Abstract
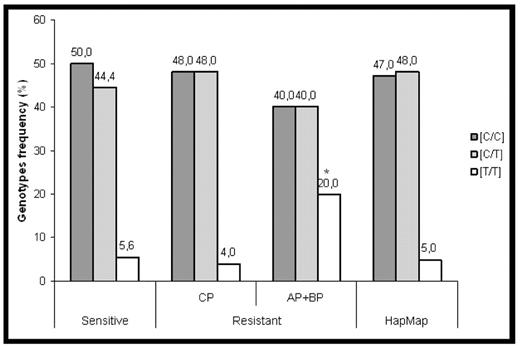
The tyrosin kinase inhibitor imatinib induces apoptosis in chronic myeloid leukaemia (CML) cells. RNA interference experiments have demonstrated that the pro-apoptotic protein BIM (Bcl-2 interacting mediator of cell death) was absolutely essential to this apoptosis. Thus, mutations in BIM sequence could lead to imatinib resistance independently of BCR-ABL mutations. In the current study, we performed BIM coding sequence analysis, using complementary DNA of leukocytes from CML patients mostly secondary resistant to imatinib. A total of 58 patients were analysed: secondary resistant to imatinib (n=40), or on the contrary sensitive to it (n=18). They were in chronic phase (CP, n=43), in accelerated phase (AP, n= 12), or in blastic phase (BP, n=3). Four patients were mutated in the tyrosin kinase domain of BCR-ABL. We did not find any mutation with amino acid change on the coding sequence of BIM. However, we observed a single nucleotide polymorphism (SNP) located in the fifth exon which encode for the BH3 alpha helix part of the protein. The impact of this previously described silent polymorphism C465T (rs724710) has never been studied before. We found the 3 following genotypes : C/C homozygous, T/T homozygous and T/C heterozygous. T/T genotype was the less frequent (It represents about 5% in the European population described in the international Hapmap project). Although not significant (10%, p=0.1468), we observed an increased T/T genotype frequency in the resistant population. In this resistant group, the T/T genotype frequency was increased in advanced phases (AP+BP, 20%) as compared to CP (4%, p=0.0016) or to sensitive cohort (5.6%, p=0.0153) (Fig.1). T/T genotype could affect RNA stability, translation speed and protein folding as described by Kimchi-Sarfaty et al. for the MDR1 gene (
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal