Abstract
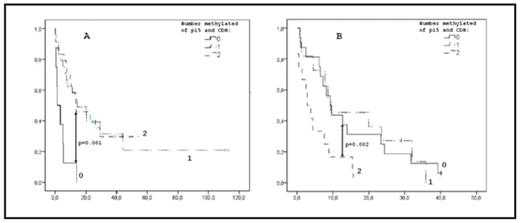
AML and high risk MDS are two myeloid malignancies with poor prognosis. Epigenetic changes such as silencing of tumor suppressor genes by promoter hypermethylation play a role in pathogenesis and disease progression. There is no consensus regarding the prognostic implications of epigenetic alterations in AML. We analyzed 108 de novo AML (median age 62 years, range 23–85), and 47 patients with MDS-related disease (19 MDS with IPSS INT-2 or High, 28 MDS-AML, median age 77 years, range 54–83), with respect to clinical and prognostic parameters and hypermethylation of the promoter region of three tumor suppressor genes; HIC, CDKN2b(p15) and CDH. All patients were eligible for and received standard induction chemotherapy. Methylation analysis was performed by Denaturing Gradient Gel Electrophoresis (DGGE) following bisulfite treatment and promoter specific PCR. A complete remission (CR) was achieved by 81 and 43 percent in the de novo AML group and the MDS group respectively. Hypermethylation of CDH or of multiple genes has previously been reported to correlate negatively to the probability of CR after intensive chemotherapy in the MDS cohort (p=0.008 and p=0.05, respectively). This was not true for the de novo AML patients (p=0.748 and p=0.681, respectively). The incidence of hypermethylation was significantly higher in de novo AML where 89% had at least one methylated gene compared to 66% in MDS (p=0.006). The mean number of methylated genes were 1.8 in de novo AML compared to 1.0 in MDS (p=0.002). In de novo AML the number of methylated genes decreased with increasing age (p=0.041) and an opposing trend was seen in MDS (p=0.15). Hypermethylation of all three genes was associated with higher age in MDS (p=0.040) whereas a trend towards the opposite was seen in de novo AML (p=0.084). In de novo AML the occurrence of FLT3-TKD was associated with hypermethylation of all three genes (p=0.011). Kaplan-Meier curves showed that hypermethylation of CDH and p15 together was associated with better overall survival (OS) in the de novo AML patients (p=0.001) (fig 1A). Median OS was 1.4 months for cases with no methylated genes and 13.5 and 14.5 months for those with one or two methylated genes, respectively. In contrast, in the MDS cohort, hypermethylation of CDH and p15 was a negative prognostic factor (p=0.002) (fig 1B). Median OS was 2.9 months for cases with two methylated genes and 12.7 and 9.4 for 0–1 methylated genes, respectively. The significance of these findings were retained after correction for age in a Cox regression analysis, HR 0.20 (p=0.001) for de novo AML and HR 2.96 (p=0.015) for MDS. We conclude that de novo AML and high risk MDS/MDS-AML show significant differences with respect to DNA promoter hypermethylation. Also, the correlation between hypermethylation and age as well as the effect on OS and complete remission rate differs between the de novo AML and the MDS cohort, respectively. This indicates epigenetic differences that may explain some of the clinical and morphological differences between the diseases. We also for the first time describe a relationship between FLT3-TKD and hypermethylation in AML.
Kaplan Meier curve showing the effect of methylation of p15 and CDH on OS (months) in de novo AML.
Fig 1B: Kaplan Meier curve showing the effect of methylation of p15 and CDH on OS (months) in high risk MDS.
Kaplan Meier curve showing the effect of methylation of p15 and CDH on OS (months) in de novo AML.
Fig 1B: Kaplan Meier curve showing the effect of methylation of p15 and CDH on OS (months) in high risk MDS.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal