Abstract
Both Toll-like receptor 4 (TLR4)– and MD-2–deficient mice succumb to otherwise nonfatal Gram-negative bacteria inocula, demonstrating the pivotal role played by these proteins in antibacterial defense in mammals. MD-2 is a soluble endogenous ligand for TLR4 and a receptor for lipopolysaccharide (LPS). LPS-bound MD-2 transmits an activating signal onto TLR4. In this report, we show that both recombinant and endogenous soluble MD-2 bind tightly to the surface of live Gram-negative bacteria. As a consequence, MD-2 enhances cellular activation, bacterial internalization, and intracellular killing, all in a TLR4-dependent manner. The enhanced internalization of MD-2–coated bacteria was not observed in macrophages expressing Lpsd, a signaling-incompetent mutant form of TLR4, suggesting that the enhanced phagocytosis observed is dependent on signal transduction. The data confirm the notion that soluble MD-2 is a genuine opsonin that enhances proinflammatory opsonophagocytosis by bridging live Gram-negative bacteria to the LPS transducing complex. The presented results extend our understanding of the role of the TLR4/MD-2 signaling axis in bacterial recognition by phagocytes.
Introduction
Recognition of bacterial lipopolysaccharides (LPSs) by mammalian cells depends upon the presence of both Toll-like receptor 4 (TLR4; CD284) and MD-2 (Ly96).1-3 TLR4 is a 110-kDa type I integral membrane glycoprotein characterized by the presence of multiple leucine-rich repeats (LRRs) on its ectodomain and a signaling Toll–interleukin-1 receptor resistance (TIR) domain on the cytoplasmic side. MD-2 is a 25- to 30-kDa secreted glycoprotein which belongs to the MD-2–lipid binding (ML) family of single-domain lipid-binding secreted proteins.4 MD-2 binds noncovalently to the extracellular domain of TLR4,5 and is tethered on the surface of cells expressing TLR4.6 The apparent Kd of this interaction, as determined for human MD-2/TLR4, is approximately 12 nM.7 In addition to binding to TLR4, MD-2 binds specifically to the lipid portion of LPS (lipid A). Lipid A from enterobacteriaceae is a collection of phosphorylated glucosamine-based saccharolipids with up to 7 acyl moieties. This highly hydrophobic structure anchors LPS to the outer leaflet of the cell wall of Gram-negative bacteria,8 and is the epitome of an innate immune stimulator. The crystal structures of MD-2 bound to 2 antagonistic tetracylated lipid A structural analogs (lipid IVa and eritoran) reveal that MD-2 interacts with its ligands via a hydrophobic pocket that can accommodate at least 4 acyl chains without entailing any conformational change.5,9-11 When complexes of MD-2 and TLR4, but not TLR4 alone, are incubated with purified LPS, they produce stable dimers.5,12 Consistently, cells expressing TLR4 are activated by purified LPS only when MD-2 is provided as a recombinant protein or a transgene, suggesting that TLR4 activation is downstream from an MD-2–dependent signal that induces its aggregation.11,13,14 The most likely scenario for TLR4 activation is that LPS moves from the surface of Gram-negative bacteria to soluble or TLR4-bound MD-2. This movement is promoted by the lipotransferases LPS-binding protein (LBP) and CD14. Although LBP and CD14 together may enhance the sensitivity to LPS, neither protein is absolutely required for LPS responses, as cellular activation is reliably observed when a sufficiently high concentration of LPS is used.15 When a phagocyte encounters a Gram-negative cell wall, such high concentrations of LPS are likely. Hence, we speculated that MD-2 might bind to live bacteria and affect the activity of TLR4 on the surface of engaged phagocytes
In an article recently published in this journal, Tissieries et al reported that MD-2 is an acute phase reactant and binds to heat killed Escherichia coli.16 In the present work, we confirm and extend these observations by presenting a thorough analysis of the binding of MD-2 to live Gram-negative bacteria, and by elucidating the role of the MD-2/TLR4 signaling axis during internalization and killing.
Methods
Miscellaneous reagents, recombinant proteins, and antibodies
Unless otherwise specified, reagents were purchased from Sigma-Aldrich (St Louis, MO). LPS (E coli 0111:B4) was repurified to remove lipopeptides.17 MD-26xHis and TLR4-Fc were expressed and purified as described.7,18 Commercial recombinant MD-2 was from R&D Systems (Minneapolis, MN). The antibodies used in this work were α-6xHis mAb (Novagen-EMD Biosciences, San Diego, CA), α–MD-2 mAb (clone 9B4; eBioscience, San Diego, CA), rat α-mouse CD11b mAb (Serotec, Oxford, United Kingdom), gold-labeled α-mouse mAb (Jackson ImmunoResearch Laboratories, West Grove, PA), horseradish peroxidase (HRP)–conjugated α-mouse pAb (Bio-Rad, Hercules, CA), Alexa 647–labeled α-6xHis mAb (Qiagen, Valencia, CA), FITC-labeled α-mouse antiserum (Sigma-Aldrich), and Alexa 647–labeled α-rat antiserum (Molecular Probes, Eugene, OR).
Cells
All mammalian cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). 293 and 293TLR4-CFP cells were described previously.19 Mouse thioglycollate elicited peritoneal exudate cells (pφ) and human peripheral blood mononuclear cells (PBMCs) were harvested and maintained as described in Visintin et al.7,20 The Neisseria meningitidis (Nm) strains MC58,21 H44/76-lpxA,22 and green fluorescent protein (GFP)–expressing Nm (MC58) were grown on chocolate agar (Remel, Lenexa, KS). An acapsular form of Streptococcus agalactiae (strain 51523 ) was grown on blood agar plates at 37°C. Yersinia pestis (Yp24 ) was cultured at 26°C in tryptose-beef extract (TB) medium with 2.5 mM CaCl2. The yellow fluorescent protein (YFP)–expressing strain of Yp was generated by electroporation of the Kim5 strain with pYFP (Clontech, Mountain View, CA) and was maintained in ampicillin.
MD-2 opsonization, cytofluorimetry, and SEM
A total of 106 live bacteria were incubated for 30 minutes at 37°C with 20 ng of MD-26xHis in Hanks balanced salt solution/bovine serum albumin (HBSS/BSA; 0.1% wt/vol) or with pooled human serum (PHS; 8% for Nm and 30% for Yp). Cells were washed three times, and bound MD-2 was detected by fluorescence-activated cell sorter (FACS) using either the Alexa 647–labeled α-6xHis mAb, the α–MD-2 mAb, or TLR4-Fc (1 μg/sample), followed by an FITC-labeled α-mouse antiserum. When indicated, PHS was depleted of MD-2 and reconstituted with MD-26xHis (20 ng/mL) as described.7 For scanning electron microscopy (SEM), MD-26xHis was detected with an α-6xHis mAb and a gold-labeled α-mouse mAb (18-nm gold particles). Cells were fixed and processed for imaging using standard procedures.11,25 Images were obtained with a JSM-7401F field emission SEM (JOEL, Peabody, MA) using a backscatter retractable detector.
Serum MD-2 isolation and Western blotting
A total of 15 mL of serum from healthy donors was precleared of immunoglobulins by protein A–sepharose (PAS) bead capture, passed through centrifugal concentrators (50-kDa cut-off; Millipore, Billerica, MA) and incubated with TLR4-Fc–coupled PAS beads. Beads were washed with phosphate-buffered saline (PBS) and subjected to SDS-PAGE under reducing conditions. Gels were electroblotted onto nitrocellulose membranes, blocked in PBS/dry milk/0.05% Tween-20, and probed with the α-human MD-2 mAb (1:1000), followed by an HRP-conjugated α-mouse pAb and enhanced chemiluminescence (ECL; GE Healthcare, Little Chalfont, United Kingdom). All incubations were done for 1 hour at room temperature.
Bacteria ELISAs and MD-2 quantitation
Liquid overnight cultures of Yp were diluted in PBS to the indicated counts and plated in 96-well high-protein-binding plates. Live bacteria were allowed to adhere for 2 hours at room temperature prior to blocking with PBS/1% BSA/5% sucrose/0.01 Tween-20. A total of 100 μL human serum (Figure 2C) or MD-26xHis (20 ng/mL; Figure 1C) was then added to each well. After washing with PBS–Tween-20, MD-2 on the surface of bacteria was detected with an α-human MD-2 mAb followed by an HRP-conjugated α-mouse pAb and chromogenic reaction. In Figure 1C, MD-26xHis was titrated on bacteria that were plated at a constant concentration (2 × 107 cells/well). In some experiments, TLR4-Fc (10 μg/mL) was adsorbed on plastic and used to capture soluble MD-2. The concentration of sMD-2 was determined by comparing the absorbance value to a standard curve generated using a commercial recombinant MD-2 (R&D Systems). All incubations were done for 1 hour at room temperature unless stated differently. Results are expressed as average of triplicate stimulations plus SD.
Determination of MD-2 binding sites on the surface of bacteria
MD-26xHis (20 ng) or whole human sera (input) were incubated with titrated amounts of live bacteria (0-2 × 107 cells) in a final volume of 1 mL for 30 minutes at room temperature. Bacteria were centrifuged, and MD-2 in the postcellular supernatants was quantitated by enzyme-linked immunosorbent assay (ELISA). The concentration values were used to generate binding isothermes. The number of MD-2 binding sites on the surface of live bacteria was determined by dividing the number of input MD-2 molecules by the number of bacteria that depleted MD-2 from the input. We assumed that bacteria have about 106 LPS molecules on the cell surface (C. R. Raetz, written personal communication, November 2007; Galloway and Raetz26 ).
Opsonophagocytosis assay
RAW cells, pMφ, or PBMCs were seeded in triplicate onto 96-well culture plates (5 × 104 cells/well) and infected with fluorescent protein (GFP or YFP)–expressing bacteria (multiplicity of infection [MOI] = 40) that were opsonized with MD-26xHis (20 ng/mL) or PHS (1 mL) as a source of endogenous MD-2. Plates were centrifuged (200g for 3 minutes) in order to ensure proper exposure of phagocytes to the bacteria, and incubated for 30 minutes at 37°C. Extracellular bacteria were removed, and the fluorescence was quantified in the wells. The phagocytosis index was determined by setting the cellular autofluorescence to 0 and plotting the average plus SD of all the readings in an arbitrary scale (arbitrary fluorescent units [AFU]). In similar experiments, phagocytes were infected with YFP-Yp, fixed, and imaged with a Leica TCS SP2 AOBS laser scanning confocal microscope (Leica, Wetzlar, Germany). Cell membranes and nuclei were stained with an α-mouse CD11b mAb (100 ng/sample), followed by an Alexa 647–labeled α-rat antiserum and Hoechst 33258 (Invitrogen, Carlsbad, CA), respectively.
Intracellular bactericidal activity
Opsonophagocytosis was performed as described in the preceding section. Extracellular bacteria were killed by gentamicin treatment (7.5 μg/mL) for 1 hour, and cells were cultured in fresh serum-free DMEM at 37°C. At 6 hours later, cells were lysed with water, and 1/30 of the lysate was plated for colony count (CFU).
NF-κB luciferase reporter assay
293 and 293TLR4CFP cells bearing a NF-κB-Luc reporter plasmid were infected with the indicated bacteria in DMEM. Luciferase activity was determined as in Visintin et al.7 LPS (0.1 μg/mL) served as a control.27 The readings were normalized by the untreated control and plotted as averages plus the range of duplicate readings (relative luciferase units [RLU]).
Cytokines and nitrite quantitation
Mouse pMφ were plated onto 96-well culture plates and infected with live Yp for 24 hours. TNF-α and RANTES were quantified using a kit from R&D Systems. Nitrite concentration was determined by Griess reaction using chemicals from Sigma-Aldrich. Results are expressed as average of triplicate stimulations plus SD.
Results
Recombinant MD-2 binds to LPS on the surface of live Gram-negative bacteria
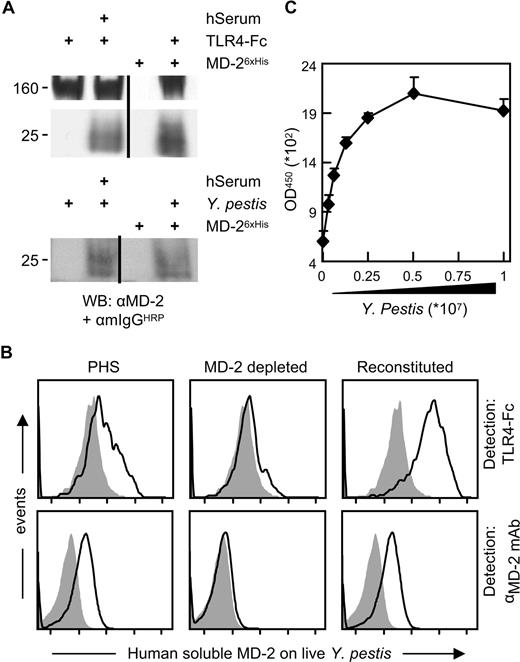
Since MD-2 physically binds to the hydrophobic portion of LPS,9-11 we hypothesized that MD-2 binds to the surface of live Gram-negative bacteria due to the high concentration of LPS there. In order to test this hypothesis, we treated 2 representative Gram-negative organisms, Nm and Yp, with a His-tagged recombinant human MD-2 (MD-26xHis). Bacteria-bound MD-26xHis was then detected by cytofluorimetry. As shown in the top 2 panels of Figure 1A, MD-26xHis bound to the surface of both organisms without affecting their viability (data not shown). As negative controls for MD-2 specificity, we used an LPS-negative mutant of Nm, H44/76-lpxA−, and a noncapsulated Gram-positive bacterium, S agalactiae (Figure 1A bottom panels). In both cases, MD-26xHis did not bind to the live organisms, suggesting that MD-2 binds specifically to LPS. In addition to its epitope tag, MD-26xHis could be detected using both a chimeric protein composed of the extracellular domain of TLR4 and the Fc portion of mouse IgG2a (TLR4-Fc11 ) and a monoclonal α–MD-2 antibody28 (Figure 1B), suggesting that bacteria-bound MD-2 is not embedded in the bacterial cell wall. The binding of MD-26xHis to the surface of live bacteria is specific and saturable, as demonstrated by the titration curves shown in Figure 1C. In these experiments, we either titrated MD-26xHis on a fixed amount of adhered bacteria (Figure 1C left panel) or detected titrated bacteria with a fixed amount of MD-2 (20 ng/mL; Figure 1C right panel). MD-26xHis appears to be homogeneously distributed on the surface of live Gram-negative bacteria, as shown by the scanning electron micrographs of MD-26xHis–treated Yp (Figure 1D). Based on our structural knowledge of lipid A, it is reasonable to assume that MD-2 is anchored to the bacterial surface by binding to at least some (1 to 4) of the lipid A's acyl moieties, leaving the others lipids or other LPS-related structures free to anchor the whole LPS/MD-2 complex to the bacterial surface. These results indicate that the binding of recombinant MD-2 to the cell surface of live Gram-negative bacteria is tight (ie, it resists extensive washing), and can directly bridge the bacterial cell surface to the extracellular domain of TLR4, here in the form of the soluble TLR4-Fc fusion protein.
Recombinant MD-2 binds to live bacteria. (A) The indicated live bacteria were left untreated (shaded profiles) or were opsonized with MD-26xHis (solid line) and subjected to cytofluorimetry using an Alexa 647–labeled α-6xHis mAb. The Nm lpxA− strain is LPS−. (B) Live Yp cells were incubated with MD-26xHis, and surface-bound MD-2 was detected by cytofluorimetry using either TLR4-Fc or an α–MD-2 mAb followed by a FITC-labeled α-mouse antiserum (solid-line profiles). The shaded profiles represent the binding of the secondary reagent without MD-2 coating. Fluorescence intensity is plotted in a log scale (x-axis, 10-105). (C) Live Yp were adsorbed to high-protein-binding plates in either fixed amounts (2 × 107/well; left panel) or in 2-fold dilutions (right panel). MD-26xHis was then applied to the wells in 2-fold dilutions (left panel) or in a fixed amount (20 ng/mL; right panel). MD-2 bound to adsorbed bacteria was detected by ELISA using an α–MD-2 mAb. Results are shown as the average plus SD of triplicate absorbance readings at 450 nm. (D) Yp (Kim5) was left untreated (left panel) or was incubated with MD-26xHis as in panel A (right panel). MD-26xHis was stained with an α-6xHis mAb and imaged by SEM. Shown are the back scatter images of 2 random fields acquired at 8000×. White bar equals 1 μm.
Recombinant MD-2 binds to live bacteria. (A) The indicated live bacteria were left untreated (shaded profiles) or were opsonized with MD-26xHis (solid line) and subjected to cytofluorimetry using an Alexa 647–labeled α-6xHis mAb. The Nm lpxA− strain is LPS−. (B) Live Yp cells were incubated with MD-26xHis, and surface-bound MD-2 was detected by cytofluorimetry using either TLR4-Fc or an α–MD-2 mAb followed by a FITC-labeled α-mouse antiserum (solid-line profiles). The shaded profiles represent the binding of the secondary reagent without MD-2 coating. Fluorescence intensity is plotted in a log scale (x-axis, 10-105). (C) Live Yp were adsorbed to high-protein-binding plates in either fixed amounts (2 × 107/well; left panel) or in 2-fold dilutions (right panel). MD-26xHis was then applied to the wells in 2-fold dilutions (left panel) or in a fixed amount (20 ng/mL; right panel). MD-2 bound to adsorbed bacteria was detected by ELISA using an α–MD-2 mAb. Results are shown as the average plus SD of triplicate absorbance readings at 450 nm. (D) Yp (Kim5) was left untreated (left panel) or was incubated with MD-26xHis as in panel A (right panel). MD-26xHis was stained with an α-6xHis mAb and imaged by SEM. Shown are the back scatter images of 2 random fields acquired at 8000×. White bar equals 1 μm.
Human sMD-2 binds to the surface of Gram-negative bacteria
We next sought to determine whether soluble serum-derived endogenous MD-2 (sMD-2) binds to live Gram-negative bacteria in a manner similar to its recombinant counterpart. Human serum from healthy donors contains sMD-2.7,10,28 Using a capture scheme based on either PAS-immobilized TLR4-Fc or live Yp cells, we isolated and visualized endogenous sMD-2 by Western blotting. The pherograms of a representative isolation from one donor are shown in Figure 2A. Consistent with the results obtained with MD-26xHis, serum derived sMD-2 which was captured using TLR4-Fc migrates with 2 main glycoforms27 (Figure 2A top panel). Even though MD-2 harbors 2 N-glycosylation sites, no glycoform appears to bind preferentially to the surface of Yp (Figure 2A bottom panel), which is consistent with the marginal role that MD-2 glycans play in LPS recognition (refer to Visintin et al6 for a thorough discussion on this topic). In order to further characterize the binding of serum-derived MD-2 to the surface of live Yp, we treated the live bacteria with PHS and detected bacteria-bound MD-2 by FACS analysis using either TLR4-Fc or an α–MD-2 mAb. As shown in Figure 2B, both TLR4-Fc and the mAb bound to MD-2 on the surface of live Yp (Figure 2B left panels). The signal was lost when we treated bacteria with PHS which was depleted of sMD-2 using TLR4-Fc bound to PAS beads7 (Figure 2B middle panels). Reactivity was reinstated by reconstituting depleted PHS with MD-26xHis (Figure 2B right panels) and, similar to MD-26xHis, the binding of human sMD-2 to the surface of live Yp was specific and saturable (Figure 2C).
Human serum–derived MD-2 binds to bacteria. (A) PAS-conjugated TLR4-Fc or live Yp cells were used to precipitate MD-2 from the serum of a healthy individual or 15 mL of baculoviral supernatants containing MD-26xHis as a positive control. The pellets were washed and Western blotted for the presence of MD-2 using a commercial α–MD-2 mAb, followed by an HRP-conjugated α-mouse antiserum and ECL. The 160-kDa band in the uppermost panel is TLR4-Fc, which is recognized by the α-mouse antiserum. Similar results were obtained with 2 other healthy donors. Vertical lines have been inserted to indicate repositioned gel lanes. (B) Live Yp cells were stained with PHS (left panels), PHS that was depleted of MD-2 using TLR4-Fc (middle panels), and depleted PHS that was reconstituted with recombinant MD-2 (20 ng/mL; right panels). Bound MD-2 was detected by FACS using either TLR4-Fc (top panels) or an α–MD-2 mAb followed by an FITC-labeled α-mouse pAb. The shaded profiles represent the binding of the secondary reagent without MD-2 coating. Fluorescence intensity is plotted in a log scale (x-axis; 10-105). (C) Bacteria were adhered to plastic in 2-fold dilution and incubated with PHS. Human serum–derived soluble MD-2 bound to the surface of the bacteria was revealed by ELISA as in Figure 1C. Data are averages of triplicates plus SD.
Human serum–derived MD-2 binds to bacteria. (A) PAS-conjugated TLR4-Fc or live Yp cells were used to precipitate MD-2 from the serum of a healthy individual or 15 mL of baculoviral supernatants containing MD-26xHis as a positive control. The pellets were washed and Western blotted for the presence of MD-2 using a commercial α–MD-2 mAb, followed by an HRP-conjugated α-mouse antiserum and ECL. The 160-kDa band in the uppermost panel is TLR4-Fc, which is recognized by the α-mouse antiserum. Similar results were obtained with 2 other healthy donors. Vertical lines have been inserted to indicate repositioned gel lanes. (B) Live Yp cells were stained with PHS (left panels), PHS that was depleted of MD-2 using TLR4-Fc (middle panels), and depleted PHS that was reconstituted with recombinant MD-2 (20 ng/mL; right panels). Bound MD-2 was detected by FACS using either TLR4-Fc (top panels) or an α–MD-2 mAb followed by an FITC-labeled α-mouse pAb. The shaded profiles represent the binding of the secondary reagent without MD-2 coating. Fluorescence intensity is plotted in a log scale (x-axis; 10-105). (C) Bacteria were adhered to plastic in 2-fold dilution and incubated with PHS. Human serum–derived soluble MD-2 bound to the surface of the bacteria was revealed by ELISA as in Figure 1C. Data are averages of triplicates plus SD.
MD-2 binding to bacterial surfaces is independent of other serum components
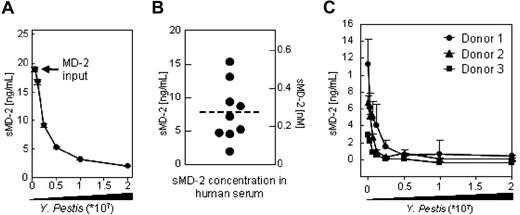
In order to determine whether serum components influence the binding of MD-2 to the surface of Gram-negative bacteria, we calculated the number of MD-2 binding sites on the surface of Yp using recombinant MD-2 in HBSS, and compared it with the number of MD-2 binding sites obtained by using whole sera as a source of MD-2. The number of binding sites was estimated by quantifying MD-2 in the postcellular supernatants of binding reactions in which the amount of MD-2 was kept constant and bacteria were titrated. Increasing amounts of bacteria were predicted to eventually deplete MD-2 from the postcellular supernatants. As shown in Figure 3A, about 1.5 × 107 live bacteria were required to deplete 20 ng of MD-26xHis. A total of 20 ng of MD-2 contains about 4 × 1011 molecules of MD-2, which corresponds to approximately 30 000 MD-2 binding sites/bacterium. Using a similar approach, we calculated the number of MD-2 binding sites when whole serum was used as a source of sMD-2. In order to provide an accurate determination of the initial concentration of sMD-2 in serum, we developed an MD-2 ELISA in which plastic-bound TLR4-Fc was used to capture sMD-2, and a commercial α–MD-2 mAb was used to detect the captured molecule. To further assure interexperimental and interlaboratory reproducibility, we used a commercially available MD-2 as a reference for concentration. As shown in Figure 3B, the concentration of sMD-2 in sera from human healthy donors ranged from 2 to 15 ng/mL. These values are consistent with values reported previously10,28 (T. G. Wolfs, personal communication, August 2007) and reflect the variability of sMD-2 concentration in human serum. From binding profiles similar to the ones shown in Figure 3C, we calculated that the number of MD-2 binding sites on the surface of bacteria incubated in the presence of serum averages 30 000 (50 000 ÷ 24 000).
Binding of MD-2 to the surface of Yp is independent of serum components. (A) The indicated amounts of live bacteria (x-axis) were incubated in HBSS with a fixed amount of MD-26xHis (input). MD-2 in the postcellular supernatants (y-axis) was quantitated by ELISA (B) and plotted as a function of the number of bacteria used in the binding reaction (x-axis). About 107 bacteria were necessary to deplete 20 ng of MD-26xHis input, which corresponds to about 40 000 MD-2 binding sites on the surface of Yp, assuming that bacteria have 106 molecules of LPS on their surfaces. Plotted values are the average of 2 independent experiments plus or minus the range. (B) Soluble MD-2 form the serum of healthy donors (n = 10) was determined by ELISA using plastic adsorbed TLR4-Fc to capture sMD-2 and a commercial α–MD-2 mAb as detection reagent. (C) Binding isothermes were generated as in panel A, but here human serum was used as a source of sMD-2. Plotted are sera from 3 representative donors. Error bars are the SD of triplicate determinations.
Binding of MD-2 to the surface of Yp is independent of serum components. (A) The indicated amounts of live bacteria (x-axis) were incubated in HBSS with a fixed amount of MD-26xHis (input). MD-2 in the postcellular supernatants (y-axis) was quantitated by ELISA (B) and plotted as a function of the number of bacteria used in the binding reaction (x-axis). About 107 bacteria were necessary to deplete 20 ng of MD-26xHis input, which corresponds to about 40 000 MD-2 binding sites on the surface of Yp, assuming that bacteria have 106 molecules of LPS on their surfaces. Plotted values are the average of 2 independent experiments plus or minus the range. (B) Soluble MD-2 form the serum of healthy donors (n = 10) was determined by ELISA using plastic adsorbed TLR4-Fc to capture sMD-2 and a commercial α–MD-2 mAb as detection reagent. (C) Binding isothermes were generated as in panel A, but here human serum was used as a source of sMD-2. Plotted are sera from 3 representative donors. Error bars are the SD of triplicate determinations.
Surface-bound MD-2 enhances bacterial internalization by activating the TLR4/MD-2 signaling axis
As the MD-2/LPS complex is the only reported activating ligand for TLR4, we next sought to determine whether bacteria-bound MD-2 affects the biology of TLR4-bearing cells. We established a phagocytosis assay in which YFP-expressing live bacteria were incubated with the phagocytes for 30 minutes at 37°C in serum-free medium (SFM). A phagocytic index was defined as the fluorescence associated with the wells containing infected cells minus the autofluorescence of uninfected cells (Figure 4A white bars). As shown in Figure 4A, coating live Nm and Yp with MD-26xHis (Figure 4A black bars) markedly enhanced the phagocytic index in mouse thioglycollate elicited peritoneal exudate cells (pφ), RAW macrophages, and human PBMCs. In order to determine whether the increase in the phagocytic index reflected an increased internalization of fluorescent bacteria, and not only an improvement in surface binding, we performed the confocal experiments shown in Figure 4B. Confocal microscopy allows for the detection of fluorescent particles (bacteria) in a 2D slice taken from the entire cell body, thus allowing the discrimination of its intracellular or extracellular localization. Both RAW macrophages and pMφ accumulated an appreciably higher number of bacteria in their cytoplasm when they were infected with MD-26xHis–treated bacteria.
MD-2 enhances opsonophagocytosis. (A) Adherent pMφ, RAW macrophages, or PBMCs were infected for 30 minutes with untreated (▩) or MD-26xHis–opsonized (20 ng/mL; ■) fluorescent protein expressing live Nm and Yp. Shown is the average fluorescence of triplicate wells plus SD. Fluorescence units were plotted on an arbitrary scale (AFU), and cellular autofluorescence (□) was set to 0 (phagocytic index). (B) RAW macrophages or mouse pMφ were incubated with YFP-Yp as in panel A and were subjected to confocal microscopy. Nuclei were stained with Hoescht 33258 (blue), and the cell surface was stained with an α-CD11b (red). Magnification 40×.
MD-2 enhances opsonophagocytosis. (A) Adherent pMφ, RAW macrophages, or PBMCs were infected for 30 minutes with untreated (▩) or MD-26xHis–opsonized (20 ng/mL; ■) fluorescent protein expressing live Nm and Yp. Shown is the average fluorescence of triplicate wells plus SD. Fluorescence units were plotted on an arbitrary scale (AFU), and cellular autofluorescence (□) was set to 0 (phagocytic index). (B) RAW macrophages or mouse pMφ were incubated with YFP-Yp as in panel A and were subjected to confocal microscopy. Nuclei were stained with Hoescht 33258 (blue), and the cell surface was stained with an α-CD11b (red). Magnification 40×.
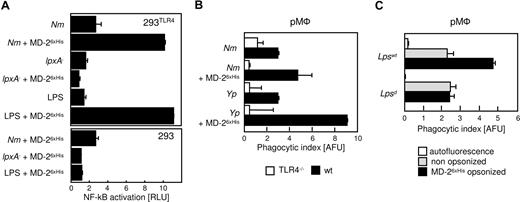
Since clustering of MD-2 activates TLR4 in 293 cells,11 we questioned whether the enhancing effect of bacteria-bound MD-2 was mediated by TLR4 signaling. In order to test this hypothesis, we first used 293 cells stably transfected with human TLR4 along with an NF-κB-luciferase reporter gene.19 These cells do not activate NF-κB in response to LPS, unless MD-2 is added as a conditioned supernatant, a transgene, or a purified protein.27 As shown in Figure 5A, 293TLR4 cells that were incubated with Nm that was coated with MD-26xHis markedly induced NF-κB activity (Nm + MD-26xHis), an effect not seen in untransfected 293 cells (Figure 5A; bottom panel). Similar results were obtained with Yp (data not shown). As expected, the LPS deficient strain of Nm, which does not bind to MD-2, failed to activate the TLR4-expressing cells. Since all the incubations were performed in DMEM without FCS or any other additional serum component, we infer that MD-2 that was bound to the bacterial cell wall aggregated TLR4 on the surface of the reporter cells, thus activating them.
The prophagocytic effect of MD-2 is mediated by TLR4. (A) 293 cells expressing TLR4 and a NF-κB-luciferase reporter plasmid were stimulated with live Nm (MC58) or with Nm lpxA− that had been treated as indicated (y-axis). Luciferase readings were normalized by the activity of unstimulated cells (RLU = 1). Shown are the averages of duplicate points plus the range. LPS served as control. A similar experiment was performed with 293 cells (bottom panel). (B) pMφ from TLR4−/− (□) or wild-type (■) mice were infected with GFP-Nm or YFP-Yp that were processed for the phagocytosis assay as in Figure 4A. Note that MD-2 coating exerts no effect on TLR4−/− cells. (C) pMφ from the BALB/C (Lpswt) and C3H/HeJ (Lpsd) mice were infected with nonopsonized (▩) or MD-26xHis–coated YFP-Yp and subjected to fluorescent opsonophagocytosis assay as in panel B. These experiments were repeated at least 3 times with similar results. Data are averages of triplicates plus SD.
The prophagocytic effect of MD-2 is mediated by TLR4. (A) 293 cells expressing TLR4 and a NF-κB-luciferase reporter plasmid were stimulated with live Nm (MC58) or with Nm lpxA− that had been treated as indicated (y-axis). Luciferase readings were normalized by the activity of unstimulated cells (RLU = 1). Shown are the averages of duplicate points plus the range. LPS served as control. A similar experiment was performed with 293 cells (bottom panel). (B) pMφ from TLR4−/− (□) or wild-type (■) mice were infected with GFP-Nm or YFP-Yp that were processed for the phagocytosis assay as in Figure 4A. Note that MD-2 coating exerts no effect on TLR4−/− cells. (C) pMφ from the BALB/C (Lpswt) and C3H/HeJ (Lpsd) mice were infected with nonopsonized (▩) or MD-26xHis–coated YFP-Yp and subjected to fluorescent opsonophagocytosis assay as in panel B. These experiments were repeated at least 3 times with similar results. Data are averages of triplicates plus SD.
In order to determine the contribution of the TLR4/MD-2 signaling axis in the phagocytic process, we compared pMφ derived from TLR4-deficient and -sufficient mice. As shown in Figure 5B, MD-2 coating enhanced phagocytosis significantly in wild-type cells, where the TLR4/MD-2 axis is intact. On the contrary, MD-2 coating had no effect on the internalization of bacteria by the TLR4-deficient cells. A similar result was obtained with macrophages from the TRIF/MyD88 double-knockout mice, in which TLR4 signaling is nearly, if not entirely, abrogated (data not shown) despite the presumed normal expression of surface TLR4. In order to determine the precise role of signal transduction in MD-2–mediated opsonophagocytosis, we examined the uptake of MD-2–coated Yp by macrophages from BALB/C mice expressing Lpsd, a mutant form of TLR4 in which a point mutation in the TIR domain of the receptor that renders the receptor nonfunctional. Although Lpsd will not signal in response to LPS, the ectodomain of the molecule is expressed and is unaffected by the cytoplasmic mutation. Lpsd mutant macrophages failed to exhibit the enhanced phagocytosis observed in wild-type BALB/C macrophages (Figure 5C). Taken together, these results suggest that the opsonic activity of serum MD-2 requires signal transduction downstream from TLR4 in order to be observed, and involves at least one other surface receptor on macrophages.
MD-2 opsonization enhances inflammatory phagocytosis via the TLR4/MD-2 signaling axis
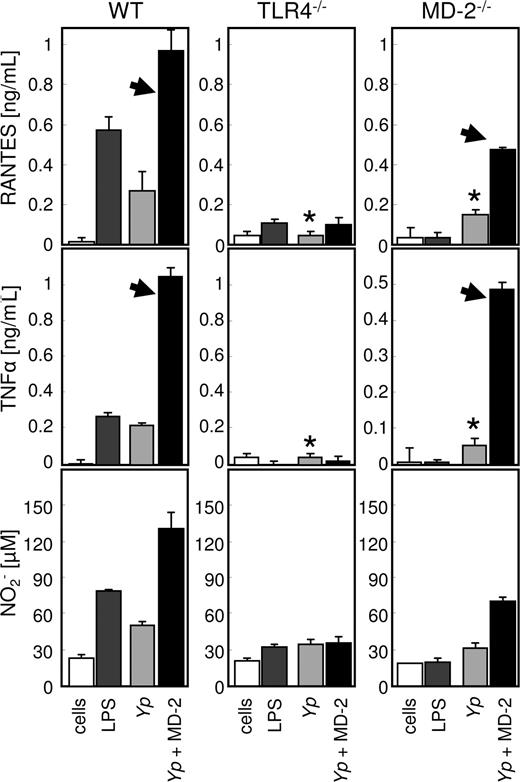
Since MD-2 treatment affected opsonophagocytosis in a TLR4-dependent manner, we speculated that the internalization of live bacteria would be accompanied by the activation of the phagocytes. In order to test this hypothesis, we determined the content of nitrite (as a measure of bactericidal potential29 ), RANTES (as a representative leukocyte chemoattractant), and TNF-α (as a representative proinflammatory cytokine) in the supernatant of infected pMφ from wild-type, TLR4−/−, and MD-2−/− mice. As shown in Figure 6, all the cellular responses examined were markedly enhanced by MD-2 opsonization in the cells derived from the wild-type mouse. The response of TLR4-deficient pMφ was modest and not affected by MD-2 opsonization, indicating that the TLR4/MD-2 signaling axis is pivotal in the cellular reaction to live Gram-negative bacteria such as Yp.30 Not surprisingly, MD-2–deficient cells resembled the TLR4−/− phenotype when bacteria were not opsonized with MD-2 (Figure 6 asterisks), but reacted to the presence of recombinant MD-2 when it was provided on the bacterial surface as opsonin (Figure 6 arrowheads). In contrast to RANTES and TNF-α, nitric oxide production in response to live bacteria appeared to be almost entirely dependent on MD-2 opsonization and therefore on TLR4 signaling (Figure 6 bottom left panel).
MD-2 opsonization enhances cytokine and nitrite production. pMφ from wild-type (left column), TLR4 (middle column), and MD-2 mice (right column) were left untreated (□) or treated with LPS (■), live Yp ( ), or MD-2–coated Yp (
), or MD-2–coated Yp ( ). RANTES, TNF-α, and nitrite concentrations were determined from the same supernatants by ELISA. The arrowheads indicate the restoration of the “wild-type” phenotype in MD-2−/− cells, when MD-2 is provided on the surface of bacteria. *In the absence of MD-2 treatment, MD-2–deficient pMφ resemble the TLR4 knockout phenotype. Results are the average of triplicate readings plus SD, and the experiment is representative of 3.
). RANTES, TNF-α, and nitrite concentrations were determined from the same supernatants by ELISA. The arrowheads indicate the restoration of the “wild-type” phenotype in MD-2−/− cells, when MD-2 is provided on the surface of bacteria. *In the absence of MD-2 treatment, MD-2–deficient pMφ resemble the TLR4 knockout phenotype. Results are the average of triplicate readings plus SD, and the experiment is representative of 3.
MD-2 opsonization enhances cytokine and nitrite production. pMφ from wild-type (left column), TLR4 (middle column), and MD-2 mice (right column) were left untreated (□) or treated with LPS (■), live Yp ( ), or MD-2–coated Yp (
), or MD-2–coated Yp ( ). RANTES, TNF-α, and nitrite concentrations were determined from the same supernatants by ELISA. The arrowheads indicate the restoration of the “wild-type” phenotype in MD-2−/− cells, when MD-2 is provided on the surface of bacteria. *In the absence of MD-2 treatment, MD-2–deficient pMφ resemble the TLR4 knockout phenotype. Results are the average of triplicate readings plus SD, and the experiment is representative of 3.
). RANTES, TNF-α, and nitrite concentrations were determined from the same supernatants by ELISA. The arrowheads indicate the restoration of the “wild-type” phenotype in MD-2−/− cells, when MD-2 is provided on the surface of bacteria. *In the absence of MD-2 treatment, MD-2–deficient pMφ resemble the TLR4 knockout phenotype. Results are the average of triplicate readings plus SD, and the experiment is representative of 3.
MD-2 opsonization enhances intracellular killing
In order to test whether the enhancement in cellular activation translates into improved bactericidal activity, we performed gentamicin protection assays in which the viability of ingested bacteria was assessed after opsonophagocytosis. As shown in Figure 7A, PBMCs killed both Yp and Nm that had been opsonized with MD-26xHis more efficiently. In order to exclude any artifactual contribution of gentamicin in the killing of ingested bacteria, we performed opsonophagocytosis experiments with both PBMCs and pMφ in which extracellular bacteria were washed and no antibiotic was added with comparable results (data not shown). Thus, TLR4 engagement during phagocytosis not only drives a proinflammatory phenotype in the involved cells, but also enhances their bactericidal potential. Opsonization almost doubled bacterial internalization (Figure 4A), yet it reduced bacteria viability by more than 60% in the same cells.
MD-2 opsonization enhances phagocytosis and intracellular killing. (A) Human PBMCs were incubated for 30 minutes with Yp and Nm that were left untreated or were opsonized with MD-26xHis. Extracellular bacteria were killed by gentamicin treatment, and cells were chased for 6 additional hours in DMEM. Viability of internalized bacteria was assessed by colony count (CFU) and plotted as the average of duplicate determinations plus the range. Similar results were obtained with PBMCs from 2 other unrelated donors. (B) Adherent PBMCs were infected with either untreated or MD-26xHis–opsonized YFP-Yp in the absence (SFM) or presence of 30% PHS. Samples were processed and plotted as in Figure 4A. Note the scale difference between the 2 panels. (C) pMφ from the MD-2−/− mouse were treated as in panel A. (D) pMφ from the MD-2−/− mouse were infected with untreated or MD-26xHis–opsonized Yp in the presence (□) or absence (■) of PHS. Colony count and plotting of live phagocytosed bacteria was performed as in Figure 7A. These experiments were repeated at least 3 times with similar results. Data are averages of triplicates plus SD.
MD-2 opsonization enhances phagocytosis and intracellular killing. (A) Human PBMCs were incubated for 30 minutes with Yp and Nm that were left untreated or were opsonized with MD-26xHis. Extracellular bacteria were killed by gentamicin treatment, and cells were chased for 6 additional hours in DMEM. Viability of internalized bacteria was assessed by colony count (CFU) and plotted as the average of duplicate determinations plus the range. Similar results were obtained with PBMCs from 2 other unrelated donors. (B) Adherent PBMCs were infected with either untreated or MD-26xHis–opsonized YFP-Yp in the absence (SFM) or presence of 30% PHS. Samples were processed and plotted as in Figure 4A. Note the scale difference between the 2 panels. (C) pMφ from the MD-2−/− mouse were treated as in panel A. (D) pMφ from the MD-2−/− mouse were infected with untreated or MD-26xHis–opsonized Yp in the presence (□) or absence (■) of PHS. Colony count and plotting of live phagocytosed bacteria was performed as in Figure 7A. These experiments were repeated at least 3 times with similar results. Data are averages of triplicates plus SD.
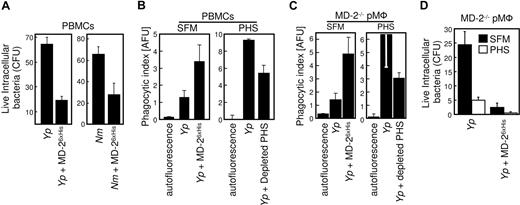
In order to study the effect of serum-derived sMD-2 in both opsonophagocytosis and intracellular bactericidal activity, we infected PBMCs with Yp in the presence of PHS (Figure 7B right panel; Yp. Compared with the internalization of bacteria in protein-free conditions (Figure 7B left panel), PHS greatly promoted bacterial internalization of untreated Yp organisms. This enhancing effect was substantially reduced when PHS was depleted of MD-2 with TLR4-Fc (Figure 7B right panel; + depleted PHS), an MD-2 dependent outcome that reflects the binding experiments shown in Figure 2. Since phagocytosis was not completely abrogated under conditions known to deplete the activity of MD-2,7 we surmised that MD-2 produced by the phagocytes themselves contributed to the phagocytic process. In order to address this point, we used macrophages from MD-2−/− mice and performed similar opsonophagocytosis experiments (Figure 7C). MD-2–deficient cells were efficiently complemented by both MD-26xHis (Yp + MD-2; Figure 7C left panel) and serum-derived MD-2 (Yp; Figure 7C right panel). MD-2 depletion from PHS reduced substantially the internalization of live bacteria (Yp + depleted PHS; Figure 7C last bar). Taken together, these results suggest that both human sMD-2 present in PHS and recombinant MD-2 contribute to the phagocytic process. In addition, endogenous sMD-2 (PHS) appeared to enhance the killing of internalized bacteria, as shown in Figure 7D. In these experiments, MD-2–deficient phagocytes were infected with Yp (or Nm; not shown) in the presence of PHS (Figure 7D open bars) or in serum free conditions (Figure 7D black bars). The amount of intracellular killing which was mediated by either sMD-2 containing PHS or MD-26xHis opsonization was consistently higher than that obtained by infecting the cells in SFM.
Discussion
The main novel findings presented in this paper are as follows: (1) approximately 30 000 molecules of MD-2 (recombinant or serum-derived) bind specifically to LPS on the surface of live Gram-negative bacteria; (2) MD-2 binding to Gram-negative bacteria is resistant to extensive washing; and (3) phagocytosis and intracellular killing of MD-2–opsonized bacteria depend on TLR4 signaling. Considering the soluble nature and the distribution of sMD-2 (approximately 10 ng/mL or 0.3 nM in human serum), it is reasonable to assume that MD-2 is among the first components that a microbe encounters after breaching the body's physical barriers. Based on the data presented here, sMD-2 is anticipated to be a central player in the antibacterial defense program in mice and humans. However, our studies raise the issue of whether the pro-opsonophagocytic effect of sMD-2 is physiologically relevant, since the reaction to a pathogen in vivo relies on many complex dynamic systems in interaction with each other. In order to establish a reciprocal connection between the in vitro experimental findings and the biological role of MD-2 in the whole organism, one would need to study individuals in which MD-2 activity or expression levels are impaired. Since there is no reported condition linked to such deficiencies, this question cannot be answered directly in humans. Nevertheless, traditional reductionistic laboratory methods are the best way of testing simple causal theories because they allow for the manipulation of a limited set of interdependent variables that influence both the outcomes and each other. For this reason, we developed and performed opsonophagocytosis assays in protein-free conditions in order to detect only very strong effects, which are likely to be physiologically relevant. It is imperative to emphasize that under the defined conditions used in this work, MD-2 opsonization more than doubled bacterial internalization (Figures 4A,5B) and yet enhanced intracellular killing by more than 60% in the same cells (Figure 7). Therefore, the effect of MD-2 opsonization appears to be even more dramatic when it is considered in absolute terms. In addition, the PHS depletion experiments using MD-2–deficient phagocytes (Figure 7) demonstrate that sMD-2 (and not cellular MD-2) can account for about 50% of the entire phagocytic and killing activities, even in the presence of soluble serum factors. Finally, the presence of soluble MD-2 in normal human serum (and other biological fluids such as milk, tears, urine, and the serum of chimpanzee and Macacus rhesus; V.J. and A.V., unpublished observations, September 2007) further supports a physiologic role for soluble MD-2 in the antibacterial defenses of primates, rodents, and, presumably, mammals in general.
Mouse and human phagocytes are known to express simultaneously both TLR4 and MD-2. The interaction between TLR4 and MD-2 is noncovalent; hence, TLR4 interacts with MD-2 in accordance to the law of mass action. The calculated Kd for this interaction is about 12 nM.7,27 This implies that, independent of the source of MD-2 (the same TLR4-expressing cell or the interstitial/extracellular fluids), less than 50% of the total TLR4 on the cell surface is expected to be saturated with MD-2 at chemical equilibrium, because the Kd of this interaction is about 40 times higher than the concentration of sMD-2 in serum. Therefore, a large proportion of cellular TLR4 is expected to be available for the binding to bacteria-associated MD-2. Since we could not detect LPS dependent differences in the Kd of the TLR4-Fc/MD-2 equilibrium, a first-order binding kinetic of noncooperative binding appears to govern the interaction between TLR4 and LPS-ligated or -free MD-2.7 Accordingly, the binding affinity of TLR4 for MD-2 is predicted to increase if TLR4 clusters upon binding to patched MD-2, in a manner that is reminiscent of the interaction of the Fc gamma receptor with aggregated immunoglobulins.31 Consequently, it is conceivable to assume that bacteria-clustered MD-2 has the ability of displacing MD-2 that is prebound to TLR4.
The sparse and homogeneous distribution of MD-26xHis on the surface of bacteria shown in the SEM micrographs of Figure 1D is likely to reflect the harsh conditions used in the preparation of these samples. In fact, we estimated that the surface of live Yp bears about 30 000 MD-2 docking sites, roughly corresponding to one molecule of MD-2 per 33 molecules of LPS, assuming that bacteria have approximately 106 molecules of LPS on their surfaces.26 Considering the steric hindrance of MD-2 toward LPS,9 and the presence of other surface structures and proteins that require LPS solvation, the mechanism of MD-2 binding to LPS in the outer layer of the bacteria wall can only be hypothesized. We favor a model in which the bacterial surface provides a platform on which MD-2 is layered homogeneously and exposes most of its surface to TLR4. This view is consistent with the observation that 2 mAbs and TLR4-Fc combine with bacteria-bound MD-2 (Figure 1A,B). The results presented in this work also suggest that at least 2 complementary modes of LPS recognition and TLR4 activation exist, depending on whether LPS is partially embedded in the cell wall or it is delivered to MD-2 as a soluble molecule. In the first case (as presented here), the binding appears to be independent of serum components. MD-2 is likely to bind only to a fraction of the available lipids of any given lipid A moiety,5,9 probably allowing the others to maintain cell-wall anchorage. As a consequence, surface-patterned MD-2 passively aggregates TLR4, which in turn initiates an activating signal on the phagocytes. This model is consistent with the structure of the TLR4/MD-2 complex, in which the ligand-binding cavity of MD-2 appears to be directed opposite to its TLR4-interacting surface.5 In the second scenario, multiple MD-2 molecules (most likely 2; AV data not shown) gain access to the whole complement of lipid A's acyl chains, thus generating a TLR4-activating ligand. Although we could not exclude that TLR4 activation occurred as a consequence of the release of MD-2/LPS complexes from the bacteria during the phagocytic process, our results are consistent with the idea that opsonized live bacteria induce the activation of TLR4 in the developing phagosome and ensure that the “danger” signal associated with the presence of LPS is delivered in the very same phagocytic compartment in which killing, antigen processing, and presentation ought to be promoted, as proposed by Blander and Medzhitov.32,33 An additional implication of these findings is that during phagocytosis of live bacteria, which are a good source of freshly produced ATP, both ATP and pathogen-associated molecular patterns (PAMPs) are concentrated together in the phagosome and can find their way to the cytosol, thus providing optimal conditions for the activation of the inflammasome.34
We observed that both TLR4 and MD-2 are an absolute requirement for mediating proinflammatory opsonophagocytosis of Gram-negative bacteria. This is in contrast with previously published results suggesting that the effect of MD-2 is observed only at early time points (less than 20 minutes).16 Although the overall conclusions of both the works are overlapping, we believe that the difference might be due to the fact that we used live bacteria and phagocytes from gene-targeted animals, whereas Tissieries et al used lyophilized bioparticles and antibody inhibition of TLR4 for their phagocytosis assays. Thus, the 2 systems are hardly comparable.
Intuitively, it might be assumed that blocking MD-2 activity is beneficial in LPS-related conditions such as the sepsis syndrome. This notion has been supported by a variety of reports, including one from our group.7,35 At the same time, an early MD-2/TLR4–dependent “innate” reaction to the presence of Gram-negative bacteria is undisputedly a pivotal defense mechanism, since a strong TLR4 response is necessary and sufficient in order to overcome the totality of the multifaceted virulence armamentarium that an organism like Yp can deploy.30 The results presented in the present work provide a rationale for the latter observation, and demonstrate that interfering with MD-2 activity might be detrimental in the earliest stages of exposure to Gram-negative bacteria. Understanding how serum components such as MD-2 direct and modulate the fate of the phagocytic cargo might inspire novel strategies for developing efficient therapeutic and prophylactic interventions aimed at accelerating pathogen clearance and altering the course of infection.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We wish to acknowledge Drs Steeghs and van der Ley (Netherlands Vaccine Institute, Bilthoven, The Netherlands) for providing us with the Nm H44/76-lpxA strain, Dr Agarwal for the GFP-Nm strain, Dr Viriyakosol for the MD-26xHis baculovirus, and Dr Wozniak for helping with mouse experiments. We are also indebted to Drs Akira and Miyake and the Japan Science and Technology Agency for providing us with the TLR4 and MD-2 knockout mice, respectively.
This study was supported by National Institutes of Health grants AI069084 (A.V. and V.J.), RO1GM54060 (D.T.G., K.A.H., and A.H.), AI057588 (E.L.), AI054544 (S.R.), and AI052455 (M.C.-D.).
National Institutes of Health
Authorship
Contributions: A.V. supervised the project; A.V., V.J., and D.T.G. designed research and analyzed the results; V.J. performed all the experiments; A.H. helped with producing SEM and confocal micrographs; K.A.H. and A.V. produced critical reagents; E.L., M.C.-D., and S.R. provided bacterial strains; and A.V., V.J., and D.T.G. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alberto Visintin, University of Massachusetts Medical School, LRB Rm 370L, 364 Plantation St, Worcester, MA 01605; e-mail: alberto.visintin@umassmed.edu.
References
Author notes
*D.T.G. and A.V. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal