Abstract
Mutations in several ribosomal proteins (RPs) lead to Diamond-Blackfan anemia (DBA), a syndrome characterized by defective erythropoiesis, congenital anomalies, and increased frequency of cancer. RPS19 is the most frequently mutated RP in DBA. RPS19 deficiency impairs ribosomal biogenesis, but how this leads to DBA or cancer remains unknown. We have found that rps19 deficiency in ze-brafish results in hematopoietic and developmental abnormalities resembling DBA. Our data suggest that the rps19-deficient phenotype is mediated by dysregulation of deltaNp63 and p53. During gastrulation, deltaNp63 is required for specification of nonneural ectoderm and its up-regulation suppresses neural differentiation, thus contributing to brain/craniofacial defects. In rps19-deficient embryos, deltaNp63 is induced in erythroid progenitors and may contribute to blood defects. We have shown that suppression of p53 and deltaNp63 alleviates the rps19-deficient phenotypes. Mutations in other ribosomal proteins, such as S8, S11, and S18, also lead to up-regulation of p53 pathway, suggesting it is a common response to ribosomal protein deficiency. Our finding provides new insights into pathogenesis of DBA. Ribosomal stress syndromes represent a broader spectrum of human congenital diseases caused by genotoxic stress; therefore, imbalance of p53 family members may become a new target for therapeutics.
Introduction
The precise control of ribosome biogenesis and translation is vital for cell survival. Cell growth and proliferation as well as responses to genotoxic stress depend on ribosomal activity. Defective ribosomal synthesis has been associated with bone marrow failure syndromes, such as Diamond-Blackfan anemia (DBA), dyskeratosis congenita, and Shwachman-Bodian-Diamond syndrome.1 DBA is a congenital syndrome characterized by anemia, bone marrow erythroblastopenia, and increased incidence of cancer.1-3 Anemia is often accompanied by growth retardation and various malformations. Ribosomal protein S19 (RPS19) is mutated in one fourth of patients4 ; mutations of other ribosomal proteins can also lead to DBA.5,6
Red blood cells of DBA patients manifest altered transcription, translation, and activated apoptotic pathways.7-9 RPS19 is necessary for ribosome biosynthesis. Certain RPS19 mutations affect its structure and assembly into the ribosome.10,11 In yeast and human, deficiency of RPS19 impairs the processing of 18S rRNA and aberrant pre-40S particles are retained in the nucleolus.12-14 Nucleolar organization is distorted in skin fibroblasts in all DBA patients regardless of the presence of RPS19 mutations,13 suggesting that defect in ribosome biogenesis is a general feature of DBA. The nucleolus can act as a stress sensor; its disruption mediated p53 up-regulation.15 Mutations in other proteins participating in ribosome biogenesis lead to p53 up-regulation.16-18 Haploinsufficiency of RPS6 and RPL22 also leads to activation of p53.19,20 Hence, we hypothesized that up-regulation of p53 pathway is a common response to ribosomal proteins deficiency and is involved in molecular pathogenesis of DBA.
There is currently no animal model of DBA. In mouse, a homozygous deletion of rps19 was lethal before implantation; heterozygous mice, however, fully compensated for the loss of one rps19 allele.21 Deficiency of RPS19 created in cell lines with RNAi technology leads to cellular defects mimicking those in DBA.22 Nonetheless, cellular models cannot take into account the role of RPS19 during development. DBA is a congenital disease, which implies that the abnormal phenotype forms during early development. Therefore, there is still a need for a true animal model to understand the impact of RPS19 deficiency during embryogenesis. To easily follow all developmental stages, it is advantageous to use zebrafish. Antisense morpholino nucleotides have been successfully used in zebrafish to knockdown various genes.23 The use of this technology for ribosomal genes including RPS19 has been recently demonstrated.24
To this end, we created rps19-deficient zebrafish using antisense morpholinos. In this model, we evaluated how RPS19 deficiency affected early development. An advantage of the morpholino knockdown approach is that a gradual change in RPS19 deficiency can be achieved by changing the dose of injected morpholinos. Similar to the human disease, rps19 deficiency in zebrafish leads to failure of erythropoiesis and to developmental abnormalities. Our data suggest that dysregulation of p53 family contributes to DBA.
Methods
Use of zebrafish was approved by UCLA Animal Committee.
RNA synthesis
cDNA corresponding to the coding region of RPS19 was amplified with primers containing EcoRV and Spe1 sites, digested, and ligated into the expression vector T3TS. Mutations R62Q were introduced using GeneTailor Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA). Capped RNA was prepared using the Message Machine kit (Ambion, Austin, TX). RNA was purified using RNeasy kit (QIAGEN, Valencia, CA) and dissolved in water at the concentration 100 ng/μL.
Morpholinos
Sequences were: rps19 translation MO 5′-cactgttacaccacctggcatcttg; splicing MO 5′-gcttccccgacctttcaaaagacaa; splicing MO, 5 bases mismatch: 5′-gattcctcgaactctcaatagacaa; ΔNp63 MO inhibitor of translation 5′-ttggtctccaggtacaacatattgg; p53 MO inhibitor of translation, 5′-gcgccattgctttgcaagaattg25 (Gene Tools, Corvallis, OR). Morpholinos were injected at one cell stage.
Reverse-transcription polymerase chain reaction
Total RNA was prepared using Trizol (Invitrogen). cDNA was synthesized by reverse transcription of 2 μg of the total RNA with the random hexamer primers. Polymerase chain reaction (PCR) primers sequences are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Flow cytometry
A total of 300 embryos (20 hours postfertilization [hpf]) expressing green fluorescent protein (GFP) under the control of the gata1 promoter26 were dechorionated with pronase and homogenized in cold phosphate-buffered saline (PBS) adjusted to fish blood osmolarity by addition of 10% of water, and containing 30% fetal bovine serum (FBS). They were washed twice and incubated in the same buffer with 1 mg/mL collagenase/dispase (Roche Diagnostics, Indianapolis, IN), 10 mg/mL trypsin, and 10 units/mL DNAse1 at 30°C for 1 hour with agitation. Cells were washed twice, filtered through a cell strainer, and suspended in the PBS/FBS with 1 μg/mL DAPI (4′6-Diamidino-2′-phenylindole dihydrochloride (Roche Diagnostics) to label dead cells. GFP-positive cells were sorted and collected to PBS/FBS.
In situ hybridization
Results
Creating rps19-deficient zebrafish using morpholinos
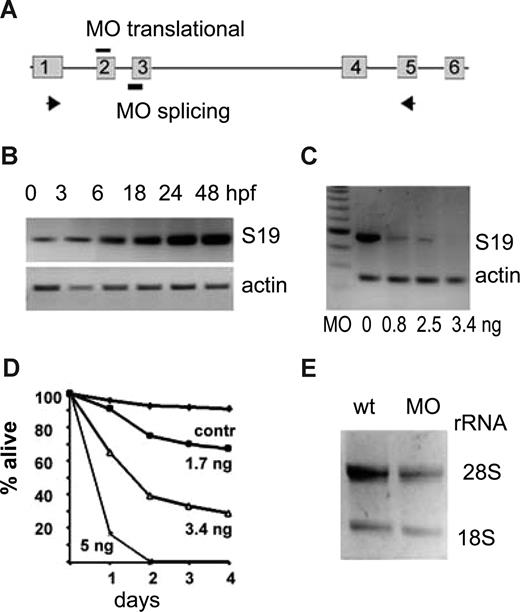
RPS19 is highly conserved with 87.7% identity between zebrafish and humans. Most residues that are mutated in DBA patients are identical in the human and zebrafish. Similar to the human gene, zebrafish rps19 has 6 exons, the first exon is untranslated, and the start codon is located at the beginning of exon 2 (Figure 1A). Zebrafish embryos start transcribing rps19 at approximately 6 hpf but at a low level (Figure 1B). Eggs are supplied with maternal ribosomes, and the embryo can proceed through initial developmental stages even if its own ribosome biosynthesis is blocked. The expression of rps19 is gradually increased during the first day of development.
Expression of rps19 in zebrafish embryos is specifically down-regulated by an antisense morpholino. (A) Zebrafish rps19 gene is composed of 6 exons. Antisense morpholinos corresponding to the sequence around the translational start site in exon 2 are designed to inhibit translation. Morpholinos derived from the intron 2 exon 3 boundary are designed to inhibit splicing. (B) Embryos start synthesizing rps19 mRNA at approximately 6 hpf; the low amount of rps19 mRNA at 0 to 3 hours postfertilization (hpf) is of maternal origin. RNA was pooled from at least 30 embryos at each time point. (C) Morpholinos targeting the splicing site of exon 3 of rps19 gene significantly decrease the level of rps19 mRNA message at 24 hpf: RT-PCR.  in panel A show the positions of primers. Amounts of injected morpholino per embryo are indicated. Actin expression is used as a load control. A total of 60 to 100 embryos were injected with each dose; RNA was prepared from at least 30 embryos. A result representative of 3 experiments is shown. (D) Survival of zebrafish embryos depends on the dose of injected morpholinos. Complete blocking of RPS19 expression at 5 ng per embryo leads to death of all embryos at 48 hpf. The dose of 3.4 ng allows embryos to survive for approximately a week. Sixty to 100 embryos were injected with each dose, a representative experiment is shown. (E) Ribosomal biogenesis is defective in morphants. At 30 hpf, the amount of rRNA is lower in morphants. Total RNA was prepared from 30 wild-type or morphant embryos; the amount of loaded RNA is equivalent to 4 embryos.
in panel A show the positions of primers. Amounts of injected morpholino per embryo are indicated. Actin expression is used as a load control. A total of 60 to 100 embryos were injected with each dose; RNA was prepared from at least 30 embryos. A result representative of 3 experiments is shown. (D) Survival of zebrafish embryos depends on the dose of injected morpholinos. Complete blocking of RPS19 expression at 5 ng per embryo leads to death of all embryos at 48 hpf. The dose of 3.4 ng allows embryos to survive for approximately a week. Sixty to 100 embryos were injected with each dose, a representative experiment is shown. (E) Ribosomal biogenesis is defective in morphants. At 30 hpf, the amount of rRNA is lower in morphants. Total RNA was prepared from 30 wild-type or morphant embryos; the amount of loaded RNA is equivalent to 4 embryos.
Expression of rps19 in zebrafish embryos is specifically down-regulated by an antisense morpholino. (A) Zebrafish rps19 gene is composed of 6 exons. Antisense morpholinos corresponding to the sequence around the translational start site in exon 2 are designed to inhibit translation. Morpholinos derived from the intron 2 exon 3 boundary are designed to inhibit splicing. (B) Embryos start synthesizing rps19 mRNA at approximately 6 hpf; the low amount of rps19 mRNA at 0 to 3 hours postfertilization (hpf) is of maternal origin. RNA was pooled from at least 30 embryos at each time point. (C) Morpholinos targeting the splicing site of exon 3 of rps19 gene significantly decrease the level of rps19 mRNA message at 24 hpf: RT-PCR.  in panel A show the positions of primers. Amounts of injected morpholino per embryo are indicated. Actin expression is used as a load control. A total of 60 to 100 embryos were injected with each dose; RNA was prepared from at least 30 embryos. A result representative of 3 experiments is shown. (D) Survival of zebrafish embryos depends on the dose of injected morpholinos. Complete blocking of RPS19 expression at 5 ng per embryo leads to death of all embryos at 48 hpf. The dose of 3.4 ng allows embryos to survive for approximately a week. Sixty to 100 embryos were injected with each dose, a representative experiment is shown. (E) Ribosomal biogenesis is defective in morphants. At 30 hpf, the amount of rRNA is lower in morphants. Total RNA was prepared from 30 wild-type or morphant embryos; the amount of loaded RNA is equivalent to 4 embryos.
in panel A show the positions of primers. Amounts of injected morpholino per embryo are indicated. Actin expression is used as a load control. A total of 60 to 100 embryos were injected with each dose; RNA was prepared from at least 30 embryos. A result representative of 3 experiments is shown. (D) Survival of zebrafish embryos depends on the dose of injected morpholinos. Complete blocking of RPS19 expression at 5 ng per embryo leads to death of all embryos at 48 hpf. The dose of 3.4 ng allows embryos to survive for approximately a week. Sixty to 100 embryos were injected with each dose, a representative experiment is shown. (E) Ribosomal biogenesis is defective in morphants. At 30 hpf, the amount of rRNA is lower in morphants. Total RNA was prepared from 30 wild-type or morphant embryos; the amount of loaded RNA is equivalent to 4 embryos.
To create deficiency of rps19, we designed morpholino MOs blocking splicing of rps19 mRNA at exon 3 (Figure 1A). The effect of MOs was monitored by RT-PCR. At 24 hpf, the amount of rps19 mRNA message was specifically, dose-dependently reduced in embryos injected with the morpholino (Figure 1C). The reduction in mRNA paralleled the viability of morphants (Figure 1D). rps19 mutations identified in DBA include allele deletion, internal deletions, premature stop codons, and missense mutations, some of which are inherited as dominant and some as recessive. Morpholino knockdown most closely mimics allele deletion, although splicing morpholino may lead to aberrant truncated messages similar to premature stop codon mutations. After injection of 0.8 ng MOs, we observed approximately 50% decrease of rps19 mRNA level, similar to DBA. At a dose of 3.4 ng, only a trace amount of mRNA is produced, and we used this dose to reveal the consequences of rps19 deficiency at conditions harsher than in DBA. This dose lets embryos survive up to 7 to 8 days. Some morpholinos can cause off-target effects.36 To confirm specificity of our morpholino, we designed another morpholino, MOt, blocking translation of rps19. Injection of both morpholinos resulted in a similar phenotype, and the simultaneous injection of 0.8 ng of MOs and MOt had an additive effect approximately equal to the injection of 5 ng of each morpholino (data not shown). As an additional control, we designed a morpholino with 5 bases mismatch to MOs. Its injection in doses 0.8 to 13 ng per embryo had no effects on development (data not shown).
Phenotype of rps19-deficient embryos
We compared the effects of rps19 deficiency in zebrafish with clinical manifestations of DBA. Developmental defects are found in approximately 40% of DBA patients with mutations in RPS19. They include such dissimilar malformations as microcephaly and macrocephaly, low ear set, small lower jaw, glaucoma, heart septal defects, spine defects, kidney aplasia, and anogenital fistula.38 Short stature is frequently observed in DBA. Another trait is duplication of thumbs and other structures. A particular defect can be restricted to one side of the body, such as left kidney aplasia. This feature suggests defective left-right patterning. Some patients have multiple defects, whereas in others only erythropoiesis is impaired. Indeed, individuals who are carriers for RPS19 mutations may be asymptomatic. For example, an R94Stop identified in 2 sisters and their mother led to thumb malformations and duplicated ureter in one sister and congenital glaucoma in the other but had no effect in their mother.4 Thus, humans can compensate for a defective RPS19 allele and other factors modulate the phenotypic manifestation of the primary genetic defects.
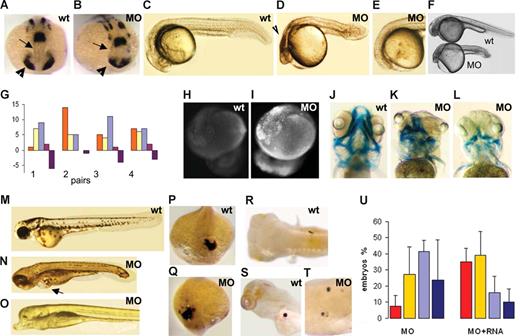
Embryos injected with a low dose (0.8 ng) of morpholino, which reduces the rps19 level by 50%, are morphologically normal and have defects only in erythropoiesis. The phenotype of embryos with higher rps19 deficiency resembled overall the phenotype reported previously in zebrafish with overactivated p53 pathway.25,36 The effect of morpholino injection was not apparent until gastrulation, at which point a delay in the development of the morphants was observed in comparison to noninjected embryos. It may be that at this stage maternal ribosomes cannot fulfill the embryo's needs anymore, and it starts assembling its own ribosomes. Morphologic defects in morphants became evident during somatogenesis. rps19-deficient embryos had reduced forebrain and eye field as was revealed by in situ hybridization with pax2a probe (Figure 2A,B). Anterior movement of ventral diencephalic precursors, which separates the eye field into 2 bilateral eyes,39 is defective in morphants. In a number of embryos, the eye field was separated incompletely, resulting in defective eyes. At 18 to 24 hpf, morphants had dark regions in their forebrain (Figure 2C,D). In 0% to 5% of embryos, the forebrain and eyes were absent (Figure 2E). Overall, rps19-deficient embryos are shorter than the wild-type and have smaller head and eyes (Figure 2F) similar to the phenotype of Drosophila ribosomal mutants Minute.40 Short stature is also a common feature in DBA. Another feature resembling DBA is sensitivity of zebrafish response to rps19 deficiency manifested between different fish lines, between the progeny of different pairs, and even between siblings (Figure 2G).
Phenotype of rps19-deficient embryos. (A,B) Forebrain ( ) and eye field (▶) are reduced in morphants. In situ hybridization, pax2a probe, 16 hpf. In situ hybridization was performed on 30 morphant or wild-type embryos. (C,D) At 24 hpf, morphants have small head and eyes, pinched midbrain-hindbrain boundary, and necrotic regions in brain (
) and eye field (▶) are reduced in morphants. In situ hybridization, pax2a probe, 16 hpf. In situ hybridization was performed on 30 morphant or wild-type embryos. (C,D) At 24 hpf, morphants have small head and eyes, pinched midbrain-hindbrain boundary, and necrotic regions in brain ( ). A typical morphant embryo is shown. (E) In some morphants, forebrain and eyes are absent. The frequency of this phenotype was 0% to 5%. (F) rps19-deficient embryos are smaller than wild-type. (G) Phenotypic manifestations of rps19 deficiency vary between the progeny of different pairs. Red represents normal looking; yellow, mild malformations; blue, strong malformations; purple, forebrain and eyes are absent; violet, dead. Twenty-five healthy looking morphants from each pair were selected at 6 hpf and compared at 20 hpf. Progeny of 4 pairs are shown as an example. (H,I) The level of apoptosis is higher in morphants. Acridine orange staining, 1 μg/mL, 30 minutes, 20 hpf. A representative embryo from at least 30 is shown. (J-L) Cartilage of the head is malformed in morphants. Alcian blue, 120 hpf. (M-O) The morphant fish shown in panel N has edema (
). A typical morphant embryo is shown. (E) In some morphants, forebrain and eyes are absent. The frequency of this phenotype was 0% to 5%. (F) rps19-deficient embryos are smaller than wild-type. (G) Phenotypic manifestations of rps19 deficiency vary between the progeny of different pairs. Red represents normal looking; yellow, mild malformations; blue, strong malformations; purple, forebrain and eyes are absent; violet, dead. Twenty-five healthy looking morphants from each pair were selected at 6 hpf and compared at 20 hpf. Progeny of 4 pairs are shown as an example. (H,I) The level of apoptosis is higher in morphants. Acridine orange staining, 1 μg/mL, 30 minutes, 20 hpf. A representative embryo from at least 30 is shown. (J-L) Cartilage of the head is malformed in morphants. Alcian blue, 120 hpf. (M-O) The morphant fish shown in panel N has edema ( ); eyes are present but abnormal. The fish shown in panel O has no eyes and forebrain. Note an abnormally large ventral fin in this fish, 120 hpf. (P,Q) Heart field's position and orientation are reversed in some morphants. In situ hybridization, lft2 probe, 19 hpf. An embryo representative of the group of 30 embryos used for in situ hybridization is shown. (R-T) Position of visceral organs is also randomized in morphants. Pancreas is located on the right side in wild-type fish (R), whereas in some morphants it is on the left (S) or bilateral. (T) 120 hpf, insulin probe. (U) rps19-deficient embryos are rescued by injection of 200 pg of rps19 mRNA. The proportion of normal-looking embryos among RPS19-deficient morphants is higher in the group injected with the RNA; the proportion of embryos with severe defects is reduced as is the death rate. No effect was observed after injection of GFP mRNA: 48 hpf, 3.4 ng MOs/embryo. Red represents morphologically normal embryos; yellow, mild defects; blue, severe defects; black, death. Eighty embryos were used per group. Shown are the results representative of 3 independent experiments.
); eyes are present but abnormal. The fish shown in panel O has no eyes and forebrain. Note an abnormally large ventral fin in this fish, 120 hpf. (P,Q) Heart field's position and orientation are reversed in some morphants. In situ hybridization, lft2 probe, 19 hpf. An embryo representative of the group of 30 embryos used for in situ hybridization is shown. (R-T) Position of visceral organs is also randomized in morphants. Pancreas is located on the right side in wild-type fish (R), whereas in some morphants it is on the left (S) or bilateral. (T) 120 hpf, insulin probe. (U) rps19-deficient embryos are rescued by injection of 200 pg of rps19 mRNA. The proportion of normal-looking embryos among RPS19-deficient morphants is higher in the group injected with the RNA; the proportion of embryos with severe defects is reduced as is the death rate. No effect was observed after injection of GFP mRNA: 48 hpf, 3.4 ng MOs/embryo. Red represents morphologically normal embryos; yellow, mild defects; blue, severe defects; black, death. Eighty embryos were used per group. Shown are the results representative of 3 independent experiments.
Phenotype of rps19-deficient embryos. (A,B) Forebrain ( ) and eye field (▶) are reduced in morphants. In situ hybridization, pax2a probe, 16 hpf. In situ hybridization was performed on 30 morphant or wild-type embryos. (C,D) At 24 hpf, morphants have small head and eyes, pinched midbrain-hindbrain boundary, and necrotic regions in brain (
) and eye field (▶) are reduced in morphants. In situ hybridization, pax2a probe, 16 hpf. In situ hybridization was performed on 30 morphant or wild-type embryos. (C,D) At 24 hpf, morphants have small head and eyes, pinched midbrain-hindbrain boundary, and necrotic regions in brain ( ). A typical morphant embryo is shown. (E) In some morphants, forebrain and eyes are absent. The frequency of this phenotype was 0% to 5%. (F) rps19-deficient embryos are smaller than wild-type. (G) Phenotypic manifestations of rps19 deficiency vary between the progeny of different pairs. Red represents normal looking; yellow, mild malformations; blue, strong malformations; purple, forebrain and eyes are absent; violet, dead. Twenty-five healthy looking morphants from each pair were selected at 6 hpf and compared at 20 hpf. Progeny of 4 pairs are shown as an example. (H,I) The level of apoptosis is higher in morphants. Acridine orange staining, 1 μg/mL, 30 minutes, 20 hpf. A representative embryo from at least 30 is shown. (J-L) Cartilage of the head is malformed in morphants. Alcian blue, 120 hpf. (M-O) The morphant fish shown in panel N has edema (
). A typical morphant embryo is shown. (E) In some morphants, forebrain and eyes are absent. The frequency of this phenotype was 0% to 5%. (F) rps19-deficient embryos are smaller than wild-type. (G) Phenotypic manifestations of rps19 deficiency vary between the progeny of different pairs. Red represents normal looking; yellow, mild malformations; blue, strong malformations; purple, forebrain and eyes are absent; violet, dead. Twenty-five healthy looking morphants from each pair were selected at 6 hpf and compared at 20 hpf. Progeny of 4 pairs are shown as an example. (H,I) The level of apoptosis is higher in morphants. Acridine orange staining, 1 μg/mL, 30 minutes, 20 hpf. A representative embryo from at least 30 is shown. (J-L) Cartilage of the head is malformed in morphants. Alcian blue, 120 hpf. (M-O) The morphant fish shown in panel N has edema ( ); eyes are present but abnormal. The fish shown in panel O has no eyes and forebrain. Note an abnormally large ventral fin in this fish, 120 hpf. (P,Q) Heart field's position and orientation are reversed in some morphants. In situ hybridization, lft2 probe, 19 hpf. An embryo representative of the group of 30 embryos used for in situ hybridization is shown. (R-T) Position of visceral organs is also randomized in morphants. Pancreas is located on the right side in wild-type fish (R), whereas in some morphants it is on the left (S) or bilateral. (T) 120 hpf, insulin probe. (U) rps19-deficient embryos are rescued by injection of 200 pg of rps19 mRNA. The proportion of normal-looking embryos among RPS19-deficient morphants is higher in the group injected with the RNA; the proportion of embryos with severe defects is reduced as is the death rate. No effect was observed after injection of GFP mRNA: 48 hpf, 3.4 ng MOs/embryo. Red represents morphologically normal embryos; yellow, mild defects; blue, severe defects; black, death. Eighty embryos were used per group. Shown are the results representative of 3 independent experiments.
); eyes are present but abnormal. The fish shown in panel O has no eyes and forebrain. Note an abnormally large ventral fin in this fish, 120 hpf. (P,Q) Heart field's position and orientation are reversed in some morphants. In situ hybridization, lft2 probe, 19 hpf. An embryo representative of the group of 30 embryos used for in situ hybridization is shown. (R-T) Position of visceral organs is also randomized in morphants. Pancreas is located on the right side in wild-type fish (R), whereas in some morphants it is on the left (S) or bilateral. (T) 120 hpf, insulin probe. (U) rps19-deficient embryos are rescued by injection of 200 pg of rps19 mRNA. The proportion of normal-looking embryos among RPS19-deficient morphants is higher in the group injected with the RNA; the proportion of embryos with severe defects is reduced as is the death rate. No effect was observed after injection of GFP mRNA: 48 hpf, 3.4 ng MOs/embryo. Red represents morphologically normal embryos; yellow, mild defects; blue, severe defects; black, death. Eighty embryos were used per group. Shown are the results representative of 3 independent experiments.
The dark regions in the forebrains of rps19-deficient embryos suggest that the embryos had increased apoptosis. Apoptosis is a part of normal development. Apoptotic cells appear in zebrafish embryo at the tail bud stage, peak at approximately 17 hpf and decline afterward, so that only few apoptotic cells are seen after 19 hpf. We visualized apoptotic cells by staining with the vital dye acridine orange.41 In contrast to control embryos, apoptosis was abundant at 20 hpf in morphants even when we used 0.8 ng morpholino per embryo, a dose that did not lead to obvious morphologic defects (Figure 2H,I).
At day 5, morphologic abnormalities became more evident in morphants (Figure 2J-O). Their head cartilage had not formed properly. Sometimes only rudiments of the jaw arches were present, whereas posterior pharyngeal arches were completely absent (Figure 2K,L). The head cartilage is formed by neural crest cells. Defects in cartilage formation indicate that neural crest cells could not form, migrate to their destinations, or proliferate. Cartilage malformations varied between the embryos and gave them diverse phenotypes (Figure 2K). Many morphants developed edema pointing to cardiovascular or blood problems (Figure 2N). Eyes are usually fully developed in zebrafish at day 5 but morphants often had underdeveloped eyes, sometimes asymmetric. In fish without forebrain, we noticed overgrowth of the fins (Figure 2O).
Because asymmetric defects are common in DBA, we studied left-right patterning in rps19-deficient embryos by analyzing the expression of asymmetrically expressed genes. The lft2 gene is expressed on the left side of the body in the heart field. In a fraction of morphants, lft2 staining was found on the right side or medially (Figure 2P,Q). During development, the heart undergoes looping to the right from the atrium to the ventricle. In our sample, of 45 wild-type embryos 42 were right-looping and 3 were left looping at 48 hpf, whereas of 37 of their rps19-deficient siblings, 24 were right looping, 9 were left looping, and 4 were nonlooping. Reversed or bilateral position of visceral asymmetric organs, such as pancreas, was also found in morphants. Pancreas localized by staining with an insulin probe was found on the right side in 22 wild-type embryos. In their rps19-deficient siblings, 10 had pancreas on the right side, 6 on the left side, 3 in the middle, and one had bilateral dispersed staining (Figure 2R-T). This finding indicates that rps19-deficient embryos exhibit cardiac and visceral heterotaxia in agreement with the presence of asymmetric defects in DBA patients. Abnormalities in left-right pattering can also account for frequent cardiac septal defects described with DBA.
The rps19-deficient phenotype was rescued by injection of zebrafish rps19 mRNA (Figure 2U). In the group coinjected with MOs and mRNA, the whole phenotypic spectrum shifted in comparison to the group receiving only the morpholino. The number of embryos with normal morphology at 48 hpf increased from the average 7.3% to 35%, the number of embryos with serious defects dropped from 41% to 16%, and the death rate decreased from 24% to 10%. These changes clearly illustrate the specificity of the phenotype. No rescue was observed after injection of mRNA encoding GFP protein. Next, we questioned whether RPS19 mRNA could rescue embryos injected with the translation-inhibiting morpholino. Zebrafish rps19 mRNA cannot be used for this purpose because its translation would be blocked by the morpholino. We therefore synthesized a human RPS19 mRNA. It is only approximately 50% identical to the zebrafish mRNA in nucleotide sequence around the initiation site, and its translation, therefore, cannot be blocked by MOt. Injection of the human mRNA rescued rps19 deficiency caused by MOt. In embryos injected with 3.4 ng of MOt, 20% had normal morphology, 28% had mild defects, 15% had severe defects, and 37% were dead at 48 hpf. In embryos coinjected with MOt and human RPS19 mRNA, the corresponding numbers were 42/34/3/21.
To determine whether introduction of a missense mutation into rps19 would cause a phenotype similar to that of rps19 down-regulation, we injected embryos with 200 to 400 pg of zebrafish or human RPS19 mRNA carrying R62/Q mutation, typical in DBA. We observed, in approximately 25% of embryos, phenotypic changes similar to those observed in rps19-deficient embryos, such as shortening of the body and craniofacial defects although milder than in rps19-deficient fish (data not shown). These results are in accord with human data that various types of RPS19 mutations lead to a similar phenotype.
Erythropoiesis is compromised in rps19-deficient zebrafish
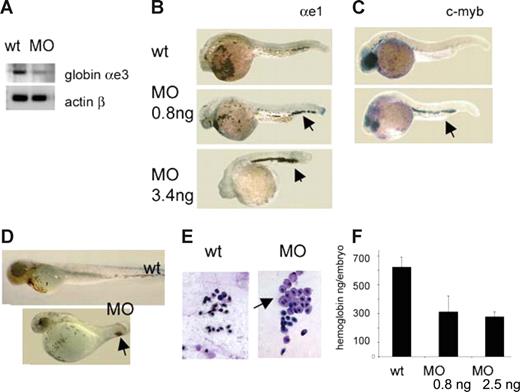
The hallmark of the DBA syndrome is deficiency of hematopoietic progenitors, in particular, of the erythroid lineage. Many patients are diagnosed with anemia at birth or within the first year of life, which suggests an early onset of erythropoietic defects. Similar to other vertebrates, the zebrafish experiences 2 waves of blood formation: primitive, producing erythrocytes and macrophages; and definitive, producing all blood lineages.32 Zebrafish rps19-deficient embryos have normal or slightly increased expression of gata1 in blood progenitors at 18 hpf (data not shown). Erythroid progenitors proliferate and differentiate into mature erythrocytes in the intermediate cell mass (ICM), a region between the dorsal aorta and axial vein. We injected rps19 morpholino into transgenic zebrafish expressing GFP protein under the control of the gata1 promoter26 and, at 20 hpf, sorted the GFP-expressing cells from controls and morphants. Approximately 3% of cells in control embryos were GFP-positive, whereas in RPS19-deficient siblings 2% were GFP-positive, which may indicate a lower rate of proliferation in cells committed to erythroid lineage in morphants. The expression of the globin αe3 was lower in morphant cells, also indicating a defect in erythroid lineage (Figure 3A). Circulation establishes in zebrafish at approximately 24 hpf, when cells leave the ICM. In morphants, however, more red cell progenitors remained in the ICM as evidenced by in situ hybridization with globin αe1 probe (Figure 3B). This effect was dose-dependent; the higher the dose of the morpholino, the fewer erythroid cells entered the circulation. At higher doses, where cardiac and other defects are common, this effect could be partially explained by circulation defects, but it is observed even in embryos without morphologic defects injected with 0.8 ng of morpholino. C-myb is highly expressed in erythroid progenitors; and when erythrocytes mature, c-myb expression declines. In morphants, however, cells continued to express c-myb. These features suggest that differentiation of erythroid lineage was defective in morphants (Figure 3C).
RPS19-deficient embryos have defects in erythropoiesis. (A) RT-PCR, red blood progenitors sorted out from embryos at 19 to 20 hpf. Globin expression is lower in morphants. The cells were purified from a pool of 300 embryos. (B) The differentiation of erythroid cells is delayed in morphants: at 30 hpf almost all erythroid cells are still in ICM region in morphants. In situ hybridization, globin αe1 probe, 30 hpf, 30 embryos per group. (C) Expression of c-myb in the tail regions in morphants ( ) points to immaturity of red blood cells in these embryos. In situ hybridization, 30 hpf. (D) At 72 hpf, there are fewer blood cells in morphants; they are localized mostly in the posterior of the embryo (
) points to immaturity of red blood cells in these embryos. In situ hybridization, 30 hpf. (D) At 72 hpf, there are fewer blood cells in morphants; they are localized mostly in the posterior of the embryo ( ), and there are only few cells in the heart region: 2.5 ng morpholino per embryo, O-dianizidine staining, 30 embryos per group. (E) Erythroid cells from the blood of a morphant embryo are variable in size; some look like erythroblasts (
), and there are only few cells in the heart region: 2.5 ng morpholino per embryo, O-dianizidine staining, 30 embryos per group. (E) Erythroid cells from the blood of a morphant embryo are variable in size; some look like erythroblasts ( ). Zebrafish erythrocytes differ from mammalian cells in that both primitive and definitive cells are nucleated. Blood smears were prepared from individual embryos; the results are representative of 6 to 10 embryos. (F) The level of hemoglobin is lower in morphants at day 5 in dependence with morpholino dose. Hemoglobin levels were measured in pools of 80 embryos in triplicates. Five-day-old embryos (wild-type, morphants, or cloche mutants, which have no blood cells) were homogenized in 500 μL of Drabkin reagent (Sigma-Aldrich, St Louis, MO), incubated for 1 hour at room temperature, centrifuged 10 minutes at 16 000g, and the absorbance of supernatants was measured at 540 nm. Absorbance of cloche mutants was subtracted from the wild-type and morphant samples. In this way, only the absorbance derived from red blood cells was taken into consideration. It was plotted on a calibration curve obtained from a standard sample with known concentration of hemoglobin (Drew Scientific, Dallas, TX), and the amount of hemoglobin per embryo was calculated.
). Zebrafish erythrocytes differ from mammalian cells in that both primitive and definitive cells are nucleated. Blood smears were prepared from individual embryos; the results are representative of 6 to 10 embryos. (F) The level of hemoglobin is lower in morphants at day 5 in dependence with morpholino dose. Hemoglobin levels were measured in pools of 80 embryos in triplicates. Five-day-old embryos (wild-type, morphants, or cloche mutants, which have no blood cells) were homogenized in 500 μL of Drabkin reagent (Sigma-Aldrich, St Louis, MO), incubated for 1 hour at room temperature, centrifuged 10 minutes at 16 000g, and the absorbance of supernatants was measured at 540 nm. Absorbance of cloche mutants was subtracted from the wild-type and morphant samples. In this way, only the absorbance derived from red blood cells was taken into consideration. It was plotted on a calibration curve obtained from a standard sample with known concentration of hemoglobin (Drew Scientific, Dallas, TX), and the amount of hemoglobin per embryo was calculated.
RPS19-deficient embryos have defects in erythropoiesis. (A) RT-PCR, red blood progenitors sorted out from embryos at 19 to 20 hpf. Globin expression is lower in morphants. The cells were purified from a pool of 300 embryos. (B) The differentiation of erythroid cells is delayed in morphants: at 30 hpf almost all erythroid cells are still in ICM region in morphants. In situ hybridization, globin αe1 probe, 30 hpf, 30 embryos per group. (C) Expression of c-myb in the tail regions in morphants ( ) points to immaturity of red blood cells in these embryos. In situ hybridization, 30 hpf. (D) At 72 hpf, there are fewer blood cells in morphants; they are localized mostly in the posterior of the embryo (
) points to immaturity of red blood cells in these embryos. In situ hybridization, 30 hpf. (D) At 72 hpf, there are fewer blood cells in morphants; they are localized mostly in the posterior of the embryo ( ), and there are only few cells in the heart region: 2.5 ng morpholino per embryo, O-dianizidine staining, 30 embryos per group. (E) Erythroid cells from the blood of a morphant embryo are variable in size; some look like erythroblasts (
), and there are only few cells in the heart region: 2.5 ng morpholino per embryo, O-dianizidine staining, 30 embryos per group. (E) Erythroid cells from the blood of a morphant embryo are variable in size; some look like erythroblasts ( ). Zebrafish erythrocytes differ from mammalian cells in that both primitive and definitive cells are nucleated. Blood smears were prepared from individual embryos; the results are representative of 6 to 10 embryos. (F) The level of hemoglobin is lower in morphants at day 5 in dependence with morpholino dose. Hemoglobin levels were measured in pools of 80 embryos in triplicates. Five-day-old embryos (wild-type, morphants, or cloche mutants, which have no blood cells) were homogenized in 500 μL of Drabkin reagent (Sigma-Aldrich, St Louis, MO), incubated for 1 hour at room temperature, centrifuged 10 minutes at 16 000g, and the absorbance of supernatants was measured at 540 nm. Absorbance of cloche mutants was subtracted from the wild-type and morphant samples. In this way, only the absorbance derived from red blood cells was taken into consideration. It was plotted on a calibration curve obtained from a standard sample with known concentration of hemoglobin (Drew Scientific, Dallas, TX), and the amount of hemoglobin per embryo was calculated.
). Zebrafish erythrocytes differ from mammalian cells in that both primitive and definitive cells are nucleated. Blood smears were prepared from individual embryos; the results are representative of 6 to 10 embryos. (F) The level of hemoglobin is lower in morphants at day 5 in dependence with morpholino dose. Hemoglobin levels were measured in pools of 80 embryos in triplicates. Five-day-old embryos (wild-type, morphants, or cloche mutants, which have no blood cells) were homogenized in 500 μL of Drabkin reagent (Sigma-Aldrich, St Louis, MO), incubated for 1 hour at room temperature, centrifuged 10 minutes at 16 000g, and the absorbance of supernatants was measured at 540 nm. Absorbance of cloche mutants was subtracted from the wild-type and morphant samples. In this way, only the absorbance derived from red blood cells was taken into consideration. It was plotted on a calibration curve obtained from a standard sample with known concentration of hemoglobin (Drew Scientific, Dallas, TX), and the amount of hemoglobin per embryo was calculated.
The development of the definitive wave of erythrocytes in rps19-deficient embryos was also impaired. During normal development, primitive erythrocytes stay in circulation for several days to be replaced by adult-type cells starting from days 4 to 5. Zebrafish adult-type erythrocytes start to develop from approximately 24 hpf in the region of the dorsal aorta. They then migrate to the caudal region where they proliferate and differentiate before entering the circulation.42 It seems that in morphants primitive cells were eliminated from circulation, but they were not replaced by adult-type erythrocytes and the total number of cells in circulation gradually dropped to very low numbers (Figure 3D). Erythroid precursors appeared to be present in rps19-deficient embryos but did not mature and were retained in the tail blood island. This pattern again points to differentiation defect in these embryos. Morphologically, erythrocytes from morphants differed from wild-type cells at day 5 (Figure 3E). They varied in size and some cells retained a blastlike phenotype. Immature erythrocytes in rps19-deficient embryos are reminiscent of that of DBA patients, in which the cells have higher mean corpuscular volume. The level of hemoglobin is decreased in DBA patients; therefore, we analyzed hemoglobin levels in morphants. We used MOs doses that caused no (0.8 ng) or few (2.5 ng) morphologic abnormalities. We found that the hemoglobin level was decreased even after injection of 0.8 ng of MOs (Figure 3F). These findings indicate that rps19 deficiency in zebrafish results in defective proliferation and differentiation of erythroid progenitors similar to that found in humans.
The development of other blood lineages was less affected in morphants. The expressions of myeloid and lymphoid markers do not significantly differ between wild-type and rps19-deficient embryos (Figure S1).
p53 network up-regulation in rps19-deficient embryos
In our morphants, we observed headless phenotype in the progeny of some fish pairs. Partial loss of the forebrain was previously noted in zebrafish after forced overexpression of zebrafish and human p53 and p73.43 Interestingly, this phenotype is also observed after overexpression of ΔNp63.34 In the latter case, the headless phenotype was rescued by coinjection of p53 mRNA. These data indicate that headless phenotype can be caused by p53/ΔNp63 imbalance regardless of the absolute amount of these messages.
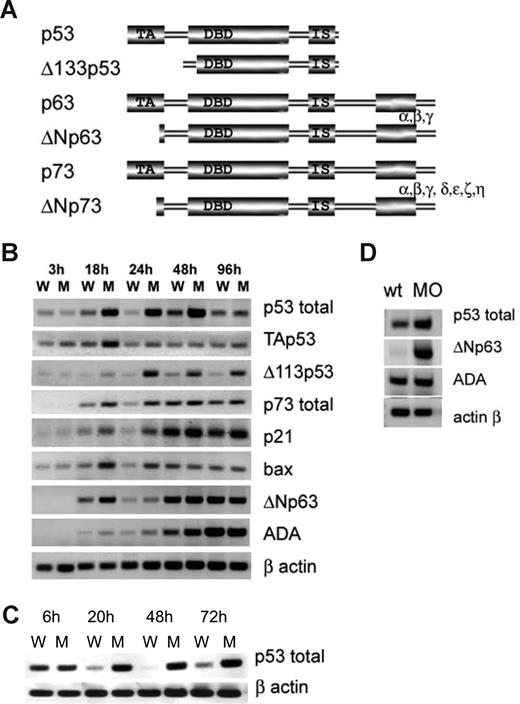
Therefore, we analyzed the expression of p53 family genes in RPS19-deficient embryos. p53, p63, and p73 are expressed as full-length isoforms, capable of inducing cell-cycle arrest and apoptosis or as N-terminally truncated isoforms acting as dominant-negative regulators of the full-length proteins44 (Figure 4A). Complex regulatory loops connect the full-length and dominant-negative isoforms. At some conditions, p53 drives the expression of its inhibitor, ΔNp63, and down-regulation of p53 abolishes the expression of ΔNp63.45 On the other hand, p53 can associate with ΔNp63 and target it to degradation.46 Both members of this loop receive inputs from other pathways. Specifically, ΔNp63 is stimulated by growth factors,47 whereas in response to some genotoxic stressors it mediates silencing of its own promoter.45 UV irradiation results in up-regulation of p53 but down-regulation of ΔNp63. Therefore, p53 network in a developing organism experiencing stress would receive contradicting inputs.
p53 family expression is dysregulated in rps19-deficient zebrafish. (A) Proteins of the p53 family have similar structure. They are expressed as full-length protein or truncated isoforms lacking all (Δ113p53) or most part of transactivation domain (ΔNp63 and ΔNp73). p63 and p73 can also be alternatively spliced at the C end. ΔNp63 and ΔNp73 can inhibit all full-length p53 family members. (B) Expression of p53 network genes in whole embryos received 3.4 ng of splicing blocking morpholino at different time points. RT-PCR, RNA was pooled from 30 embryos. W indicates wild-type; M, morphant embryos. (C) p53 is transcriptionally up-regulated in embryos injected with 3.4 ng of MOt morpholino inhibiting rps19 translation: RT-PCR. (D) p53 is up-regulated in red blood cell purified from 20 hpf embryos (pool of 300 embryos). ΔNp63 is not expressed in wild-type but induced in morphant cells. ADA is slightly up-regulated in morphant cells. Vertical lines have been inserted to indicate a repositioned gel lane.
p53 family expression is dysregulated in rps19-deficient zebrafish. (A) Proteins of the p53 family have similar structure. They are expressed as full-length protein or truncated isoforms lacking all (Δ113p53) or most part of transactivation domain (ΔNp63 and ΔNp73). p63 and p73 can also be alternatively spliced at the C end. ΔNp63 and ΔNp73 can inhibit all full-length p53 family members. (B) Expression of p53 network genes in whole embryos received 3.4 ng of splicing blocking morpholino at different time points. RT-PCR, RNA was pooled from 30 embryos. W indicates wild-type; M, morphant embryos. (C) p53 is transcriptionally up-regulated in embryos injected with 3.4 ng of MOt morpholino inhibiting rps19 translation: RT-PCR. (D) p53 is up-regulated in red blood cell purified from 20 hpf embryos (pool of 300 embryos). ΔNp63 is not expressed in wild-type but induced in morphant cells. ADA is slightly up-regulated in morphant cells. Vertical lines have been inserted to indicate a repositioned gel lane.
We have found that rps19 deficiency induced by MOs changes the transcription of p53 family (Figure 4B). Both full-length and truncated p53 was up-regulated in rps19-deficient embryos but with different dynamics. Transcriptional activation of full-length p53 peaked around 18 to 24 hpf. A truncated form of p53, Δ113Np53, followed p53 up-regulation with a delay and remained strongly up-regulated until 96 hpf. Overexpression of this isoform has previously been shown to have no obvious effects on development.36 In zebrafish embryos, p63 has only been found in the truncated form, ΔNp63. It plays a crucial role in specification of nonneural ectoderm during gastrulation and in development of epidermis and its derivatives.34,48 p73 in zebrafish embryos has only been found in the full-length form, TAp73, expressed in a restricted pattern in olfactory and other tissues.49 In RPS19-deficient embryos, ΔNp63 up-regulation, although lower and less consistent than Δ113Np53 up-regulation, was detected from 18 to 24 hpf. Full-length p73 was up-regulated during the first day of development. Other isoforms of p63 and p73 were not detected, in accord with previous reports. Up-regulation of p73 was modest in comparison to p53 and probably does not contribute much to p53 effects. The expression of p53 targets, such as p21 responsible for induction of cell-cycle arrest and bax, a mediator of apoptosis, followed the pattern of p53 expression. The expression of ada enzyme, a target of p63/ΔNp63,50 followed the pattern of ΔNp63 expression. In human DBA, ADA is up-regulated in erythrocytes of approximately 80% of patients.38 In embryos injected with MOt, we also found up-regulation of p53 (Figure 4C), which confirms that the observed effect is a specific response to rps19 deficiency.
In red blood cell progenitors, purified from embryos, p53 was modestly up-regulated, whereas ΔNp63 up-regulation was more dramatic (Figure 4D). Overexpression of p53 increases cell sensitivity to apoptosis, whereas ΔNp63 overexpression supports proliferation vs differentiation. Imbalance between p53 and ΔNp63 may affect the ability of blood cell progenitors to proliferate and differentiate into erythrocytes.
Our findings show that ribosomal stress resulting from RPS19 deficiency in zebrafish leads to up-regulation of p53 network. To verify our hypothesis that up-regulation of p53 family may be a common response to ribosomal protein deficiency, we examined expression of p53 family genes in zebrafish with mutations in rps8, rps11, and rps18, which have been recently reported among other RP mutations, all having similar phenotypes.51 We found a pattern of p53 family protein expression in these mutants similar to rps19 morphants. Specifically, p53, Δ113p53, as well as p53 targets p21 and bax were strongly up-regulated in mutants at 22 hpf (Figure S2) with modest up-regulation of TAp53, ΔNp63, and p73 (∼1.5 times). Up-regulation was also observed in siblings, a mixture of wild-type (∼33%) and heterozygous for RP mutations fish (∼67%).
Role of ΔNp63 dysregulation
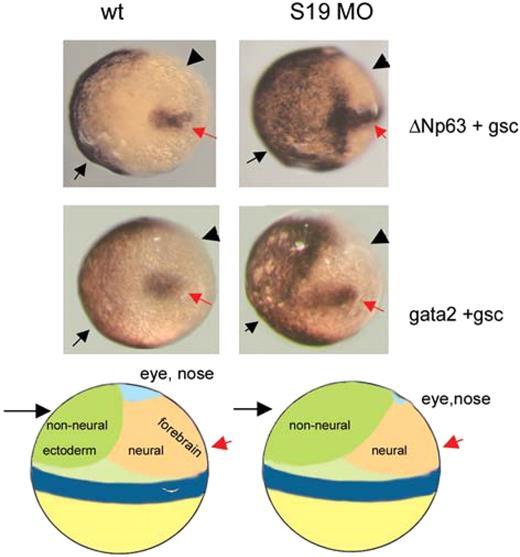
It is known that ΔNp63 overexpression during gastrulation leads to expansion of nonneural ectoderm and suppression of neural structures.34 Because a fraction of rps19 morphant phenotype resembled that of ΔNp63 overexpression, we questioned whether morphants also had expansion of nonneural ectoderm during gastrulation. We did not detect increase in the expression of ΔNp63 at 80% epiboly by RT-PCR, but by in situ hybridization with ΔNp63 probe we found an expansion of the area of ΔNp63 expression in approximately 25% of morphants (Figure 5). In wild-type embryos, ΔNp63 expression is localized in the ventral side of the embryo, whereas in this fraction of morphants, ΔNp63 positive area is expanded to the dorsal side. This expansion corresponded to the expansion of nonneural ectoderm into the neural field as confirmed by hybridization with a marker of nonneural ectoderm gata2. Shrinkage of the neural field affects the eye and forebrain more than other structures. This may explain why in RPS19-deficient embryos the forebrain is reduced. Suppression of the neural field can affect all aspects of neurogenesis from formation of the neural tube and ventricles to neural crest cells development and migration. In our morphants, reduction of the forebrain was observed in the progeny of not all fish pairs and it differed between fish lines. It could be that up-regulation of p53 causes disproportional up-regulation of ΔNp63 only in a particular genetic background. Later in development during limb formation, ΔNp63 is expressed in epidermal stem cells in the apical ectodermal ridge and its up-regulation may explain the overgrowth of epidermal derivatives, such as enlarged fins in zebrafish and perhaps extra digits in humans.
ΔNp63 dysregulation contributes to the rps19-deficient phenotype. Nonneural ectoderm ( ) is expanded in rps19-deficient embryos at the expense of neural one (◀). In situ hybridization with p63 and gata2 probes. Hybridization with gsc probe (red arrow) was used to mark the dorsal side. Tilted dorsal view. Thirty wild-type or morphant embryos were used in the reaction; 7 or 8 from the morphant group had visible expansion of the nonneural ectoderm markers. Representative embryos are shown. Schema of zebrafish fate map at 6 hpf shows side view.
) is expanded in rps19-deficient embryos at the expense of neural one (◀). In situ hybridization with p63 and gata2 probes. Hybridization with gsc probe (red arrow) was used to mark the dorsal side. Tilted dorsal view. Thirty wild-type or morphant embryos were used in the reaction; 7 or 8 from the morphant group had visible expansion of the nonneural ectoderm markers. Representative embryos are shown. Schema of zebrafish fate map at 6 hpf shows side view.
ΔNp63 dysregulation contributes to the rps19-deficient phenotype. Nonneural ectoderm ( ) is expanded in rps19-deficient embryos at the expense of neural one (◀). In situ hybridization with p63 and gata2 probes. Hybridization with gsc probe (red arrow) was used to mark the dorsal side. Tilted dorsal view. Thirty wild-type or morphant embryos were used in the reaction; 7 or 8 from the morphant group had visible expansion of the nonneural ectoderm markers. Representative embryos are shown. Schema of zebrafish fate map at 6 hpf shows side view.
) is expanded in rps19-deficient embryos at the expense of neural one (◀). In situ hybridization with p63 and gata2 probes. Hybridization with gsc probe (red arrow) was used to mark the dorsal side. Tilted dorsal view. Thirty wild-type or morphant embryos were used in the reaction; 7 or 8 from the morphant group had visible expansion of the nonneural ectoderm markers. Representative embryos are shown. Schema of zebrafish fate map at 6 hpf shows side view.
Alleviation of rps19-deficient phenotype by down-regulation of p53 and ΔNp63
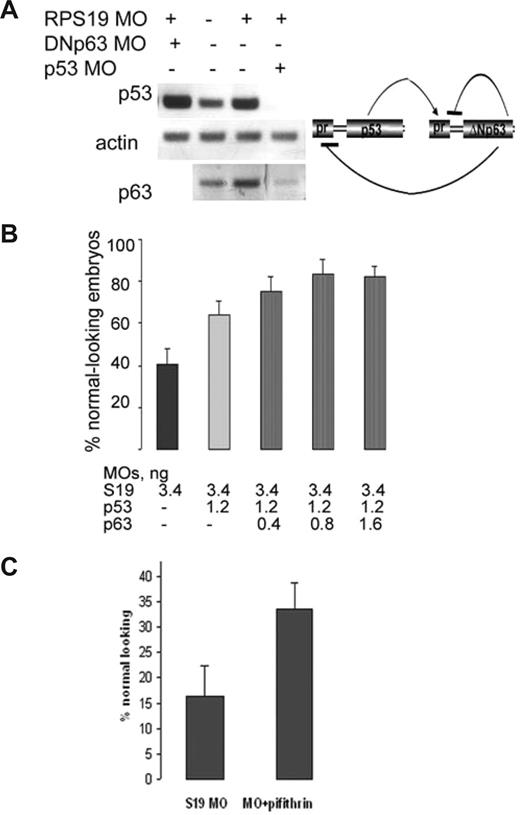
Given that rps19-deficient phenotype is partially mediated by p53 pathway up-regulation, one could expect alleviation of the phenotype with down-regulation of p53. Indeed, when we injected p53−/− zebrafish embryos with a lethal dose of 3.4 ng rps19 MOs, reducing rps19 mRNA level by more than 90%, morphants were smaller than wild-type embryos but had no obvious morphologic defects and no neural apoptosis at day 1 (Figure S3). These embryos survived approximately 4 days longer than the regular morphants. Some of them, however, developed edema by day 5, suggesting a shortage of erythroid cells. A lower dose of MOs (0.8 ng) that resulted in 50% reduction of rps19 mRNA had no effect on the phenotype and survival of p53−/− zebrafish embryos, suggesting that they compensated fully for rps19 deficiency of this magnitude (data not shown). Down-regulation of p53 by injection of 3 ng/embryo of p53 antisense morpholino as in the earlier reports,25 also improved the morphants' survival rates and phenotype (data not shown). We noticed that down-regulation of p53 led to coordinated down-regulation of ΔNp63, which might be responsible for the rescue of forebrain development in fish deficient both in rps19 and p53 (Figure 6A). Interestingly, a lower (1.2 ng) dose of p53 morpholino acted synergistically with 0.4 to 0.8 ng morpholino targeting ΔNp63 in alleviating the morphant phenotype (Figure 6B), whereas injected alone ΔNp63 morpholino led to a stronger up-regulation of p53 and to increase in the death rate at 24 hpf. These data illustrate again that the right balance of the p53-ΔNp63 loop is important for normal development.
Down-regulation of p53 and ΔNp63 alleviates rps19-deficient phenotype. (A) p53 and ΔNp63 regulate each other expression. Expression of ΔNp63 is decreased in rps19-deficient embryo injected with 8 ng of p53-specific morpholinos. On the contrary, expression of p53 in RPS19-deficient embryo injected with high dose (8 ng/embryo) of ΔNp63-specific morpholino is increased. RNA was prepared from 30 embryos at each time point. (B) Coinjection of low doses of p53-specific and ΔNp63-specific morpholinos increases the number of morphologically normal embryos among morphants. Eighty embryos were injected in each group. A result representative of 3 independent experiments is shown. (C) Pharmacologic inhibition of p53 pathway by a small molecule pifithrin μ improves phenotype of rps19-deficient embryos. Eighty injected embryos were used for the control and experimental group.
Down-regulation of p53 and ΔNp63 alleviates rps19-deficient phenotype. (A) p53 and ΔNp63 regulate each other expression. Expression of ΔNp63 is decreased in rps19-deficient embryo injected with 8 ng of p53-specific morpholinos. On the contrary, expression of p53 in RPS19-deficient embryo injected with high dose (8 ng/embryo) of ΔNp63-specific morpholino is increased. RNA was prepared from 30 embryos at each time point. (B) Coinjection of low doses of p53-specific and ΔNp63-specific morpholinos increases the number of morphologically normal embryos among morphants. Eighty embryos were injected in each group. A result representative of 3 independent experiments is shown. (C) Pharmacologic inhibition of p53 pathway by a small molecule pifithrin μ improves phenotype of rps19-deficient embryos. Eighty injected embryos were used for the control and experimental group.
We next questioned whether compounds inhibiting p53 pathway can improve the phenotype and survival of rps19-deficient zebrafish. Pifithrin μ inhibits p53 binding to mitochondria by reducing its affinity to antiapoptotic proteins Bcl-xL and Bcl-2.52 It specifically suppresses the mitochondria branch of the p53 pathway but does not affect other p53 functions. We found that pifithrin μ had no effect of the p53 mRNA level in zebrafish embryos but was efficient in improving rps19-deficient phenotype (Figure 6C). These data suggest that suppression of p53 activity might be a new direction for development of novel drugs for DBA treatment.
Discussion
Zebrafish response to rps19 deficiency has many features of DBA, including impaired ribosome biogenesis, increased apoptosis, developmental abnormalities, and defective hematopoiesis. It is accompanied by up-regulation of p53 family. Such up-regulation may result from the distortion of nucleolar organization reported in RPS19 deficiency,13 which may lead to p53 activation.15,53 On the other hand, several ribosomal proteins can activate p53 through inhibition of MDM2-mediated ubiquitination,54 which makes it probably that several pathways converge on p53. We observed up-regulation of p53 pathway not only in rps19 morphants but also in rps8, rps11, and rps18 mutants. Similar to rps19 and many other RPs, Rps8, Rps11, and Rps18 are necessary for pre-rRNA processing.55 These data suggest that up-regulation of p53 pathway may be a common response to deficiency of essential ribosomal proteins.
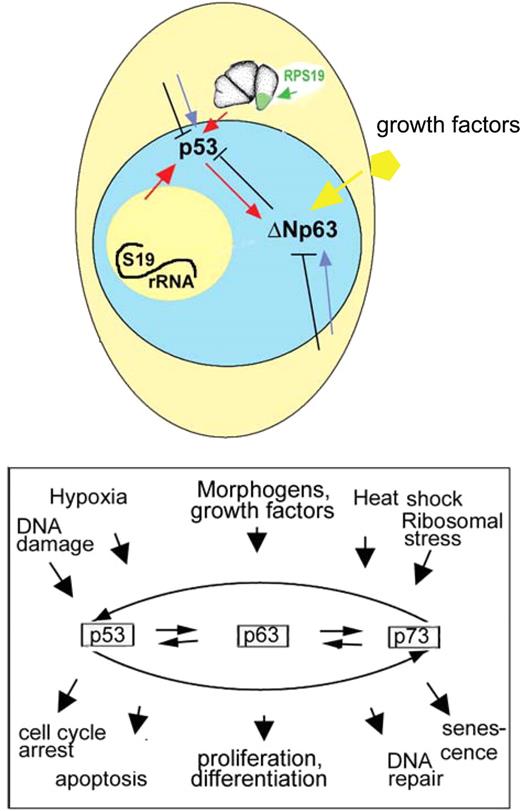
p53 family is instrumental in development for control of proliferation and differentiation. p53 has been implicated in suppression of stem cells renewal and promotion of differentiation.56 On the other hand, ΔN isoforms can be directly regulated by growth factors and support proliferation.47,57 Our data illustrate that a fine balance between p53 and the ΔNp63 is required for proper development. In blood cell progenitors, which have a high rate of proliferation, the pattern of p53 family activation differs from the whole embryo as indicated by a stronger activation of ΔNp63 than p53. Therefore, differential regulation of p53 and delta isoforms may depend on tissue type and extracellular signals (Figure 7).
Up-regulation of p53 and ΔNp63 are interconnected but separately regulated events. Some p53 network activating signals may originate from nucleolus resulting from defective ribosome biogenesis or from cytoplasm resulting from defective translation. p53 and ΔNp63 may also integrate other signals from the cell interior as well as from the environment (eg, from growth factors). Both p53 and ΔNp63 proteins act not in isolation but as part of p53 family network.
Up-regulation of p53 and ΔNp63 are interconnected but separately regulated events. Some p53 network activating signals may originate from nucleolus resulting from defective ribosome biogenesis or from cytoplasm resulting from defective translation. p53 and ΔNp63 may also integrate other signals from the cell interior as well as from the environment (eg, from growth factors). Both p53 and ΔNp63 proteins act not in isolation but as part of p53 family network.
The importance of ΔNp63 in development is well recognized. Deficiency of p63 in humans results in developmental defects characterized by limb malformations, orofacial clefts, and ectodermal dysplasia.58 Less attention has been paid to the effect of ΔNp63 overactivation. It may have a dual signature. Suppression of the neural field during gastrulation could affect the eye, brain, and neural crest derivatives. Overexpression of ΔNp63 at the stage when epidermal appendages are formed would most probably be manifested in duplication of structures, such as digits.
ΔNp63 up-regulation in erythroid progenitors may keep them in a nondifferentiated state. ΔNp63 is a BMP4 target and BMP4 regulates the expansion of the erythroid progenitors termed stress-BFU-E in response to acute anemia.59 One possibility is that in response to ribosomal stress BMP4-ΔNp63 pathway is activated constitutively. Downstream targets of ΔNp63 include Shh60 and continuous SH activation leads to hematopoietic stem cells exhaustion.61 Therefore, ΔNp63 may contribute to the RPS19-deficient blood phenotype in zebrafish and human. Overexpression of ΔNp63 increases growth of transformed cells,62 which may contribute to high risk of hematopoietic malignancies in DBA.
The p53 family network is very complex with hundreds of proteins involved. It may explain variable penetrance and expressivity of RPS19-deficient phenotype. Both environmental and genetic factors may modulate phenotypic manifestations. We observed, for example, that if rps19-deficient zebrafish embryos developed at room temperature instead of 28°C, the phenotype was less penetrant. Genetic background may significantly influence the p53 network. It is known that some strains of p53−/− mice are normal developmentally,63 whereas in other strains, females have brain overgrowth, extra digits, and eye defects.64,65
Dysregulation of p53 family may be a mechanism underlying many congenital abnormalities. The p53 family monitors all crucial cellular processes. Defects in genes responsible for ribosomal biogenesis, chromatin maintenance, DNA repair, RNA splicing, metabolism, destruction of oxygen species, protein folding, and other processes can potentially trigger p53 pathway as has been already demonstrated for some genes.16,17,66,67 The generality of this phenomenon may be underestimated. Excessive neural cell death was observed in more than half of mutants found in a recent mutagenic screen, most of them in the aforementioned categories.51 Neural cell death and “off-target” p53 activation have been reported in more than 20% of cases in knockdown technologies, such as morpholino or RNAi.36 Developmental phenotypes resulting from p53-mediated generalized stress response would depend on a development stage, when a deficiency of a particular gene triggers stress response. If stress occurs early in gastrulation, global defects in establishment of body axis or neural system patterning are probable. On the other hand, when the affected gene is expressed later in development, the defects may be milder.
In conclusion, our data together with previous findings suggest that many human congenital syndromes, probably a majority of them, develop not according to the linear scheme, a gene defect causes certain downstream phenotype, but according to a more systemic process, where defects in multiple genes can trigger the same master network leading to a spectrum of cognate abnormal phenotypes. Excessive activity of p53 and imbalance between p53 family members may be an underlying cause of many developmental anomalies in humans. Strong modulation of a phenotype by the genetic background may be a hallmark of p53 family-dependent disorders, taking into account the number of genes involved in the p53 network. New approaches to treatment and prevention of such diseases may involve the p53 family.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Amsterdam for providing the RP mutants embryos, J. Werk for help with the hemoglobin assay, N. Federman for critical review, and members of S.L.'s laboratory for helpful discussions and reagents.
This study was supported by the Diamond Blackfan Anemia Foundation.
Authorship
Contribution: N.D. designed and performed research, analyzed data, and wrote the paper; and K.M.S. and S.L. designed research, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shuo Lin, Department of Molecular, Cell & Developmental Biology, University of California, Los Angeles, 615 Charles E. Young Drive South BSRB 490B, Los Angeles, CA 90095-1606; e-mail: shuolin@ucla.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal