse, n = 4) mice. (B) Bulk splenocytes were harvested from mice fed chow containing LY 411575 at the dose previously described for 3 months, and were cultured for 72 hours with or without αCD3ϵ stimulation. Whole-cell lysates were prepared, and Notch1IC and GAPDH expression was detected by immunoblot. Data are representative of n = 4 (control) and n = 5 (GSI). (C) Graphic representation of band intensities from panel B. Expression of Notch1IC was normalized to GAPDH expression.
Reduced in vivo Notch signaling results in reduced Foxp3 expression within the Treg pool and spontaneous lymphocyte infiltration of the liver. For 3 months, wild-type mice were fed either normal rodent chow (control, n = 4) or chow formulated with LY 411575 (GSI, n = 5) to deliver 5 mg/kg per day. (A) Bulk splenocytes were stained for flow cytometry with antibodies specific for CD4 and Foxp3. Values represent mean percentages. (B) Histogram representing level of Foxp3 expression, gated on live CD4+Foxp3+ cells. Data are representative of data collected from 2 independent experimental groups (group 1: control, n = 2 and GSI, n = 3; group 2: control, n = 2 and GSI, n = 2). (C) Graphic representation of mean fluorescence intensity of Foxp3 staining in all experimental groups from panel B. Values represent the mean. (D-G) H&E staining of livers from mice with normal (D,F) or reduced (E,G) levels of Notch1 signaling. One representative liver section from each group is shown: (D) control mouse, (E) GSI chow-fed mouse, (F) wild-type mouse (n = 6), (G) Notch1 antisense mouse (n = 6). A Spot digital camera (Diagnostic Instruments, Sterling Heights, MI) mounted on a Zeiss Axioscope microscope (Carl Zeiss, Jena, Germany) using a 20× objective was used to acquire figures.
Reduced in vivo Notch signaling results in reduced Foxp3 expression within the Treg pool and spontaneous lymphocyte infiltration of the liver. For 3 months, wild-type mice were fed either normal rodent chow (control, n = 4) or chow formulated with LY 411575 (GSI, n = 5) to deliver 5 mg/kg per day. (A) Bulk splenocytes were stained for flow cytometry with antibodies specific for CD4 and Foxp3. Values represent mean percentages. (B) Histogram representing level of Foxp3 expression, gated on live CD4+Foxp3+ cells. Data are representative of data collected from 2 independent experimental groups (group 1: control, n = 2 and GSI, n = 3; group 2: control, n = 2 and GSI, n = 2). (C) Graphic representation of mean fluorescence intensity of Foxp3 staining in all experimental groups from panel B. Values represent the mean. (D-G) H&E staining of livers from mice with normal (D,F) or reduced (E,G) levels of Notch1 signaling. One representative liver section from each group is shown: (D) control mouse, (E) GSI chow-fed mouse, (F) wild-type mouse (n = 6), (G) Notch1 antisense mouse (n = 6). A Spot digital camera (Diagnostic Instruments, Sterling Heights, MI) mounted on a Zeiss Axioscope microscope (Carl Zeiss, Jena, Germany) using a 20× objective was used to acquire figures.
To examine the presence of lymphocyte infiltration, we harvested multiple tissues from GSI-treated and control animals. We found minor periportal lymphocytic infiltration in the liver of all GSI-treated animals, but not in control animals (Figure 5D,E). Lymphocyte infiltration of the liver is a common side effect of drug toxicity, however, we did not identify any changes in intestinal cells that have been associated with GSI toxicity (data not shown). Further examination of livers from Notch1 AS animals revealed a much more striking infiltration, which is to be expected since these animals have had reduced Notch1 signaling throughout their entire life span (Figure 5F,G). Lymphocyte infiltration of the liver was present in all Notch1 AS animals that we examined, but not in any of the WT controls. These data indicate that chronic in vivo GSI treatment results in reduced Foxp3 expression within the peripheral Treg pool and lymphocyte infiltration of the liver. In addition, the fact that infiltration is present in the livers of Notch1 antisense mice suggests that the infiltration seen is due to reduced Notch1 signaling, and is not a result of drug toxicity.
Transgenic mice expressing reduced levels of Notch1 have fewer peripheral, but not thymic, regulatory T cells
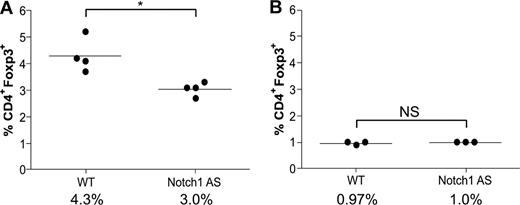
Transgenic mice deficient in TGFβ signaling have a normal number of thymic Tregs, but none in the spleen, indicative of a defect in the maintenance of the peripheral Treg pool. These data have led to the hypothesis that TGFβ1-induced Foxp3 up-regulation is necessary to maintain the peripheral Treg pool, which has previously been shown to be maintained throughout the life span of both humans and mice, despite having a faster doubling time, shorter telomeres, and reduced telomerase activity.31,32 Because lymphocyte infiltration was present in Notch1 AS mice (Figure 5F,G), we hypothesized they might have impaired peripheral Treg maintenance, similar to that of TGFβ-deficient mice. Bulk splenocytes and thymocytes were harvested from 6-week-old wild-type (WT) or Notch1 AS mice for analysis by flow cytometry. Indeed, we found a greater than 30% reduction in the number of splenic CD4+Foxp3+ regulatory T cells (4.3% vs 3.0%) in the AS mice compared with WT controls, whereas the number in the thymus was the same (0.97% versus 1.0%) (Figure 6A,B). These data indicate that Notch1 AS mice exhibit defective maintenance of their peripheral regulatory T-cell pool similar to that of mice deficient in TGFβ1 signaling, and this may contribute to the symptoms of autoimmune infiltration of the liver.
Defective regulatory T-cell maintenance in mice expressing reduced levels of Notch1. Bulk splenocytes (A) or thymocytes (B) were harvested from WT or Notch1 antisense (AS) mice and then stained with antibodies specific for CD4 and Foxp3 for analysis by flow cytometry. *P < .05. NS indicates not significant.
Defective regulatory T-cell maintenance in mice expressing reduced levels of Notch1. Bulk splenocytes (A) or thymocytes (B) were harvested from WT or Notch1 antisense (AS) mice and then stained with antibodies specific for CD4 and Foxp3 for analysis by flow cytometry. *P < .05. NS indicates not significant.
Discussion
There is previous evidence of genetic interactions between Notch, TGFβ signaling, and Fox-family proteins. Differentiation of the osteoblast cell line, MC3T3, with the TGFβ superfamily member, bone morphogenic protein-2 (BMP-2), induces the up-regulation of TGFβ1, Smad1, Foxm1, Foxf2, as well as the Notch-target gene Hey1.33 Furthermore, Notch has been shown to function downstream of Fox-family protein activity. Overexpression of Foxc1 and Foxc2 induces the expression of Notch1 and the Notch ligand Delta-like4 (Dll4).34 Foxf1 heterozygous mice have reduced Notch2 signaling, as defined by reduced Notch2 receptor and Hes1 expression, that results in lung hemorrhage-induced death.35 In addition, both Smad2/3/4 and Notch1IC are able to bind FoxO family members, which leads to target gene activation and repression, respectively.36,37 Recent studies have also indicated that Notch1 is required for TGFβ1-mediated immunosuppression by regulatory T cells, though a direct mechanism by which this occurs has not yet been defined.38,39
It was unexpected to find that GSI treatment inhibited Smad binding to the foxp3 promoter. Although another study has identified that there is no TGFβ1-responsive element in the foxp3 promoter using a luciferase reporter assay in the EL4 cell line, we have found that Smad does in fact bind to a Smad binding element in primary CD4+ T cells.25 Previous studies have found that Smad3 can bind putative CSL binding sites only in the presence of Notch1 and CSL, which led to the hypothesis that CSL binds DNA and Notch1 binds both CSL and Smad.21 Smad proteins have weak affinity for their consensus binding site and, thus, it is possible their affinity for the putative binding site in the foxp3 promoter is not sufficient to promote a stable interaction in the absence of a Notch1/CSL complex. Furthermore, both Notch1 and Smad have been shown to bind the coactivator CBP/p300, an interaction that could further stabilize the protein complex.40,41 Acetylation of Smad3, mediated by p300, is essential for its function. Smad1, which is phosphorylated in the context of BMP-2 signaling, can interact with Notch1IC only in the presence of p300 and PCAF (p300/CBP associated-factor), and the entire complex is required to actively transcribe Hes5.41,42 Together, these data indicate either that Notch1 is interacting directly with Smad3 to recruit and stabilize its presence at the foxp3 promoter or, perhaps, that their interaction results from binding to mutual coactivators. Although our chromatin immunoprecipitation data may appear to conflict with reports that CSL is bound with corepressors to Notch1-target gene promoters when Notch1IC is absent from the nucleus, a recent study using the Drosophila S2 cell line indicates that Notch activation does, in fact, induce a dramatic increase in CSL binding to target gene promoters,43 similar to what we observed with the foxp3 promoter. Furthermore, it is also possible that CSL remains bound to the foxp3 promoter in the presence of GSI treatment, but the binding of corepressors masks the epitope recognized by the antibody used for our ChIP assay.
The finding that GSI treatment does not inhibit acetylation of the foxp3 promoter indicates a dual role for Smad in foxp3 induction. Previous studies have found that, at early time points, Smad binds the foxp3 enhancer to induce acetylation both of the enhancer and promoter regions,25 an event that we found was not abrogated in the presence of GSI. At later time points, however, we found that Smad binds the foxp3 promoter in a Notch-dependent manner, indicating a role for TGFβ signaling both in the initiation as well as in the induction phase of foxp3 transcription. There is previous evidence of TGFβ functioning in an autocrine loop in T cells.44 Thus, it is entirely possible that while exogenous TGFβ1 is required for acetylation of the promoter and enhancer regions immediately following stimulation, it induces a feed-forward loop, which provides CD4+ T cell–intrinsic TGFβ1 that activates Smad3, which, once Notch1 is up-regulated, interacts with Notch1IC and CSL to bind the promoter region and directly induce transcription.
The presence of periportal lymphocyte infiltration in the liver, seen in mice with chronically reduced levels of Notch activation, is similar to that which is seen in mice deficient in Foxp3 and TGFβ1 signaling. Scurfy mice, which have a mutation in foxp3 leading to a premature stop codon, and TGFβ1- or TGFβRII-deficient animals display severe disease, characterized by splenomegaly, widespread autoimmunity, and lymphocyte infiltration of the skin, lung, pancreas, and liver.16-18,45 Mice with reduced Notch1 activation display normal organ pathology, except in the liver, where Notch1 AS mice display a disease slightly more severe than that of in vivo GSI-treated animals. Compared with scurfy mice and those deficient in TGFβ1 signaling, however, mice with reduced Notch1 signaling, either by pharmacologic or genetic means, display a much less severe disease, and remain viable with no physical symptoms of disease.
It is unclear why chronically reduced Notch1 activation results in infiltration in only the liver, but it is possible this is the tissue most sensitive to decreased Foxp3 expression, and global Notch levels were not reduced enough to elicit more widespread autoimmunity. Complete inactivation of Notch signaling using a conditional deletion in CD4+ T cells, or treatment with GSI either at a higher dose or for a longer period of time, may help shed light on this. Along these lines, mice with reduced presenilin gene dosage, which is the catalytic core of the γ-secretase complex, also display severe autoimmunity characterized by splenomegaly and infiltration both of the kidney and liver.46 Disease develops in these mice as adults, and it would be interesting to determine whether they exhibit a defect in Foxp3 expression and regulatory T-cell maintenance. Furthermore, it is also possible that, although GSI treatment did reduce Foxp3 expression, it also inhibited the TH1 and TH2 responses that would have been necessary to elicit an autoimmune response.
There is also a close link between TGFβ, Foxp3 expression, and autoimmune hepatitis. Mice with a conditional knockout of TGFβRII in CD4+ T cells or a complete knockout of Smad3 are more susceptible to disease induction in a mouse model of autoimmune hepatitis, compared with wild-type controls, and reduced expression of TGFβRII on peripheral blood mononuclear cells (PBMCs) in patients with autoimmune hepatitis at least partially contributes to disease pathology.47-49 Patients with autoimmune hepatitis also have reduced FOXP3 expression within the Treg pool, and diminished Foxp3 protein expression in murine cells has been associated with the onset of autoimmune disease.50,51
In summary, this study provides evidence that Foxp3 is a novel downstream target of Notch signaling both in vitro and in vivo. We propose a model whereby upon interaction with ligand, γ-secretase catalyzes the cleavage of the active form of Notch1, which translocates to the nucleus and recruits Smad3 and CSL to the foxp3 promoter to activate transcription in the context of maintaining the regulatory T-cell pool in the periphery. In the presence of GSI, however, Foxp3 expression is not induced, resulting in defective Treg maintenance and lymphocyte infiltration of the liver. This study brings to light a potential side effect of chronic GSI therapy, and will require extensive validation in a clinical setting to completely and confidently identify the risks and benefits of long-term GSI treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge members of the Osborne laboratory for helpful discussions, Brooke Bentley and Matthew Carter (Pioneer Valley Life Sciences Institute, Springfield, MA) for assistance in tissue embedding and histology, Justine Roderick for assistance with the in vivo GSI studies, and Stephen P. Naber MD, PhD (New England Medical Center, Boston, MA) for evaluating tissue pathology.
This work was supported by grants R01 AI49361 and PO1 AG/CA025531 (B.A.O.) and PO1 AG/CA025531 (T.E.G., A.F., and L.M.) from the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: J.B.S. designed and performed the experiments, analyzed the data, and wrote the paper; A.C. helped with the chromatin immunoprecipitation assay experiments; L.M. generated the Notch1 antisense transgenic mice; A.F. synthesized the γ-secretase inhibitors; P.D., T.E.G., and J.C.T. provided advice for experiments and edited the paper; and L.M.M. and B.A.O. supervised the project and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara A. Osborne, Department of Veterinary and Animal Sciences, University of Massachusetts Amherst, 311 Paige Laboratory, 161 Holdsworth Way, Amherst, MA 01003; e-mail: osborne@vasci.umass.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal