Abstract
Background: Multiple Myeloma (MM) is an incurable plasma cell malignancy. Recently, there have been major therapeutic advances in the treatment of MM, including the use of immunomodulatory drugs. Thalidomide alone or in combination represents an effective treatment strategy for newly diagnosed, relapsed and refractory MM patients. The identification of novel biomarkers could lead to more effective, individualized therapeutic strategies with improved patient outcomes.
Patients, Method & Material: Serum samples of sixteen newly diagnosed multiple myeloma patients, who had had initial treatment with thalidomide based regimens were analyzed. Based on D100 re-staging, 8 responders and 8 non responders to thalidomide were identified. Samples were analysed using 2D-DIGE, a technique based on pre-electrophoretic labelling of samples with one of three spectrally resolvable fluorescent CyDyes (Cy2, Cy3, and Cy5) allowing multiplexing of samples into the same gel. Initially serum samples were immunodepleted, which specifically removes 14 high-abundant proteins representing approximately 94% of total protein mass. This allowed for easier analysis of low abundance proteins, which are more likely to be a source of potential biomarkers. All 2D-DIGE images were scanned and collected on a Typhoon Fluorescent Imager. Pooled samples were used as an internal standard to quantify expression changes with statistical significance. Statistics and quantitation of protein expression were carried out initially using DeCyder Biological Variation Analysis (BVA) software before performing subsequent Extended Data Analysis (EDA).
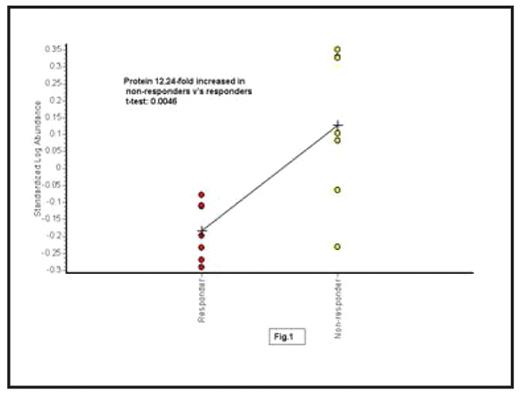
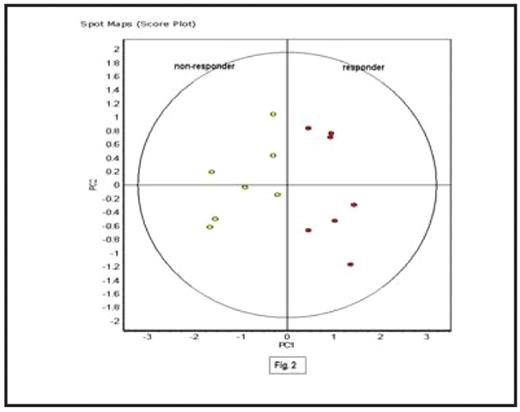
Results: 18 proteins have been identified to be differentially expressed in non-responders compared to responders: 13 were up-regulated and 5 were down-regulated (t-test ≤ 0.02). All 18 proteins were >1.25-fold differentially expressed, with changes up to 6.62-fold. For example, Fig.1 shows statistical analysis of protein 1 using DeCyder BVA software. This protein was increased 2.24-fold in the immuno-depleted serum from non-responders compared to responders, (t-test 0.0046). Once the 18 panel proteins were established, further statistical analysis was performed using DeCyder EDA software. Principal Components Analysis (PCA) was used to separate the responders from the non-responders based on the panel of 18 statistically significant differentially expressed proteins (Fig.2). Each dot on this plot represents a clinical sample; clinical samples from the same experimental groups are located in the same distinct areas, i.e. contained in one half of the plot, confirming consistency of results.
Conclusion: Accurate prediction of an individual patient’s drug response is an important prerequisite of personalized medicine. Using a panel of proteomic biomarkers, we have demonstrated the feasibility of predicting sensitivity and response to thalidomide in previously untreated myeloma. We are in the process of identification of theses proteins and plan to confirm their predictive value in a larger group of patients.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal