Abstract
Acute myelogenous leukemia (AML) is a heterogenous group of myeloid malignancies that are characterized by the clonal outgrowth of immature myeloid progenitor cells. For most subtypes of AML, mutations that give rise to the leukemic phenotype occur in the hematopoietic stem/progenitor cell (HSC) subset as demonstrated by studies showing that only primitive CD34+CD38− bone marrow cells could function as leukemia-initiating cells (LSC) when transferred into immunodeficient NOD-SCID mice. One rather significant challenge has been that LSC share many of the same cell-surface markers as their normal counterparts in bone marrow, thus making it difficult to functionally characterize and purify this important subset of leukemic cells from bulk bone marrow samples. To rapidly identify novel antigens that are mutation-specific and induced specifically on LSC and not on normal HSC, we have transduced highly purified HSC isolated from mouse bone marrow with retroviral vectors that co-expressed AML1-ETO along with a green fluorescent protein (GFP) reporter gene. HSC of the cell-surface phenotype c-Kit+Lin−Sca-1+Flt3− (KLSF) were transduced for 18 hours in vitro and then were resorted for GFP+ cells by flow cytometry (FACS). Purified cells were then lysed for mRNA isolation and cDNA synthesis for the generation of a probe that was then hybridized to Affymetrix oligonucleotide arrays (430 2.0 GeneChip arrays). HSC transduced with a retroviral vector that only expressed GFP were used as controls to identify genes that would normally be expressed in the HSC subset. Importantly, since retroviral vectors only integrate into cycling cells, all sorted GFP+ cells from the independent transductions would represent cycling cells, which minimizes any gene expression differences due to differential frequencies of resting versus actively cycling HSC. Changes in expression of cell-surface proteins observed at the mRNA level were then validated at the protein level using FACS. Bone marrow cells were isolated from an animal that was transplanted with cells expressing AML1-ETO and GFP from a retroviral vector. Cells were stained for the HSC/progenitor cell phenotype (KLS) as well as for the cell-surface marker of interest. For one marker, CD55, we noted a 100-fold increase in cell-surface expression specifically on HSC that express AML1-ETO and not on normal HSC. These results indicate that short-term retroviral expression of specific AML-associated mutations in HSC followed by microarray analysis of transduced cells may provide a rapid means of prospectively identifying leukemia-initiating cells in bulk patient bone marrow samples and that CD55 may be a useful therapeutic and diagnostic marker for patient samples that express the AML1-ETO chromosomal translocation.
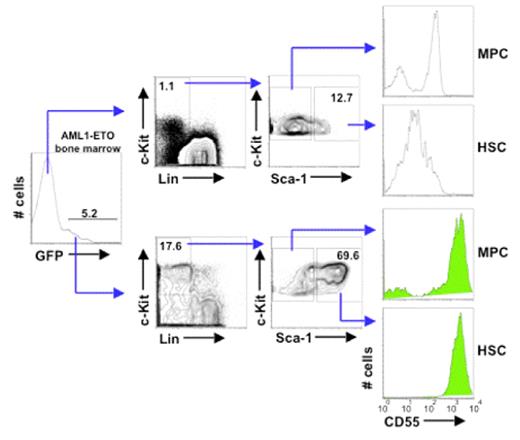
CD55 expression distinguishes normal HSC from HSC with theAML1-ETOtranslocation. Bone marrow cells were isolated from an animal that was transplanted with cells expressing AML1-ETO and the green fluorescent protein (GFP) from a retroviral vector. Cells were stained for the HSC/progenitor cell phenotype (KLS) as well as for CD55. Cells that expressed AML1-ETO are shown below as GFP+ gated cells. Note that cells that express AML1-ETO express CD55 at approximately 100-fold greater levels on the cell-surface than GFP-negative (AML1-ETO-negative) bone marrow cells. MPC=myeloid progenitor cells of the phenotype c-Kit+ Lin− Sea-1− cells. HSC are defined c-Kit+ Lin− Sca-1+ cells.
CD55 expression distinguishes normal HSC from HSC with theAML1-ETOtranslocation. Bone marrow cells were isolated from an animal that was transplanted with cells expressing AML1-ETO and the green fluorescent protein (GFP) from a retroviral vector. Cells were stained for the HSC/progenitor cell phenotype (KLS) as well as for CD55. Cells that expressed AML1-ETO are shown below as GFP+ gated cells. Note that cells that express AML1-ETO express CD55 at approximately 100-fold greater levels on the cell-surface than GFP-negative (AML1-ETO-negative) bone marrow cells. MPC=myeloid progenitor cells of the phenotype c-Kit+ Lin− Sea-1− cells. HSC are defined c-Kit+ Lin− Sca-1+ cells.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal