Abstract
Epstein-Barr virus (EBV) persists in healthy virus carriers within the immunoglobulin (Ig)D−CD27+ (class-switched) memory B-cell compartment that normally arises through antigen stimulation and germinal center transit. Patients with X-linked lymphoproliferative disease (XLP) lack such class-switched memory B cells but are highly susceptible to EBV infection, often developing fatal symptoms resembling those seen in EBV-associated hemophagocytic syndrome (EBV-AHS), a disease caused by aberrant virus entry into the NK- or T-cell system. Here we show that XLP patients who survive primary EBV exposure carry relatively high virus loads in the B-cell, but not the NK- or T-cell, compartment. Interestingly, in the absence of conventional class-switched memory B cells, the circulating EBV load was concentrated within a small population of IgM+IgD+CD27+ (nonswitched) memory cells rather than within the numerically dominant naive (IgM+IgD+CD27−) or transitional (CD10+CD27−) subsets. In 2 prospectively studied patients, the circulating EBV load was stable and markers of virus polymorphism detected the same resident strain over time. These results provide the first definitive evidence that EBV can establish persistence in the B-cell system in the absence of fully functional germinal center activity and of a class-switched memory B-cell compartment.
Introduction
Epstein-Barr virus (EBV), a predominantly B-lymphotropic herpesvirus, is the causative agent of infectious mononucleosis (IM), a self-limiting lymphoproliferative disease seen in some cases of primary infection, and is linked to a range of malignancies that arise as rare accidents of long-term virus carriage.1 Despite its pathogenic potential, EBV is widespread in human populations, is usually acquired subclinically, and is carried by the vast majority of individuals as a latent, asymptomatic infection of the recirculating B-cell pool. Fractionation of B-cell subsets in the blood of such carriers has shown that the virus is harbored selectively within IgD−CD27+ cells, that is, within the classical isotype-switched memory B-cell population.2,3 Physiologically, this population is produced from naive (IgD+IgM+CD27−) B cells by antigen stimulation and subsequent transit through a germinal center (GC) reaction. Within the GC, diversification of IgV gene sequences by somatic hypermutation, coupled with Ig class switching, leads to the selection of high-affinity antibody variants and the appearance of IgD−CD27+ memory cells expressing either IgG or IgA.4,5 Because EBV infects and transforms both naive and memory B cells apparently equally well in vitro,6 how EBV achieves selective colonization of the memory B-cell pool in vivo remains unclear. One hypothesis envisages preferential entry of the virus into naive B cells which, as a result of a transient growth-transforming infection mimicking antigen stimulation, are driven to form a GC; thereafter the physiologic processes of GC transit come into play and deliver latently infected GC progeny cells into the long-lived memory compartment.3,7-10 Another hypothesis, based largely on the study of individual infected cells in IM lymphoid tissues, questions the involvement of the GC reaction in this context and envisages either the preferential infection of memory cells or the preferential expansion/survival of memory cells after infection, compared with their naive counterparts.11-13
To inform this debate, one approach is to examine EBV infection in patients with immune deficiencies characterized by an inability to generate class-switched memory B-cell responses. Typically, the circulating B-cell pool of such patients is dominated by IgD+CD27− naive B cells. The remaining cells are immature bone marrow–derived transitional B cells (distinguished by CD10 expression and normally present in very low numbers in healthy donor blood) and a small population of IgM+IgD+CD27+ cells that express mutated IgV sequences14-16 ; these latter cells have been referred to as IgM-memory or nonswitched memory B cells.17,18 The origin of these atypical memory cells is unclear but, given their presence in such immune deficient patients, they may arise independently of a conventional GC pathway.18-20 Interestingly, such IgM+IgD+CD27+ memory B cells are also present in the blood and spleen of healthy individuals21,22 but, like the naive cell fraction, they are reportedly not a reservoir for EBV persistence.3
Here we focus on one such immune deficiency characterized by memory B-cell impairment, X-linked lymphoproliferative disease (XLP). This is associated with mutations in SH2D1A that encode SAP,23-25 a protein involved in the regulation of B cell–T cell interactions mediated by members of the signalling lymphocytic activation molecule (SLAM) family of cell surface receptors.26-28 SAP is selectively expressed in T and NK cells, rather than B cells,15 and SAP deficiency abrogates the delivery of CD4+ T-cell help to B cells necessary for the generation of fully functional GCs and hence for the production of class-switched memory B cells.14,29,30 One of the clinical symptoms shown by XLP, therefore, is hypogammaglobulinemia and an inability to mount IgG or IgA antibody responses; moreover, from the histologic analysis of splenic tissues, this is associated with an inability to form recognizable GC structures.15
Virus carriage in boys with XLP has not previously been analyzed in detail, not least because another consequence of SAP deficiency is an extreme susceptibility to primary EBV infection, such that many patients do not survive into the carrier state.31,32 Thus, exposure to EBV during childhood or adolescence often (but not always) leads to severe IM-like symptoms, with high fever, lymphadenopathy, and acute expansions of NK- and CD8+ T-cell populations in blood. Thereafter, in many cases the disease progresses to a fatal aplastic phase, first involving macrophage activation (thought to be initiated by NK and T cell–derived cytokines), then hemophagocytosis, and eventually bone marrow failure.28,33,34 At this point the clinical symptoms of XLP resemble those of another very rare complication of primary EBV infection, the EBV-associated hemophagocytic syndrome (EBV-AHS).35,36 Very interestingly, it now appears that EBV-AHS stems from the aberrant infection and virus-driven expansion of NK- and/or T-cell subsets in the blood, leading to a cytokine storm and ensuing macrophage activation and hemophagocytosis37 ; survivors of the acute infection thereafter harbor EBV in circulating NK or T cells.38 The aim of the present work, therefore, was to examine XLP patients who had survived their primary EBV infection and to ask 3 questions: (1) Is a stable virus carrier state established in these individuals? (2) If so, does this reflect virus persistence in the B-cell pool in the absence of a class-switched memory compartment, or is the virus elsewhere? (3) If there is persistence within B cells, is the virus colonizing all available B-cell subsets in the blood or just the “nonswitched,” potentially GC-independent, memory pool?
Methods
Donors
Peripheral blood samples were collected with informed consent obtained in accordance with the Declaration of Helsinki from healthy donors, from XLP patients, and from an EBV-AHS patient. All studies were approved by the Central Sydney Area Health Service Human Research Ethics Committee (Royal Prince Alfred Hospital, Sydney, Australia) and the South Birmingham (United Kingdom) Research Ethics Committee.
Cell surface staining and cell sorting
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Isopaque centrifugation. In initial experiments to isolate the B-cell and non–B-cell fractions from PBMCs, peripheral blood B cells were positively selected from PBMCs by immunomagnetic cell isolation using M-450 CD19 (Pan B) Dynabeads (Dynal Biotech, Oslo, Norway) followed by bead detachment. In subsequent experiments to identify transitional, naive and memory B cell populations, PBMCs were labeled with. FITC-conjugated anti-CD20 and PE-conjugated anti-CD27 monoclonal antibodies (mAbs, BD PharMingen, San Diego, CA) and with one of the following antibodies, either APC-conjugated anti-CD10 or biotinylated anti-IgM, anti-IgD, anti-IgG, or anti-IgA mAbs followed by streptavidin (SA)–peridinin chlorophyll-a protein (BD Biosciences PharMingen) as previously described.14 Data were collected on a FACScalibur flow cytometer (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (TreeStar, Ashland, OR). PBMCs triple-stained for CD20, CD10, and CD27 as above were then sorted by FACS to isolate transitional (CD20+CD10+CD27−), naive (CD20+CD10−CD27−), and total memory (CD20+CD10−CD27+) populations using a FACSAria (BD Biosciences) cell sorter. In further experiments to identify naive, nonswitched memory, and class-switched memory B cells, PBMCs were labeled with FITC-conjugated anti-CD20, PE-conjugated anti-IgD, PE-conjugated anti-IgM (BD PharMingen) and APC-conjugated anti-CD27 (eBioscience, San Diego, CA) mAbs and subsequently FACS-sorted to isolate naive (CD20+IgM+IgD+CD27−) and nonswitched memory (CD20+IgM+IgD+CD27+) populations.
Quantitative PCR assays for viral load
EBV genome load was assayed by quantitative real-time polymerase chain reaction (PCR) as described.39,40 Briefly, total genomic DNA was extracted from PBMCs or isolated B-cell subsets using a DNeasy Tissue kit (Qiagen, Valencia, CA) and triplicate aliquots of DNA (300 ng) were then subjected to a multiplex PCR using primer/probe combinations specific for the EBV BALF5 (POL) and the human β2 microglobulin (β2m) sequences. In parallel, serial DNA dilutions prepared from the Namalwa Burkitt lymphoma (BL) cell line (known to contain 2 integrated EBV copies per cell41 ) were also amplified and used to generate standard curves for the absolute quantitation of POL and β2m copy numbers in the starting samples. In each case, the number of POL copies per sample was normalized using the corresponding β2m value, and the final result expressed as EBV genomes per 106 cells.
Quantitative reverse-transcriptase PCR assays for viral gene expression
RNA extraction, cDNA synthesis and quantitative reverse-transcriptase (RT)–PCR assays for the EBV lytic gene transcripts BZLF1, BVRF2 and BLLF1 were performed essentially as described,42 with the following modifications. To ensure that RNA preparations were free of residual genomic DNA (which might generate false positive signals for the unspliced BVRF2 and BLLF1 transcripts), total RNA was subjected to a second DNAse I treatment (DNA-free; Ambion, Austin, TX). cDNA was prepared using a pool of primers specific for the viral transcripts BZLF1, BVRF2 and BLLF1, and the cellular GAPDH mRNA. In addition to the previously described primers and probe to amplify BZLF1 transcripts,42 we also designed new primer/probe combinations to amplify BVRF2 (forward primer 5′-CCACGGCAGTCTACGGTACA-3′, reverse primer 5′-GCGGCATTGGCGTCAT-3′, Taqman probe 5′-(FAM)-ACCTTGCGTGGGTCCTGAAGCACTT-(TAMRA)-3′) and BLLF1 (forward primer 5′-AGAATCTGGGCTGGGACGTT-3′, reverse primer 5′-ACATGG-AGCCCGGACAAGT-3′, Taqman probe 5′-(FAM)-AGCCCACCACAGATTACGGCGGT-(TAMRA)-3′) lytic gene transcripts. PCR reactions were prepared in a final volume of 25 μL containing 1x Taqman Universal PCR Mastermix (Applied Biosystems, Foster City, CA), 300 nM primers, 200 nM probe, 0.5 μL human GAPDH endogenous control (Applied Biosystems) and 5 μL cDNA. Relative quantitation of gene expression was performed as described,42 using serial dilutions of cDNA derived from a suitable EBV-positive reference line Sal tr-LCL. Template-negative and RT-negative samples served as controls and were always undetectable. In cell reconstruction experiments using RNA preparations from serial dilutions of the semipermissive B95.8 cell line (in which 10% cells were in lytic cycle) into an excess of EBV-negative BJAB cells, all 3 assays were capable of detecting the presence of a single lytically infected cell (data not shown).
Heteroduplex tracking assays
EBV isolates were characterized on the basis of sequence polymorphisms within the EBNA2 and LMP1 latent genes using established heteroduplex tracking assays (HTA).43,44 Aliquots of DNA from ex vivo PBMC samples were subjected to nested PCR amplification using primers specific for EBNA2 and LMP1. The subsequent product/probe binding reactions and heteroduplex analysis were performed as previously described.44
Results
High EBV loads in XLP patients involve the B-cell compartment
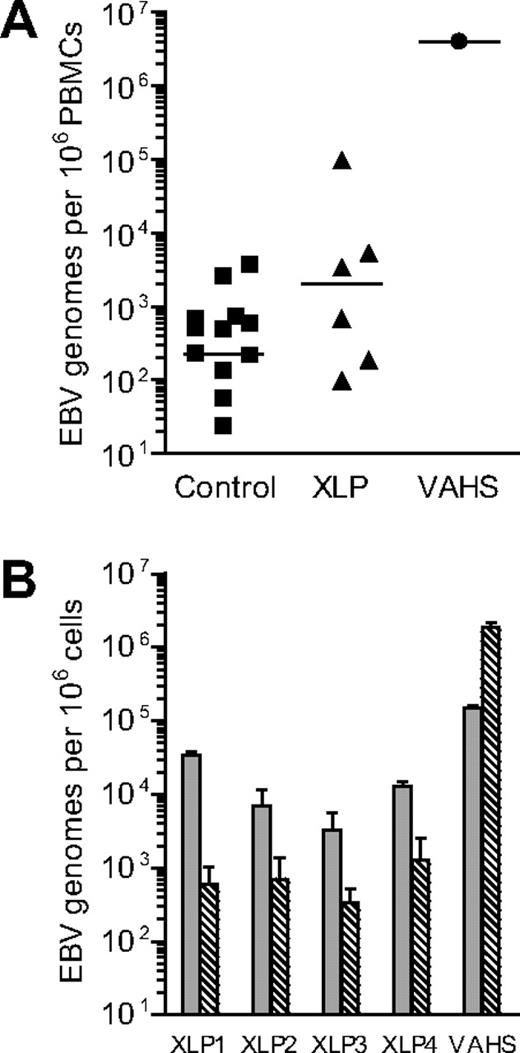
Eight XLP patients were studied during the course of this work. Table 1 gives each patient's age and clinical phenotype at time of sampling and indicates whether or not there was an earlier history of a fulminant IM episode. In an initial series of experiments, PBMCs from 6 of these patients (XLP1-6) were assayed for EBV DNA load by quantitative PCR alongside control samples from 12 healthy EBV carriers and, as a comparator, PBMCs from a rare patient who presented with acute EBV-AHS. As shown in Figure 1A, all 6 XLP patients' samples were found to be EBV-positive with levels of infection spread across an unusually wide range. Individual results are shown in Table 1. In particular, 3 of the 6 XLP patients had loads either at the top of or (for XLP5) greatly exceeding the normal donor range; the EBV-AHS patient also had a very high virus load. We then positively selected B cells from the PBMCs of 4 of these patients (XLP1-4) and of the EBV-AHS patient using CD19 antibody-coated Dynabeads, a procedure that routinely yields B cell–enriched preparations with more than 98% purity and leaves depleted preparations with less than 5% B-cell contaminants, as judged by staining for a second B-cell marker, CD20 (data not shown). These paired preparations were examined for EBV load, the data being shown in Figure 1B. Clearly the virus load in XLP patients was concentrated in the CD19+ B-cell fraction, in contrast to the situation in EBV-AHS where the dominant virus load was in non–B cells. Positive selection of CD3+ T cells or of CD56+ NK cells confirmed infection of these subsets in the EBV-AHS patient, whereas the same procedure showed no evidence of T- or NK-cell infection in the XLP patients (data not shown).
Features of X-linked lymphoproliferative disease patients
| Patient . | Age, yrs . | Clinical phenotype . | History of fulminant IM . | EBV genomes per 106 PBMCs . |
|---|---|---|---|---|
| XLP1 | 12 | Hypogammaglobulinemia | Yes | 5.2 × 103 |
| XLP2 | 10 | Hypogammaglobulinemia | No | 1.9 × 102 |
| XLP3 | 31 | T cell disorder, vasculitis | No | 1.0 × 102 |
| XLP4 | 5 | Hypogammaglobulinemia | No | 3.4 × 103 |
| XLP5 | 40 | Hypogammaglobulinemia | Yes | 9.7 × 104 |
| XLP6 | 46 | Hypogammaglobulinemia | Yes | 6.7 × 102 |
| XLP7 | 49 | Hypogammaglobulinemia, lymphoma, hemophagocytosis | Yes | NT |
| XLP8 | 28 | Eosinophilia | No | NT |
| Patient . | Age, yrs . | Clinical phenotype . | History of fulminant IM . | EBV genomes per 106 PBMCs . |
|---|---|---|---|---|
| XLP1 | 12 | Hypogammaglobulinemia | Yes | 5.2 × 103 |
| XLP2 | 10 | Hypogammaglobulinemia | No | 1.9 × 102 |
| XLP3 | 31 | T cell disorder, vasculitis | No | 1.0 × 102 |
| XLP4 | 5 | Hypogammaglobulinemia | No | 3.4 × 103 |
| XLP5 | 40 | Hypogammaglobulinemia | Yes | 9.7 × 104 |
| XLP6 | 46 | Hypogammaglobulinemia | Yes | 6.7 × 102 |
| XLP7 | 49 | Hypogammaglobulinemia, lymphoma, hemophagocytosis | Yes | NT |
| XLP8 | 28 | Eosinophilia | No | NT |
EBV indicates Epstein-Barr virus; IM, infectious mononucleosis; NT, not tested; and PBMCs, peripheral blood mononuclear cells.
Analysis of EBV genome load in healthy control donors and patients with EBV-associated disease. (A) Results of quantitative PCR analysis to determine the EBV load in PBMC samples isolated from control donors, from XLP patients or from an EBV-AHS (VAHS) patient. Data are reported as EBV genome copies per 106 PBMCs, with the median value for each group denoted by the horizontal bar. (B) Results of quantitative PCR analysis to determine the EBV load in CD19+ B-cell ( ) and CD19− non–B-cell (
) and CD19− non–B-cell ( ) fractions isolated from XLP and VAHS PBMC samples. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) fractions isolated from XLP and VAHS PBMC samples. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
Analysis of EBV genome load in healthy control donors and patients with EBV-associated disease. (A) Results of quantitative PCR analysis to determine the EBV load in PBMC samples isolated from control donors, from XLP patients or from an EBV-AHS (VAHS) patient. Data are reported as EBV genome copies per 106 PBMCs, with the median value for each group denoted by the horizontal bar. (B) Results of quantitative PCR analysis to determine the EBV load in CD19+ B-cell ( ) and CD19− non–B-cell (
) and CD19− non–B-cell ( ) fractions isolated from XLP and VAHS PBMC samples. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) fractions isolated from XLP and VAHS PBMC samples. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
While healthy individuals are known to carry EBV in the circulating B-cell pool as an exclusively latent infection,3 it remained possible that the situation in XLP patients may be different and that ongoing virus replication might be contributing to the often elevated viral loads seen in the above patients. To examine this, we developed sensitive quantitative RT-PCR assays for 3 EBV lytic cycle transcripts, the immediate early gene BZLF1, and 2 late genes BVRF2 (protease) and BLLF1 (gp350). These assays were each capable of detecting the presence of a single lytically infected B95.8 cell in in vitro reconstructed cell mixtures. However, these same assays never detected the presence of such transcripts in RNA isolated from the purified B cells of XLP patients, including B cells from XLP5, the patient with the highest viral load (data not shown). We infer from this that virus load assays are truly reflecting the latent genome load in the circulating B cells of these patients.
EBV is concentrated in the nonswitched memory B cells of XLP patients
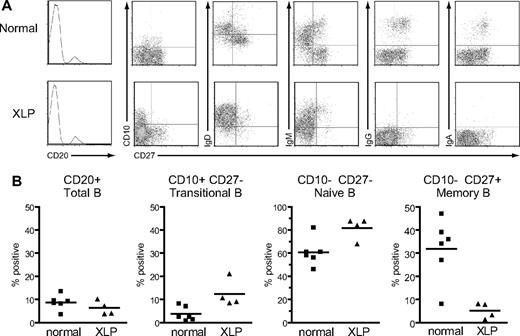
Four of the available XLP patients from whom larger blood samples could be obtained (XLP5-8) were chosen for more detailed analysis and compared with 6 healthy donor controls. Their PBMCs were costained with anti-CD20 and anti-CD27 mAbs to identify naive (ie, CD20+ CD27−) and memory (ie, CD20+ CD27+) B cells, and with a third antibody against either surface IgD, IgM, IgG or IgA, or a marker that distinguishes the transitional B-cell population, CD10.45,46 Representative FACS plots from one XLP patient and a healthy control are shown in Figure 2A and overall results are summarized in Figure 2B. In agreement with a previous study,14 total B-cell numbers were not significantly different between patients and healthy controls. However, the distribution of B-cell subsets was quite different. The percentage of circulating B cells with the CD10+ CD27− transitional phenotype was significantly higher in XLP patients than controls (mean of 12% vs 4%),16 as was the percentage of CD10−CD27− naive B cells (82% vs 61%). Conversely CD10−CD27+ memory B-cell numbers were greatly reduced in XLP patients compared with controls (5% vs 32%). Furthermore, confirming earlier observations on such patients,14,15 the vast majority of these CD27+ B cells in XLP blood were nonswitched memory cells that were IgD+IgM+ and lacked IgG or IgA expression, in contrast to the situation in the control donor (Figure 2A).
Surface phenotype of B cells from healthy control donors and XLP patients. (A) PBMCs from a healthy control donor and a representative XLP patient, XLP5, were stained with anti-CD20 mAb, and the frequency of B cells in the lymphocyte population determined (leftmost panels; frequency shown as percentage). In the remaining panels, purified CD20+ B cells were dual-stained with mAbs specific for CD27 and either CD10, IgD, IgM, IgG, or IgA to obtain the FACS profiles shown. (B) Summary of B-cell surface phenotype data obtained from 6 healthy control donors and 4 XLP patients (XLP5-8). Results are expressed (left to right) as the percentage of PBMCs that were CD20+ B cells, and the percentage of CD20+ B cells that had the transitional, naïve, or memory cell phenotypes shown. The mean value for each group is denoted by the horizontal bar.
Surface phenotype of B cells from healthy control donors and XLP patients. (A) PBMCs from a healthy control donor and a representative XLP patient, XLP5, were stained with anti-CD20 mAb, and the frequency of B cells in the lymphocyte population determined (leftmost panels; frequency shown as percentage). In the remaining panels, purified CD20+ B cells were dual-stained with mAbs specific for CD27 and either CD10, IgD, IgM, IgG, or IgA to obtain the FACS profiles shown. (B) Summary of B-cell surface phenotype data obtained from 6 healthy control donors and 4 XLP patients (XLP5-8). Results are expressed (left to right) as the percentage of PBMCs that were CD20+ B cells, and the percentage of CD20+ B cells that had the transitional, naïve, or memory cell phenotypes shown. The mean value for each group is denoted by the horizontal bar.
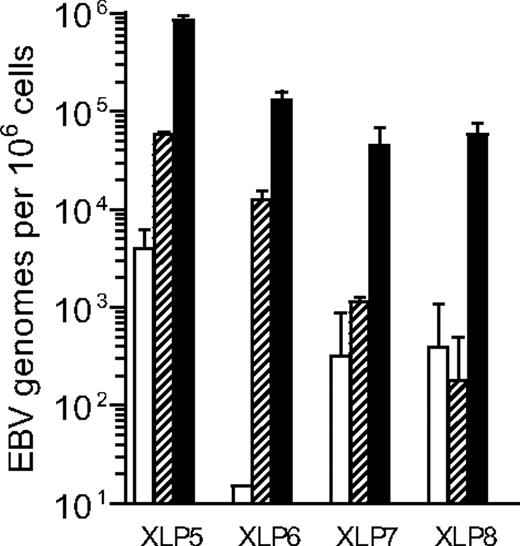
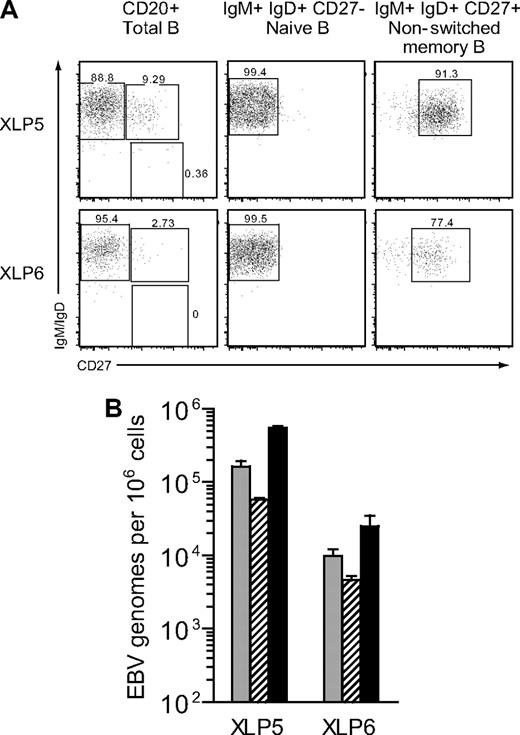
The PBMC samples from XLP patients that had been triple-stained for CD20, CD10, and CD27 were then sorted by FACS to isolate the transitional (CD20+CD10+CD27−), naive (CD20+CD10−CD27−), and memory (CD20+CD10−CD27+) cell populations; these gave typical sort purities of 95% to 98% for each of the subsets, as published.14-16 Figure 3 illustrates the data obtained when these different subsets were analyzed for EBV DNA load and shows that, in each case, the virus was highly enriched in the memory subset. In particular, the virus load in memory cells was always more than 100-fold higher than in transitional cells and, depending on the patient, was between 10- and 300-fold higher than in naive cells. As shown in earlier work,15 the CD27+ memory B-cell subset in the peripheral blood of XLP patients is dominated by cells coexpressing both IgM and IgD, with extremely small numbers of cells staining positively for IgG+ or IgA+. It therefore seemed very unlikely that such a barely detectable class-switched population could account for the very high virus loads observed in these patients. However, to address this point specifically, we obtained further blood samples from 2 of these XLP patients (XLP5, XLP6) and in this case stained their PBMCs with anti-CD20, -IgM, -IgD and -CD27 mAbs. Figure 4A confirms that, in both patients, the circulating CD20+ B-cell pool is dominated by naive (IgM+IgD+CD27−) cells, with smaller numbers of nonswitched memory (IgM+IgD+CD27+) cells and virtually no detectable class-switched (IgM−IgD−CD27+) cells. Accordingly, in subsequent cell-sorting experiments, we could not isolate a class-switched cell population from these samples, whereas we could readily obtain naive and nonswitched memory populations. With this sorting protocol, reanalysis showed that the naive cell sorts were of high purity, while the nonswitched memory cells gave purities of 91% for XLP5 and 77% for XLP6, the lower XLP6 value reflecting the difficulty of sorting a population that is at low frequency in the initial sample. In both cases, however, the vast majority of the contaminants within the nonswitched population are IgM+IgD+CD27− naive cells. Figure 4B shows the corresponding EBV DNA load data from these sorted populations compared with the load seen in unfractionated B cells from the same patient. In both cases the virus was predominantly found in the nonswitched memory population.
Analysis of EBV genome load in B-cell subsets isolated from XLP blood. Results of quantitative PCR analysis to determine the EBV load in purified CD20+ CD10+ CD27− (transitional; □), CD20+ CD10− CD27− (naïve;  ) and CD20+ CD10− CD27+ (memory; ■) B cells. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) and CD20+ CD10− CD27+ (memory; ■) B cells. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
Analysis of EBV genome load in B-cell subsets isolated from XLP blood. Results of quantitative PCR analysis to determine the EBV load in purified CD20+ CD10+ CD27− (transitional; □), CD20+ CD10− CD27− (naïve;  ) and CD20+ CD10− CD27+ (memory; ■) B cells. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) and CD20+ CD10− CD27+ (memory; ■) B cells. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
Analysis of EBV genome load in IgM memory B cells isolated from XLP blood. (A) PBMCs from 2 XLP patients, XLP5 and XLP6, were stained with mAbs specific for CD20, IgM, IgD and CD27 to identify naive (CD20+IgM+IgD+ CD27−), nonswitched memory (CD20+IgM+IgD+CD27+) and class-switched memory (CD20+IgM−IgD−CD27+) B-cell populations; numbers indicate the percentage distribution of CD20+ B cells between the 3 populations (left panel). Naive and nonswitched memory cells were subsequently isolated by FACS sorting and the isolated populations reanalyzed, giving percentage purities as shown (center and right panels). (B) The histogram shows the results of quantitative PCR analysis to determine the EBV load in unfractionated CD20+ B cells ( ), naive B cells (
), naive B cells ( ) and nonswitched memory B cells (■) isolated from the 2 donors shown in panel A above. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) and nonswitched memory B cells (■) isolated from the 2 donors shown in panel A above. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
Analysis of EBV genome load in IgM memory B cells isolated from XLP blood. (A) PBMCs from 2 XLP patients, XLP5 and XLP6, were stained with mAbs specific for CD20, IgM, IgD and CD27 to identify naive (CD20+IgM+IgD+ CD27−), nonswitched memory (CD20+IgM+IgD+CD27+) and class-switched memory (CD20+IgM−IgD−CD27+) B-cell populations; numbers indicate the percentage distribution of CD20+ B cells between the 3 populations (left panel). Naive and nonswitched memory cells were subsequently isolated by FACS sorting and the isolated populations reanalyzed, giving percentage purities as shown (center and right panels). (B) The histogram shows the results of quantitative PCR analysis to determine the EBV load in unfractionated CD20+ B cells ( ), naive B cells (
), naive B cells ( ) and nonswitched memory B cells (■) isolated from the 2 donors shown in panel A above. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
) and nonswitched memory B cells (■) isolated from the 2 donors shown in panel A above. Data (mean and SD of 3 replicates) are reported as EBV genome copies per 106 cells.
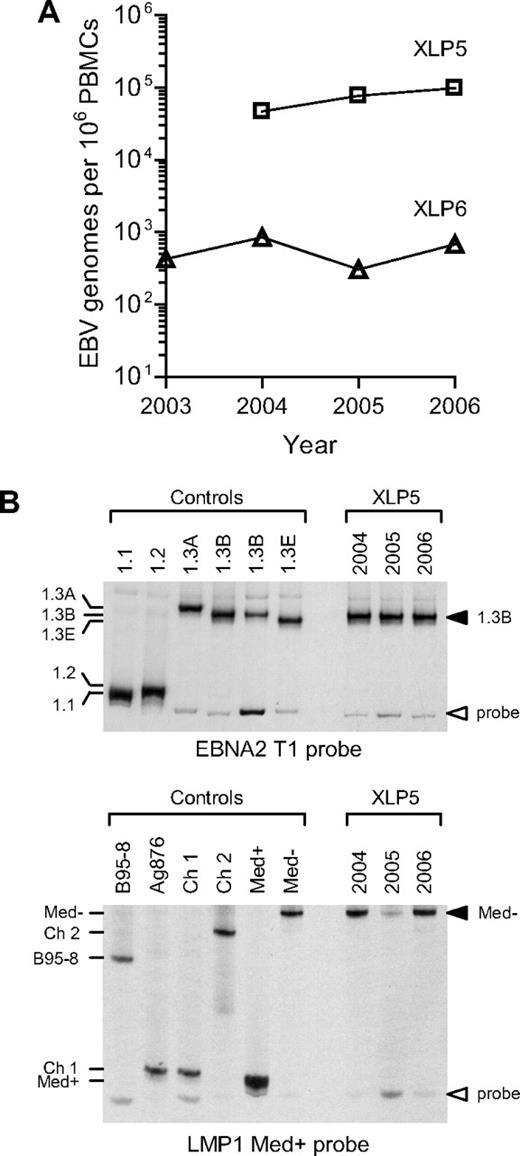
Stability of the virus load and of the resident EBV strain in XLP patients
From these 2 patients (XLP5 and XLP6), in whom the virus had been definitively localized to the nonswitched memory B-cell pool, successive bleeds had been taken and PBMCs stored over periods of 3 and 4 years, respectively. These samples were screened in parallel by quantitative PCR, and the EBV load data are shown in Figure 5A. Although the 2 patients had quite different EBV load values, each retained a unique characteristic load at a roughly stable level over the period of study. Such stability is consistent with the view that these patients are maintaining their EBV-positive status through persistence of a long-term infection rather than through a series of transient infections, each of which is cleared and then replaced by another exogenously acquired EBV strain. A more definitive resolution of the point became possible with the development of heteroduplex tracking assays to identify the resident EBV strain or strains within an individual. These assays involve amplifying viral DNA from ex vivo samples across nonlinked polymorphisms in the EBNA2 and LMP1 regions in the virus genome and in each case identifying the allelic products through the mobility of heteroduplexes that form with a standard labeled probe.44 We applied these assays to the successive blood samples from the above 2 patients. Figure 5B shows the data obtained from XLP5 DNA samples taken over a 3-year period, the amplification products being run alongside those from reference EBV strains representing 5 different alleles of the EBNA2 locus commonly seen among type 1 EBV strains and 5 of the known alleles of the LMP1 locus. At each time point, the virus strain present in patient XLP5 had a 1.3B allele at the EBNA2 locus and a Med− allele at the LMP1 locus. The correct identification of these alleles was later confirmed by DNA sequence analysis (data not shown). The second prospectively studied patient gave similar results, with a single EBNA2 and a single LMP1 allelic sequence maintained throughout (data not shown).
Analysis of EBV genome load and EBV sequences present in XLP patients. (A) The graph shows the results of quantitative PCR analysis to determine the EBV load in PBMC samples collected prospectively from patients XLP5 and XLP6 between 2003 and 2006. (B) Analysis of EBV sequences present in the same PBMC samples from patient XLP5 using EBNA2 and LMP1 HTAs. In the top panel, PBMC DNA was PCR-amplified using type 1 specific EBNA2 primers and the resulting PCR products analyzed by HTA using a 1.1 EBNA2 allele-specific probe. Also shown are the results from a panel of reference control isolates known to carry a 1.1, 1.2, 1.3A,1.3B or 1.3E EBNA2 allele. In the bottom panel, the same DNA samples were PCR-amplified using LMP1-specific primers and the resulting PCR products analyzed by HTA using a Med+ LMP1 allele-specific probe. Also shown are the results from a panel of reference control isolates known to carry a B95-8, Ag876/Ch1, Ch2, Med+ or Med− LMP1 allele. The results show that each XLP5 PBMC sample contains detectable 1.3B EBNA2 and Med− LMP1 alleles.
Analysis of EBV genome load and EBV sequences present in XLP patients. (A) The graph shows the results of quantitative PCR analysis to determine the EBV load in PBMC samples collected prospectively from patients XLP5 and XLP6 between 2003 and 2006. (B) Analysis of EBV sequences present in the same PBMC samples from patient XLP5 using EBNA2 and LMP1 HTAs. In the top panel, PBMC DNA was PCR-amplified using type 1 specific EBNA2 primers and the resulting PCR products analyzed by HTA using a 1.1 EBNA2 allele-specific probe. Also shown are the results from a panel of reference control isolates known to carry a 1.1, 1.2, 1.3A,1.3B or 1.3E EBNA2 allele. In the bottom panel, the same DNA samples were PCR-amplified using LMP1-specific primers and the resulting PCR products analyzed by HTA using a Med+ LMP1 allele-specific probe. Also shown are the results from a panel of reference control isolates known to carry a B95-8, Ag876/Ch1, Ch2, Med+ or Med− LMP1 allele. The results show that each XLP5 PBMC sample contains detectable 1.3B EBNA2 and Med− LMP1 alleles.
Discussion
The seminal observation that EBV persistence in healthy virus carriers is associated with selective colonization of the IgD−CD27+ memory B-cell pool2,3 has prompted much debate as to how that selectivity is achieved. One hypothesis envisages that EBV infection of a IgD+CD27− naive B cell can recapitulate the physiologic process of antigen-induced maturation, such that transient activation of full virus latent gene expression drives the infected cell into a GC reaction. From this arise EBV-positive progeny cells that have acquired the IgV-mutated genotype and IgD−CD27+ phenotype typical of isotype-switched memory.7 Another hypothesis denies any GC involvement in virus colonization of the B-cell system and envisages that the preexisting memory B-cell pool is either preferentially infected in vivo or is capable of preferential expansion/survival once infected, thereby outnumbering naive B- cell infections.13 The present work sought to inform this debate by studying EBV infection in patients with XLP, an immune deficiency characterized by an inability to form conventional GC structures and to mount isotype-switched antibody responses. The main conclusions are (1) that XLP patients are capable of sustaining a persistent EBV infection in the absence of an isotype-switched memory population, and (2) that this infection is associated with preferential colonization of nonswitched IgM+IgD+CD27+ memory B cells, an apparently GC-independent population that also exists in healthy individuals where, reportedly,3 it does not harbor the virus.
The genetic defect in XLP involves the SH2D1A gene encoding SAP,23-25 a protein normally expressed in T cells, NK cells and NK/T cells but, as far as is known, not in other lymphoid and myeloid lineages. SAP interacts with the cytoplasmic domains of members of the SLAM family of surface receptors.26-28 This multiplicity of interactions with different cell signaling receptors produces a complex immunodeficiency characterized by 3 main features: (1) a failure of NK-T cell development, (2) a failure to generate conventional GCs and to mount isotype-switched antibody responses in response to antigenic challenge, and (3) an unusual susceptibility to primary EBV infection, associated with aberrant polyclonal activation of CD8+ T-cell and NK-cell responses and leading to cytokine-mediated macrophage activation and an often fatal hemophagocytosis. Of these, the block in NK- and T-cell development28,47 and the impairment in eliciting long-lived humoral immune responses29,30 are reproduced in SAP-deficient mice. As to the susceptibility to primary infection, while there is no murine equivalent of EBV (a γ-1 herpesvirus), challenging SAP-deficient animals with the murine γ-2 herpesvirus MHV 68 or with a virus known to elicit strong CD8+ T-cell responses (lymphocytic choriomeningitis virus) likewise produces immunopathology from unrestrained responses of CD8+ T cells.48,49 Just as the factors influencing susceptibility to IM in immunocompetent individuals are poorly understood,50,51 it is not clear why some SAP-deficient patients die from primary EBV infection, others develop a severe IM-like illness and survive, while others appear to acquire EBV subclinically.52 The rarity of the XLP trait and the focus on understanding the pathogenesis of the fatal IM have, until now, diverted attention away from analysis of the longer-term fate of the EBV infection in surviving patients. Here we have addressed this issue in a cohort of patients with genetically confirmed SAP deficiency, all of whom lacked a detectable isotype-switched memory B-cell population in blood and displayed one or more clinical features of the XLP syndrome. Some of these patients had a clinical history of a fulminant IM-like episode after primary exposure to EBV; others did not.
We first showed by quantitative PCR for the viral genome that all patients did carry EBV in the circulating PBMC population. In some cases, the virus load was well within the normal range, while in other cases it was unusually high; although patient numbers were small, it is interesting to note that individuals with the higher loads were not necessarily those who had a prior history of fulminant IM (Table 1). As to which cells were infected, we were first prompted to compare B cells versus non-B cells because of the strong clinical parallels between the fulminant IM syndrome seen in XLP patients and primary EBV infection manifesting as EBV-AHS, a disease now shown to be caused by rare entry of the virus into T and/or NK cells.53 We found that SAP-deficient patients did indeed carry the virus in B cells and not other cell types. Furthermore, as in healthy carriers, the virus appeared to be carried as a latent infection since we could find no evidence of lytic gene expression in circulating B cells even using very sensitive quantitative RT-PCR assays. Thereafter, sorting of B-cell subsets clearly showed marked concentration of the virus in the small subset of circulating B cells with a CD10−CD27+ memory phenotype. By comparison, loads were 10- to 300-fold lower in the numerically dominant naive B-cell pool (CD10−CD27−) and consistently more than 100-fold lower in transitional B cells (CD10+CD27−), an immature population that is present as a minor subset in normal blood but is expanded in SAP-deficient patients.16,45 While the very low signals seen within sorted transitional cell populations almost certainly reflect low level contamination of these preparations by a few CD27+ cells, we cannot discount the possibility that where naive B-cell loads were only 10-fold lower than in the CD27+ subset, for example in XLP6, this could reflect some naive B-cell infection. The central point, however, is that the virus is being preferentially carried in a CD27+ B-cell population that, as earlier work has shown15 and the present staining data confirm, is dominated by nonswitched IgM+IgD+ memory B cells and is essentially devoid of isotype-switched cells. Indeed, in 2 cases we deliberately FACS-sorted this nonswitched memory population away from any possible isotype-switched contaminants and showed that it did indeed harbor the virus.
There are some parallels between the above findings and those described in an earlier study of EBV infection in patients with hyper-IgM syndrome arising from CD40L deficiency.54 These patients also are unable to form conventional GCs and lack isotype-switched memory B cells.20,55,56 In that study, virus DNA could be detected on at least one occasion in the blood and/or throat washings of 6 of 9 patients analyzed. However, the levels were low, often close to the threshold of detection, and in only 1 blood sample could viral infection be localized to the IgM+IgD+CD27+ population.54 Furthermore prospective sampling showed that, in contrast to healthy carriers included as controls, most CD40L-deficient patients did not give consistent evidence of virus infection. Only 1 patient was positive on more than 1 occasion and, even in this case, the choice of an EBNA3C gene repeat sequence as the marker of strain identity meant that persistence of the same EBV strain could not be convincingly shown. The findings from CD40L-deficient patients were therefore interpreted to suggest that infections may be occurring transiently in these individuals and that persistence within the B-cell pool requires entry into the isotype-switched memory compartment.54 By contrast, here we have used a sensitive PCR assay to analyze B-cell subsets sorted from 4 different XLP patients; the data clearly show that EBV can colonize the IgM+IgD+CD27+ population in patients lacking classic memory B cells and can do so at high levels. We also addressed the question of virus persistence using blood samples taken prospectively over 3 to 4 years from 2 different XLP patients, one with an unusually high load in IgM+IgD+CD27+ B cells, the other with a lower load. Those particular loads were maintained at roughly stable levels over the whole period of study. Moreover, using 2 independent and highly polymorphic markers of virus strain identity situated within the EBNA2 and LMP1 genes, respectively, we found both patients retained one characteristic strain throughout. Such constancy of virus load and of virus identity is indicative of a persistent rather than a recurring infection.
The present findings therefore demonstrate that, in patients who lack conventional isotype-switched memory B cells and who from histologic analysis of splenic tissues15 cannot make conventional GCs, EBV can establish a stable infection and does so by selectively colonizing an unusual B-cell subset with an IgM+IgD+CD27+ phenotype. Although such B cells are also present both in the blood and splenic marginal zone of healthy immunocompetent individuals, their presence in XLP and CD40L-deficient patients has led some investigators to propose that they are not antigen-instructed memory cells in the true sense; rather, it is suggested, they are B cells that have undergone somatic hypermutation during generation of the preimmune repertoire and that later mount antibody responses to T cell–independent antigens only.19,20 As such, these cells would be GC-independent in both their generation and function. From this viewpoint, therefore, the detection of EBV within this population in XLP patients might be used to argue against any involvement of the GC reaction in virus colonization of the B-cell system. However we would caution against unequivocal interpretations until the issues of the ontogeny, specificity, and function of IgM+IgD+CD27+ B cells are fully resolved.18,57,58 In particular, while conventional GCs are absent from the spleens of SAP-deficient patients, it remains possible that atypical transient GC-like structures (like those seen in mice responding to T-independent antigens59 ) can form in the absence of T-cell help and support a degree of somatic hypermutation without subsequent isotype switching.18 That would be consistent with the lower number of cell divisions that IgM+IgD+CD27+ cells have undergone in vivo60 and with their lower levels of IgV mutation21,22 compared with isotype-switched memory. It is, therefore, still possible that EBV infection of a naive cell might commit that cell to initiate a transient GC-like reaction which, in a SAP-deficient patient, produces EBV-positive nonswitched memory cells, but which in an immunocompetent individual goes through to completion and delivers the infected cell into the isotype-switched memory subset.
We would stress that the present findings were made in individuals with a complex immunodeficiency, XLP, where an impairment of specific cytokine production by TH2-like CD4+ T cells abrogates isotype switching in antigen-driven B-cell responses.14 The relevance of such findings to the situation in the normal healthy host remains to be seen. The evidence to date suggests that EBV selectively colonizes only class-switched (IgD−CD27+) B cells in healthy carriers.3 In light of our present findings, the EBV status of the nonswitched subset also deserves close scrutiny. Interestingly, IgM+IgD+CD27+ B-cell numbers are 3 to 5 times higher in the blood of healthy individuals than in XLP patients.14 This has led to the suggestion that there are in fact 2 types of IgM+IgD+CD27+ B cells: the minority, which arise independently of T-cell help and are the only ones seen in XLP or CD40L-deficient patients; and the majority population, which is T cell–dependent and numerically dominant in healthy individuals.18 As yet, however, there is no obvious phenotypic or functional evidence of such heterogeneity.18 Selective colonization of one population but not the other by EBV could, if confirmed, provide a means of distinguishing between these Igm+IgD+CD27+ B-cell subsets.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank all patients for their participation in this study and the following clinicians for their cooperation: Drs S. Adelstein, F. Alvaro, P. Arkwright, J. Cohen, B. Gaspar, A. Klion, S. Riminton, and P. Rohrlich.
This work was supported by grants from Cancer Research UK, the National Health and Medical Research Council (NHMRC) of Australia and Cancer Council New South Wales. S.C. was funded by a Leukemia Research Fund Clinical Research Fellowship. C.S.M. and SG.T. are recipients of Research Fellowships from the NHMRC.
Authorship
Contribution: S.C., C.S.M., A.I.B., D.C.-C., and A.D.H. performed the experiments; S.C., C.S.M., and A.I.B. analyzed the data and prepared the figures; and S.G.T. and A.B.R. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan Rickinson, Cancer Research UK Institute for Cancer Studies, University of Birmingham, Vincent Drive, Edgbaston, Birmingham, B15 2TT, United Kingdom; e-mail: a.b.rickinson@bham.ac.uk.
References
Author notes
*S.C. and C.S.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal