Abstract
Tight regulation of the balance between apoptosis and survival is essential in angiogenesis. The ETS transcription factor Erg is required for endothelial tube formation in vitro. Inhibition of Erg expression in human umbilical vein endothelial cells (HUVECs), using antisense oligonucleotides, resulted in detachment of cell-cell contacts and increased cell death. Inhibition of Erg expression by antisense in HUVECs also lowered expression of the adhesion molecule vascular endothelial (VE)–cadherin, a key regulator of endothelial intercellular junctions and survival. Using chromatin immunoprecipitation, we showed that Erg binds to the VE-cadherin promoter. Furthermore, Erg was found to enhance VE-cadherin promoter activity in a transactivation assay. Apoptosis induced by inhibition of Erg was partly rescued by overexpression of VE-cadherin–GFP, suggesting that VE-cadherin is involved in the Erg-dependent survival signals. To show the role of Erg in angiogenesis in vivo, we used siRNA against Erg in a Matrigel plug model. Erg inhibition resulted in a significant decrease in vascularization, with increase in caspase-positive endothelial cells (ECs). These results identify a new pathway regulating angiogenesis and endothelial survival, via the transcription factor Erg and the adhesion molecule VE-cadherin.
Introduction
Angiogenesis, namely the formation of new vessels from preexisting ones, is essential for normal development, as well as for pathologic conditions, including tumor development, diabetic retinopathy, atherosclerosis, and rheumatoid arthritis. In recent years, endothelial apoptosis was shown to play an important role in remodeling the vascular network. Endothelial apoptosis is observed at the initiation of angiogenesis and at the branching and regression of neovessels.1-3 Angiogenesis is controlled by the balance between proangiogenic factors and angiogenesis inhibitors, which also regulate apoptosis of endothelial cells (ECs). Indeed, the mechanism underlying many antiangiogenic therapies appears to be the induction of endothelial cell death.4
Among the transcription factors involved in the regulation of angiogenesis and vascular development are members of the ETS family. This large family of transcription factors contains approximately 30 members that share a highly conserved DNA-binding domain (ETS domain) and are involved in regulating a wide variety of biologic processes. Many ETS proteins are expressed early in the developing vasculature in several organisms, and loss-of-function studies in mice and zebrafish have shown a critical role for ETS factors in vascular development.5 In the adult, several endothelial ETS transcription factors were shown to regulate angiogenesis.6 Among these, the ETS family member Erg is constitutively expressed in ECs,7 and its expression is apparently restricted to ECs, megakaryocytes,8 and chondrocytes.9 Erg drives transcription of genes involved in endothelial homeostasis and angiogenesis, such as eNOS, HO-1, and ICAM-2.7,10,11 Data from animal models indicate that Erg is involved in endothelial differentiation and vascular development12 ; for example, Erg overexpression in Xenopus embryos resulted in ectopic endothelial differentiation.13 We have previously shown that Erg is required for angiogenesis in human ECs in vitro14 ; however, no data are available on the role of Erg in angiogenesis in the adult in vivo.
Endothelial junctions are crucial for the maintenance and regulation of vascular homeostasis and function and mediate a complex signaling network.15 A major regulator of adherent junctions is vascular endothelial (VE)–cadherin, a Ca2+-dependent cell-surface adhesion molecule that forms homophilic interactions and is required for the integrity of the endothelial monolayer, endothelial permeability, and the control of cell growth.16,17 VE-cadherin was clearly shown to regulate vascular development and angiogenesis: genetic inactivation of the VE-cadherin gene leads to early embryonic death because of vascular defects18,19 and antibodies to VE-cadherin inhibit angiogenesis both in vitro and in vivo (reviewed in Dejana et al20 ). VE-cadherin regulates a number of signaling events, by intracellular interaction with proteins of the armadillo family, including β-catenin and plakoglobin, as well as by clustering signaling molecules and growth factor receptors.15,19 In the mouse, VE-cadherin expression was shown to be regulated by ETS transcription factors, including the family founder member Ets-1.21,22 VE-cadherin was shown to protect ECs from apoptosis induced by serum starvation through pathways involving β-catenin, VEGF receptor-2, and the growth arrest–specific gene Gas-1.19,23
In this study, we investigate the mechanism by which the transcription factor Erg regulates angiogenesis and describe a new pathway involving Erg and VE-cadherin. Using a novel siRNA-mediated in vivo angiogenesis approach to inhibit Erg gene expression, we demonstrate for the first time that Erg is required for in vivo angiogenesis.
Methods
Cells
Human umbilical vein endothelial cells (HUVECs) were isolated as described by Jaffe et al24 and maintained in 1% gelatin-coated tissue-culture ware in Medium 199, supplemented with 20% fetal bovine serum (FBS; Sigma-Aldrich, Poole, United Kingdom), 30 μg/mL endothelial cell growth supplement (BD Biosciences, Oxford, United Kingdom), 10 U/mL heparin (CP Pharmaceuticals, Wrexham, United Kingdom). HeLa cells were grown in DMEM supplemented with 10% FBS. All growth media contained 2 mM l-glutamine, 100 IU/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen, Paisley, United Kingdom).
Design and delivery of Erg-specific GeneBloc and short interfering RNA
The details of synthesis and sequences of Erg and control GeneBlocs (GBs), obtained from Silence Therapeutics AG (Berlin, Germany), were already reported.14 RNA interference of Erg expression was induced with short interfering RNA (siRNA) directed against the gene at sequence 5′-AAAACTCTCCACGGTTAAT-3′, also generated by Silence Therapeutics. For the negative control, siRNA against the nonmammalian luciferase gene was used. Both the siRNA against Erg (73581DS) and the luciferase control siRNA were 2′O-methyl modified as described by Czauderna et al.25 For delivery into HUVECs, cells were seeded at 105 cells per well of a 35-mm, 6-well dish in EGM-2 medium (Lonza Wokingham, Wokingham, United Kingdom) the day before transfection. GB (final concentration, 100 nM) and lipid AtuFect01 (final concentration, 1 μg/mL; Silence Therapeutics) were mixed at 5 times concentration in OptiMEM medium (Invitrogen) at 37°C for 30 minutes in polystyrene tubes. siRNA (final concentration, 30 nM) was mixed with AtuFect01 as above. After mixing, the lipid-GB or siRNA complexes were added to each well of cells containing EGM-2 and incubated for 48 hours.
Real-time polymerase chain reaction and immunoblots
RNA and protein were prepared from HUVECs treated with GB or siRNA after 48 hours. Levels of Erg, ICAM-2, and VE-cadherin mRNA, normalized to GAPDH mRNA, were measured by real-time polymerase chain reaction (PCR). See Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) for a list of oligonucleotides used in this study. Immunoblotting of whole-cell lysates was performed with antibodies to Erg (Santa Cruz Biotechnology, Santa Cruz, CA), ICAM-2 (AbD Serotec, Oxford, United Kingdom), and VE-cadherin (BD Biosciences). Quantification of protein levels was performed by densitometry and normalized against α-tubulin (Sigma-Aldrich).
Immunofluorescence
HUVECs (5 × 104 cells) were cultured on gelatin-coated 13-mm diameter glass coverslips and treated with either Erg or control GB for 48 hours. Cells were fixed in 3% paraformaldehyde, permeabilized with 0.5% Triton-X100, and stained with antibodies to Erg (sc-354, rabbit polyclonal; Santa Cruz Biotechnology) or VE-cadherin (mouse monoclonal, clone 55-7H1; BD Biosciences). Secondary antibodies were anti–mouse AlexaFluor 555 and anti–rabbit AlexaFluor 488 (Invitrogen). Nuclei were visualized using TOPRO-3 (Invitrogen). All incubations were performed at room temperature for 15 minutes in PBS containing 3% BSA. Coverslips were mounted onto glass slides using VectorShield (Vector Laboratories, Peterborough, United Kingdom). Images were captured using a Zeiss LSM510 META confocal microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) running version 3.2 of the LSM acquisition software. Image processing was performed with Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Apoptosis assay
HUVECs, treated with Erg-specific or control GB for 48 hours, were fixed in ethanol (70%) at 4°C and stained with propidium iodide (PI; 50 μg/mL). The number of apoptotic cells was quantified by measuring the sub-G1 population by flow cytometry (Beckman Coulter Epics XL). Apoptosis was also quantified by measuring caspase 3 and 7 activation, using Caspase-Glo 3/7 Assay (Promega, Southampton, United Kingdom) on a Bio-Tek Synergy HT multidetection microplate reader.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using ChIP-IT (Active Motif, Rixensart, Belgium). HUVECs were grown to confluence in 15-cm diameter plates and fixed in formaldehyde (1%) for 10 minutes, cells were scraped from the plate, lysed, and homogenized using a Dounce homogenizer. Chromatin was sheared by enzymatic digestion for 10 minutes at 37°C. Precleared chromatin was then added to 2 μg Erg antibody (rabbit anti-Erg; Santa Cruz Biotechnology) or negative control rabbit IgG and incubated overnight on a rotator at 4°C. The antibody-chromatin mixture was then bound to Protein G beads for 1.5 hours. At the end of the incubation, beads were washed, and the immunoprecipitated DNA was eluted from the beads and purified by reversing cross-links, removal of RNA, and treatment with Proteinase K. DNA was then used as the template for PCR using primers specific for the VE-cadherin promoter sequence to amplify a region containing putative ETS-binding sites. To determine the specificity of the Erg-chromatin interactions, PCR amplification was performed with primers downstream of the promoter in a region that should not interact with transcription factors. (See Table S1 for oligonucleotide sequences.)
Reporter gene assays
The VE-cadherin promoter was PCR amplified from human genomic DNA and cloned into the pGL4 Luciferase Reporter Vector (Promega); see Table S1 for oligonucleotide sequences. Human Erg-2 (p52) cDNA was cloned into pSG5 (Stratagene, La Jolla, CA), as described previously.7 Transactivation assays were performed with the Dual-Luciferase Reporter Assay System (Promega). Cotransfection of HeLa cells was performed with GeneJuice transfection reagent (Merck Chemicals, Nottingham, United Kingdom), according to the manufacturer's conditions. Briefly, cells were seeded at 5 × 104 per well in a 24-well plate the day before transfection, incubated with 1.5 μL GeneJuice, 200 ng reporter DNA, 200 ng expression plasmid DNA (pSG5Erg or pSG5 empty vector), and 100 ng pTK-Renilla Luciferase (Promega) for 48 hours. Luciferase activities were measured with the use of a Synergy HT microplate reader.
Adenoviral infection of HUVECs with VE-cadherin–GFP or GFP
HUVECs (105 cells) grown in a 6-well dish were transduced with adenovirus (VE-cadherin [VEC]–GFP and GFP; kindly provided by F. William Luscinskas, Harvard Medical School, Boston, MA) as described previously.26 After 48 hours, cells were transfected with Erg or control GB (100 nM). Forty-eight hours later, cells were fixed in ethanol and stained with propidium iodide to determine apoptosis by flow cytometry; samples were also collected for RNA analysis. To discriminate between endogenous versus exogenous VE-cadherin, we designed a PCR strategy using primers spanning the 3′-UTR of the VE-cadherin gene, which only recognize the endogenous protein, and primers spanning the coding region, which recognize both endogenous and exogenous VE-cadherin. (See Table S1 for oligonucleotide sequences.)
In vivo angiogenesis Matrigel assay and siRNA inhibition
Mice (C57BL/6) were anesthetized with an isoflurane gas mixture and injected subcutaneously with the Matrigel Basement Membrane Matrix (BD Biosciences) mixture near the abdominal midline, to induce the formation of a single, solid gel plug. Matrigel preparation included 250 μL Matrigel, 64 U/mL heparin (CP Pharmaceuticals), 80 ng/mL basic fibroblast growth factor (FGF; R&D Systems, Abingdon, United Kingdom), 2 μM siRNA (Erg or control), and PBS to the final volume of 350 μL. Plugs were harvested after 7 days from killed mice, fixed in 4% (wt/vol) paraformaldehyde in PBS for 2 hours at room temperature, transferred to 70% ethanol, embedded in paraffin, and processed for hematoxylin and eosin staining. Experiments were performed according to the Animals Scientific Procedures Act of 1986.
Angiogenesis quantification and statistical analysis
For quantification, vessels contained in the Matrigel plug were identified by the presence of nucleated cells surrounding a lumen containing red blood cells. Vessels were counted in 4 fields of view using a 20× objective lens. Plugs from 5 mice were analyzed for each treatment. Statistical analysis was performed with an unpaired t test.
Immunofluorescence staining of Matrigel plugs
Sections (4-μm thick) of paraffin-embedded Matrigel plugs were dewaxed; antigen retrieval was performed by microwave treatment for 10 minutes in sodium citrate buffer (0.01 M, pH 6). After blocking nonspecific sites using an avidin-biotin blocking kit (Vector Laboratories), sections were incubated with anti-CD34 (AbCam, Cambridge, United Kingdom), anti-CD45 antibody (BD Biosciences), or IgG control (Santa Cruz Biotechnology). Sections were then incubated with biotinylated rabbit anti–rat antibody (Dako, Ely, United Kingdom), followed by signal amplification using Tyramine Signal Amplification Biotin System (PerkinElmer Life and Analytical Sciences, Waltham, MA). Detection of CD34 or CD45 was performed with Streptavidin-633 (Invitrogen); nuclei were counterstained using Sytox Green (Invitrogen). Images were captured using Zeiss LSM 510META.
Immunohistochemistry
Matrigel plugs were processed as described for immunofluorescence staining and incubated with rabbit anti-Erg antibody (sc-354; Santa Cruz Biotechnology), rabbit anti-Active Caspase-3 antibody (rabbit monoclonal, clone C92-605; BD Biosciences), or IgG control (Dako). Secondary antibody was biotinylated goat anti–rabbit (Vector Laboratories). Endogenous peroxidases were quenched with 3% H2O2 followed by amplification with ABC Elite Peroxidase (Vector Laboratories). Antibody complexes were detected using 3,3-diaminobenzidine tetrahydrochloride (Sigma-Aldrich). Slides were counterstained with Harris hematoxylin. Images were taken on an Olympus BX50 microscope fitted with an Olympus DP50-CU digital camera. Quantification of active caspase 3-positive cells was performed by counting the number of caspase-positive cells lining erythrocyte-containing vessels in 4 representative fields or sample, using the 20× objective. Statistical analysis was performed with an unpaired t test.
Results
Erg regulates junction stability and VE-cadherin expression
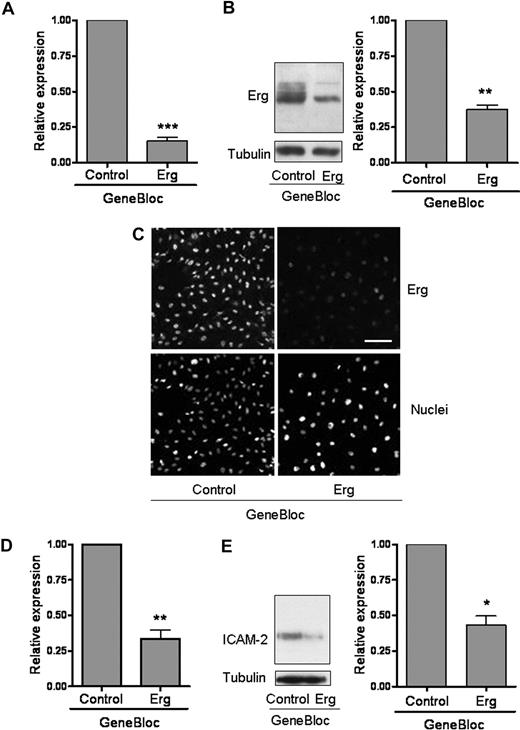
We have previously shown that the transcription factor Erg is required for in vitro tube formation on Matrigel,14 an assay based on the ability of cells to migrate, establish cell-cell and cell-matrix contact, and regulate survival. To study the role of Erg in endothelial cells, we have previously made use of modified antisense RNA oligonucleotides, called GeneBloc (GB).14 Inhibition of Erg expression by GB was achieved during the course of 24 to 78 hours, with most efficient inhibition (85% at the mRNA level, 65% at the protein level) after 48 hours (Figure 1A-C). mRNA levels of Fli-1, the ETS transcription factor most closely related to Erg,27 were not affected by GB treatment (data not shown). As expected, mRNA and protein levels of the adhesion molecule ICAM-2, a direct transcriptional target gene of Erg,7 were significantly decreased (Figure 1D,E), confirming the functional efficacy of Erg inhibition by GB. Time-lapse video microscopy of HUVECs in a confluent monolayer showed that treatment with Erg GB for 48 hours resulted in the disruption of endothelial cell-cell junctions, compared with control GB-treated cells (Figure 2A). ECs detached from each other, showing small protrusions all around the periphery of the cell body, whereas the monolayer of ECs treated with control GB retained the typical cobblestone shape. Therefore, Erg appears to be required for endothelial junction stability.
Inhibition of Erg expression in endothelial cells. HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. (A) Erg mRNA levels from RNA extracts were quantified using RT-PCR, normalized to GAPDH. Erg mRNA expression was significantly reduced after GB treatment, ***P < .001. Values are means plus or minus SEM, n = 4. (B) Whole-cell protein extracts were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) immunoblotting with antibodies to Erg and tubulin. Erg protein expression was significantly reduced in the Erg-GB–treated cells, **P < .01. Values are means plus or minus SEM, n = 3. (C) Immunofluorescence analysis of the distribution and expression of Erg before and after GeneBloc treatment. Cells grown on gelatin-coated glass coverslips were transfected with GB as above. After 48 hours, cells were labeled and visualized for Erg (top) or the nuclear marker TOPRO-3 (bottom). Scale bar, 100 μm. (D) ICAM-2 mRNA levels, measured in RNA extracts from GB-treated HUVECs using RT-PCR, normalized against GAPDH, were significantly decreased after Erg inhibition, **P < .01. Values are means plus or minus SEM, n = 3. (E) ICAM-2 protein expression was also down-regulated in Erg-GB–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to ICAM-2, normalized against tubulin, *P < .05. Values are means plus or minus SEM, n = 3.
Inhibition of Erg expression in endothelial cells. HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. (A) Erg mRNA levels from RNA extracts were quantified using RT-PCR, normalized to GAPDH. Erg mRNA expression was significantly reduced after GB treatment, ***P < .001. Values are means plus or minus SEM, n = 4. (B) Whole-cell protein extracts were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) immunoblotting with antibodies to Erg and tubulin. Erg protein expression was significantly reduced in the Erg-GB–treated cells, **P < .01. Values are means plus or minus SEM, n = 3. (C) Immunofluorescence analysis of the distribution and expression of Erg before and after GeneBloc treatment. Cells grown on gelatin-coated glass coverslips were transfected with GB as above. After 48 hours, cells were labeled and visualized for Erg (top) or the nuclear marker TOPRO-3 (bottom). Scale bar, 100 μm. (D) ICAM-2 mRNA levels, measured in RNA extracts from GB-treated HUVECs using RT-PCR, normalized against GAPDH, were significantly decreased after Erg inhibition, **P < .01. Values are means plus or minus SEM, n = 3. (E) ICAM-2 protein expression was also down-regulated in Erg-GB–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to ICAM-2, normalized against tubulin, *P < .05. Values are means plus or minus SEM, n = 3.
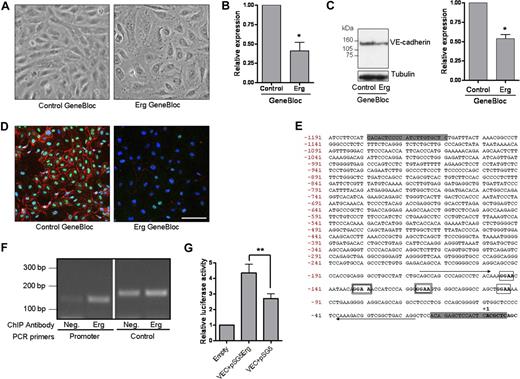
Erg regulates junctional integrity and VE-cadherin expression. (A) HUVECs (105 cells/well) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. Phase-contrast microscopy shows that the typical cobblestone monolayer shown in the control cells (left) has become disrupted in the Erg-inhibited cells (right). Original magnification 20×/0.4 Plan N objective lens (Olympus, Tokyo, Japan). (B) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. VE-cadherin mRNA levels were determined from RNA extracts from GB-treated HUVECs using RT-PCR normalized against GAPDH. VE-cadherin mRNA was significantly decreased after Erg inhibition, *P < 0.05. Values are means plus or minus SEM, n = 3. (C) VE-cadherin protein expression was also down-regulated in Erg-GB–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to VE-cadherin, normalized against tubulin. *P < .05. Values are means plus or minus SEM, n = 3. (D) HUVECs grown on gelatin-coated glass coverslips were treated with GB as above. After 48 hours, cells were labeled for Erg using a polyclonal rabbit anti-Erg antibody and for VE-cadherin using a mouse monoclonal anti–VE-cadherin antibody. Erg was visualized with AlexaFluor 488 (green), VE-cadherin with AlexaFluor 555 (red), and nuclei were stained with TOPRO-3 (blue). Scale bar, 100 μm. Objective lens used was Plan-Neofluar 20×/0.5 (Carl Zeiss, Jena, Germany). (E) Genomic DNA sequence of the proximal 5′-region of the human VE-cadherin gene. The transcription initiation site is designated as +1. The positions of the Ets binding sites (EBSs) are boxed. The 2 functional EBSs present in the murine sequence, and conserved in the human,30 are indicated by a double-lined box. Nucleotide sequences corresponding to the oligonucleotides used in the ChIP assay (see panel F) are indicated by arrows. The shaded nucleotides at the 5′ and 3′ ends of the sequence denote the position of oligonucleotides used for PCR amplification of the promoter for subsequent cloning into the pGL4 luciferase reporter plasmid. (F) Chromatin immunoprecipitation from a confluent HUVEC monolayer was performed as described in “Methods.” Genomic DNA obtained after immunoprecipitation, using a rabbit anti-Erg polyclonal antibody or a negative control rabbit IgG, was used as template in a PCR reaction with primers spanning a region of the VE-cadherin promoter containing 4 ETS-binding sites.30 The amplified product of 140 bp (base pair) was enriched in the Erg-precipitated chromatin sample (Erg) compared with the negative control IgG (Neg). To ensure the specificity of Erg binding to the VE-cadherin promoter, PCR amplification was also performed on the same samples using primers specific to the 3′ region of the VE-cadherin locus as negative control. PCR using the negative control primers gave rise to equivalent amounts of product from both samples, indicating nonspecifically captured DNA. (G) An Erg cDNA expression plasmid (pSG5Erg) was cotransfected with the VE-cadherin promoter-luciferase construct in HeLa cells, and luciferase activity was measured. Values are represented as the fold change in relative luciferase activity over the empty pGL4 vector alone. The VE-cadherin promoter was also cotransfected with an empty expression plasmid (pSG5). Erg transactivates the VE-cadherin promoter, **P < .01. Values are means plus or minus SEM, n = 5.
Erg regulates junctional integrity and VE-cadherin expression. (A) HUVECs (105 cells/well) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. Phase-contrast microscopy shows that the typical cobblestone monolayer shown in the control cells (left) has become disrupted in the Erg-inhibited cells (right). Original magnification 20×/0.4 Plan N objective lens (Olympus, Tokyo, Japan). (B) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. VE-cadherin mRNA levels were determined from RNA extracts from GB-treated HUVECs using RT-PCR normalized against GAPDH. VE-cadherin mRNA was significantly decreased after Erg inhibition, *P < 0.05. Values are means plus or minus SEM, n = 3. (C) VE-cadherin protein expression was also down-regulated in Erg-GB–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to VE-cadherin, normalized against tubulin. *P < .05. Values are means plus or minus SEM, n = 3. (D) HUVECs grown on gelatin-coated glass coverslips were treated with GB as above. After 48 hours, cells were labeled for Erg using a polyclonal rabbit anti-Erg antibody and for VE-cadherin using a mouse monoclonal anti–VE-cadherin antibody. Erg was visualized with AlexaFluor 488 (green), VE-cadherin with AlexaFluor 555 (red), and nuclei were stained with TOPRO-3 (blue). Scale bar, 100 μm. Objective lens used was Plan-Neofluar 20×/0.5 (Carl Zeiss, Jena, Germany). (E) Genomic DNA sequence of the proximal 5′-region of the human VE-cadherin gene. The transcription initiation site is designated as +1. The positions of the Ets binding sites (EBSs) are boxed. The 2 functional EBSs present in the murine sequence, and conserved in the human,30 are indicated by a double-lined box. Nucleotide sequences corresponding to the oligonucleotides used in the ChIP assay (see panel F) are indicated by arrows. The shaded nucleotides at the 5′ and 3′ ends of the sequence denote the position of oligonucleotides used for PCR amplification of the promoter for subsequent cloning into the pGL4 luciferase reporter plasmid. (F) Chromatin immunoprecipitation from a confluent HUVEC monolayer was performed as described in “Methods.” Genomic DNA obtained after immunoprecipitation, using a rabbit anti-Erg polyclonal antibody or a negative control rabbit IgG, was used as template in a PCR reaction with primers spanning a region of the VE-cadherin promoter containing 4 ETS-binding sites.30 The amplified product of 140 bp (base pair) was enriched in the Erg-precipitated chromatin sample (Erg) compared with the negative control IgG (Neg). To ensure the specificity of Erg binding to the VE-cadherin promoter, PCR amplification was also performed on the same samples using primers specific to the 3′ region of the VE-cadherin locus as negative control. PCR using the negative control primers gave rise to equivalent amounts of product from both samples, indicating nonspecifically captured DNA. (G) An Erg cDNA expression plasmid (pSG5Erg) was cotransfected with the VE-cadherin promoter-luciferase construct in HeLa cells, and luciferase activity was measured. Values are represented as the fold change in relative luciferase activity over the empty pGL4 vector alone. The VE-cadherin promoter was also cotransfected with an empty expression plasmid (pSG5). Erg transactivates the VE-cadherin promoter, **P < .01. Values are means plus or minus SEM, n = 5.
VE-cadherin is one of the critical regulators of junction assembly and stability. Disruption of VE-cadherin homophilic interaction with an anti–VE-cadherin antibody causes loss of cell-cell adhesion,28 and VE-cadherin deficiency in the mouse results in an early lethal phenotype, because of the inability of ECs to maintain junctional contacts, with consequent vascular disassembly and endothelial apoptosis.19,29 The loss of junctions in Erg-deficient HUVECs (Figure 2A) suggests that Erg may be regulating VE-cadherin expression. We tested whether inhibition of Erg expression affects VE-cadherin levels, by measuring VE-cadherin mRNA and protein levels in Erg-GB–treated cells. As shown in Figure 2B and C, inhibition of Erg expression resulted in an approximate 50% decrease in VE-cadherin mRNA and protein expression, indicating that Erg is required for VE-cadherin expression in HUVECs. VE-cadherin surface staining on Erg-GB– and control-GB–treated cells was considerably reduced after suppression of Erg expression, compared with control cells (Figure 2D). Levels of the tight junction adhesion molecule JAM-A were not affected by GB treatment (as determined by real-time PCR; data not shown). Previous detailed analysis of the VE-cadherin promoter showed that ETS transcription factors are involved in regulating its expression.22 Of the 4 ETS binding sites (EBSs) identified in the mouse VE-cadherin promoter and conserved in the human (see Figure 2E), 2 were shown to bind nuclear proteins and to be required for VE-cadherin expression.22,30 We therefore investigated whether Erg directly interacts with the VE-cadherin promoter by ChIP in HUVECs. Primers were designed across the region of the human VE-cadherin promoter containing the 2 functional EBSs30 and were used to perform PCR on nuclear extracts from confluent HUVEC monolayers, after immunoprecipitation with an anti-Erg polyclonal Ab. The results show that Erg interacts directly with the human VE-cadherin promoter (Figure 2F). To show that this interaction is functionally relevant, we tested whether Erg overexpression could enhance VE-cadherin promoter activity. HeLa cells were cotransfected with a VE-cadherin promoter-luciferase construct and Erg-2 cDNA in an expression vector. Erg overexpression resulted in an approximate 1.8-fold transactivation of VE-cadherin promoter activity (Figure 2G). Thus, our data indicate that Erg drives constitutive VE-cadherin expression in human ECs and suggest that the loss of junctional integrity observed after inhibition of Erg is secondary to the loss of VE-cadherin expression.
Erg and VE-cadherin regulate endothelial apoptosis
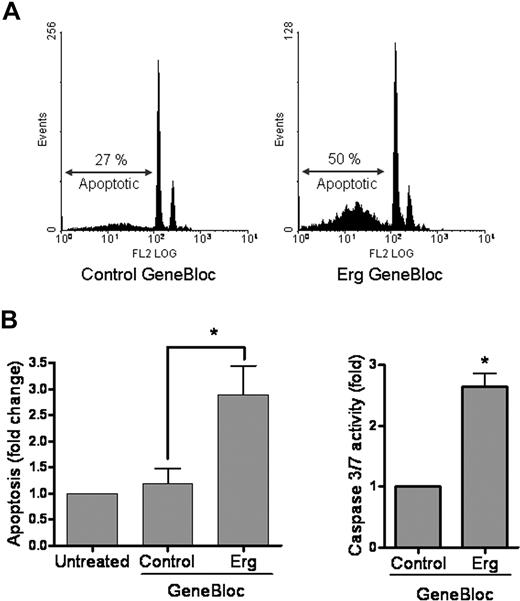
Time-lapse video microscopy of HUVECs treated with Erg GB also showed a decrease in cell density, suggesting that the cell number could be reduced because of cell death (see Figure 2A). We therefore quantified endothelial apoptosis after GB treatment, measuring the sub-G1 fraction of cells using PI staining and flow cytometry, as well as the activation of the proapoptotic proteins caspase-3 and caspase-7. As shown in Figure 3A, Erg suppression by GB for 48 hours resulted in an approximate 3-fold increase in endothelial apoptosis, compared with control GB. These results were confirmed by measurement of caspase-3 activity (Figure 3B), which was significantly increased in Erg-GB–treated cells. Thus, Erg expression appears to be required for endothelial cell survival.
Erg inhibition increases apoptosis in HUVECs. (A) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. Cells were fixed in ethanol and stained with propidium iodide. Sub-G1 apoptotic nuclei were quantified by flow cytometry. Inhibition of Erg increased apoptosis by 3-fold, *P < .05. Values are means plus or minus SEM, n = 3. (B) The regulation of apoptosis by Erg was confirmed by measuring caspase-3 or -7 activity. HUVECs (2 × 103) grown in 96-well microplates were treated with GB as above. After 48 hours, luminescence was measured using the Caspase-3 or -7 Glo Assay (Promega). *P < .05. Values are means plus or minus SEM, n = 4.
Erg inhibition increases apoptosis in HUVECs. (A) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control GB or Erg-specific GB (100 nM) for 48 hours. Cells were fixed in ethanol and stained with propidium iodide. Sub-G1 apoptotic nuclei were quantified by flow cytometry. Inhibition of Erg increased apoptosis by 3-fold, *P < .05. Values are means plus or minus SEM, n = 3. (B) The regulation of apoptosis by Erg was confirmed by measuring caspase-3 or -7 activity. HUVECs (2 × 103) grown in 96-well microplates were treated with GB as above. After 48 hours, luminescence was measured using the Caspase-3 or -7 Glo Assay (Promega). *P < .05. Values are means plus or minus SEM, n = 4.
In HUVECs, overexpression of VE-cadherin–GFP using an adenoviral construct resulted in increased survival of resting HUVECs in normal growth medium (Figure 4A,B), indicating that VE-cadherin also mediates survival signals in ECs under homeostatic conditions, in the presence of serum and growth factors. To investigate whether Erg and VE-cadherin cooperate in regulating survival in ECs, we tested whether apoptosis induced by inhibition of Erg could be reversed by overexpression of VE-cadherin–GFP. HUVECs infected with either VE-cadherin–GFP adenovirus or GFP adenovirus control were treated with Erg or control GB for 48 hours. To confirm the efficacy of Erg GB treatment in this model, levels of endogenous VE-cadherin were measured by reverse transcription (RT)–PCR using primers in the 3′ UTR of the gene; total VE-cadherin and Erg levels were also measured (see “Methods”). In VE-cadherin–GFP–overexpressing cells, levels of total VE-cadherin (endogenous plus exogenous) were 11 times higher than those in GFP-infected cells; however, the levels of endogenous VE-cadherin were not significantly different. In both the VE-cadherin–GFP cells and the GFP-infected cells, Erg GB decreased Erg expression as well as endogenous VE-cadherin levels, as expected. However, this only marginally affected total VE-cadherin levels in the VE-cadherin–GFP cells, compared with a significant decrease (45%) in GFP control cells (see Table S2). To test whether overexpression of VE-cadherin–GFP could prevent Erg GB-induced cell death, cells were stained for PI to detect apoptotic cells. As shown in Figure 4C, cell death induced by Erg inhibition was significantly decreased, but not completely reversed, by VE-cadherin overexpression despite the high levels of VE-cadherin in these cells. These results indicate that Erg regulates survival partly by the junctional molecule VE-cadherin.
VE-cadherin protects HUVECs from apoptosis. (A) HUVECs (105) grown in 6-well plates were transduced with adenovirus (VE-cadherin [VEC]–-GFP and GFP). Four days later, GFP expression levels were determined by flow cytometry, and GFP autofluorescence was visualized using confocal microscopy. Objective lens used was Plan-Neofluar 40×/1.3 oil (Carl Zeiss). VE-cadherin–GFP localizes to the endothelial cell junctions. (B) To quantify apoptosis, cells were fixed in ethanol and stained for propidium iodide. Sub-G1 apoptotic nuclei were quantified by flow cytometry. Expression of VE-cadherin–GFP significantly reduces cell death, *P < .05. Values are means plus or minus SEM, n = 7. (C) HUVECs (105) grown in 6-well dish were transduced with adenovirus (VEC-GFP and GFP). After 48 hours, cells were transfected with either control GB or Erg-specific GB (100 nM). After a further 48 hours, cells were fixed in ethanol and stained with propidium iodide to determine apoptosis. Apoptosis induced by Erg inhibition was significantly decreased in the VE-cadherin overexpressing cells, indicating that the survival pathway regulated by Erg involves VE-cadherin. *P < .05, **P < .01. Values are means plus or minus SEM, n = 7.
VE-cadherin protects HUVECs from apoptosis. (A) HUVECs (105) grown in 6-well plates were transduced with adenovirus (VE-cadherin [VEC]–-GFP and GFP). Four days later, GFP expression levels were determined by flow cytometry, and GFP autofluorescence was visualized using confocal microscopy. Objective lens used was Plan-Neofluar 40×/1.3 oil (Carl Zeiss). VE-cadherin–GFP localizes to the endothelial cell junctions. (B) To quantify apoptosis, cells were fixed in ethanol and stained for propidium iodide. Sub-G1 apoptotic nuclei were quantified by flow cytometry. Expression of VE-cadherin–GFP significantly reduces cell death, *P < .05. Values are means plus or minus SEM, n = 7. (C) HUVECs (105) grown in 6-well dish were transduced with adenovirus (VEC-GFP and GFP). After 48 hours, cells were transfected with either control GB or Erg-specific GB (100 nM). After a further 48 hours, cells were fixed in ethanol and stained with propidium iodide to determine apoptosis. Apoptosis induced by Erg inhibition was significantly decreased in the VE-cadherin overexpressing cells, indicating that the survival pathway regulated by Erg involves VE-cadherin. *P < .05, **P < .01. Values are means plus or minus SEM, n = 7.
Erg siRNA inhibits angiogenesis in vivo
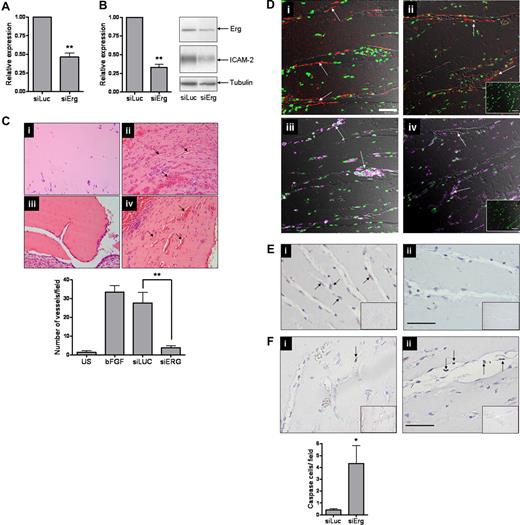
The data discussed so far suggest that Erg is an important regulator of endothelial homeostasis and angiogenesis. However, no in vivo evidence is available to show the role of Erg in angiogenesis. Therefore, we decided to suppress Erg expression in vivo in a model of angiogenesis using siRNA. Nine siRNA sequences were designed to regions of the Erg gene identical in human and mouse and were tested in vitro for their effect on Erg expression (data not shown). Of these, one (73581DS) achieved an inhibition of Erg at the protein level greater than 50%, with a concomitant decrease in ICAM-2 protein expression, and was therefore selected for further experiments (Figure 5A,B). To perform in vivo inhibition of angiogenesis with siRNA, we used a modification of the Matrigel plug model.31-33 A Matrigel mixture, containing 2 μM siRNA, heparin, and basic FGF, was injected subcutaneously into C57BL/6 mice. After 7 days, plugs were carefully harvested, fixed in 4% paraformaldehyde, paraffin embedded, and processed for staining. H&E staining showed that inhibition of Erg by siRNA resulted in a significant inhibition of vascularization in the Matrigel plugs (Figure 5C), whereas control siRNA (Luc-siRNA) had no significant effect on vessel formation. Blood vessels were visualized by immunostaining for CD34 (Figure 5Di,ii); leukocytes, identified by CD45 staining (Figure 5Diii,iv), were found scattered throughout the plug and also lining some of the new vessels, as previously observed.34 Inhibition of Erg levels in the plugs was verified by staining with an anti-Erg polyclonal antibody. This showed that in the Luc-siRNA–treated mice, Erg was expressed in the endothelial cells lining blood vessels; however, rare Erg-positive ECs were found in the Erg-siRNA–treated mice (Figure 5E). To test whether the observed increase in cell death induced by Erg inhibition in vitro was reflected in vivo, plug sections were stained with an anti–active caspase 3 Ab to detect apoptotic cells. A significant increase in caspase 3 staining of cells lining the blood vessels was observed in the Erg-siRNA–treated samples, compared with the Luc-siRNA control samples (Figure 5F). Interestingly, in the Erg-siRNA–treated samples several caspase 3–positive ECs appeared to be detached from the vascular wall and to be hanging in the lumen of the vessels (Figure 5Fii arrows), a phenotype reminiscent of that observed in the VE-cadherin−/− embryos at day 8.5.19 Therefore, these data show that Erg is an important regulator of angiogenesis in vivo, by mediating survival signals and regulating endothelial apoptosis.
Inhibition of angiogenesis in vivo by Erg siRNA. (A) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control siRNA or Erg-specific siRNA (30 nM) for 48 hours. Erg mRNA levels were quantified using RT-PCR, normalized to GAPDH. Erg mRNA expression was significantly reduced after Erg-siRNA treatment, **P < .01. Values are means plus or minus SEM, n = 3. (B) Whole-cell protein extracts were analyzed by SDS-PAGE immunoblotting with antibodies to Erg and tubulin. Erg protein expression was significantly reduced in the Erg-siRNA treated cells, **P < .01. Values are means plus or minus SEM, n = 3. ICAM-2 protein expression was also down-regulated in Erg-siRNA–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to ICAM-2. (C) Matrigel mixture containing siRNA (2 μM) with or without bFGF (80 ng/mL) was injected subcutaneously into C57BL/6 mice. Matrigel plugs were harvested 7 days after implantation, fixed, sectioned, and H&E stained. (i) Matrigel plug without bFGF. (ii) Matrigel plug plus bFGF. (iii) Matrigel plug with bFGF plus Erg siRNA. (iv) Matrigel plug with bFGF plus luciferase control siRNA. Neovessels containing red blood cells (arrows). Original magnification was 10 ×/0.25 Plan objective lens (Olympus). Vessel density was quantified by counting the number of structures containing a lumen and red blood cells. ** indicates P less than .01. Values are means plus or minus SEM, n = 5. (D) Sections from Matrigel plugs treated with siRNA for luciferase (i,iii) or Erg (ii,iv) were stained with antibodies to CD34 (i,ii) or CD45 (iii,iv) and visualized by immunofluorescence confocal microscopy, overlaid with a transmitted light DIC image to enable the vascular structure of the plug to be observed. For CD34 (red), staining is most intense among endothelial cells lining the neovessels (arrows). The pan-leukocyte CD45 antibody (purple) stains isolated cells of the inflammatory infiltrate within the Matrigel matrix and leukocytes inside the lumen of the neovessels (arrows); however. CD45+ cells are also found lining the neovessels, as previously reported.34 Cell nuclei were counterstained with Sytox Green. Inset: IgG-negative controls. Objective lens used was Plan Neofluar 20×/0.5 (Carl Zeiss). Scale bar represents 50 μm. (E) Immunohistochemistry staining with an anti-Erg Ab in the Matrigel plug sections. (i) Endothelial cells lining neovessels express Erg in Luc-siRNA–treated plugs (arrows). (ii) Plugs treated with Erg siRNA show reduced Erg staining. Insert: Control IgG. Original magnification 20×/0.4 Plan N objective lens (Olympus). Scale bar represents 50 μm. (F) (i) Rare apoptotic cells, as shown by anti-Active caspase 3 staining (arrow), are found in Luc-siRNA–treated Matrigel plug samples. (ii) Active Caspase 3–positive ECs are found in Erg siRNA-treated Matrigel plug samples and appear detached from the vessel wall (arrows). *P < .05. Values are means plus or minus SEM n = 3 Erg and n = 5 control. Scale bar represents 50 μm.
Inhibition of angiogenesis in vivo by Erg siRNA. (A) HUVECs (105) grown on gelatin-coated 6-well plates were transfected with either control siRNA or Erg-specific siRNA (30 nM) for 48 hours. Erg mRNA levels were quantified using RT-PCR, normalized to GAPDH. Erg mRNA expression was significantly reduced after Erg-siRNA treatment, **P < .01. Values are means plus or minus SEM, n = 3. (B) Whole-cell protein extracts were analyzed by SDS-PAGE immunoblotting with antibodies to Erg and tubulin. Erg protein expression was significantly reduced in the Erg-siRNA treated cells, **P < .01. Values are means plus or minus SEM, n = 3. ICAM-2 protein expression was also down-regulated in Erg-siRNA–treated HUVECs, as determined by SDS-PAGE immunoblotting using antibodies to ICAM-2. (C) Matrigel mixture containing siRNA (2 μM) with or without bFGF (80 ng/mL) was injected subcutaneously into C57BL/6 mice. Matrigel plugs were harvested 7 days after implantation, fixed, sectioned, and H&E stained. (i) Matrigel plug without bFGF. (ii) Matrigel plug plus bFGF. (iii) Matrigel plug with bFGF plus Erg siRNA. (iv) Matrigel plug with bFGF plus luciferase control siRNA. Neovessels containing red blood cells (arrows). Original magnification was 10 ×/0.25 Plan objective lens (Olympus). Vessel density was quantified by counting the number of structures containing a lumen and red blood cells. ** indicates P less than .01. Values are means plus or minus SEM, n = 5. (D) Sections from Matrigel plugs treated with siRNA for luciferase (i,iii) or Erg (ii,iv) were stained with antibodies to CD34 (i,ii) or CD45 (iii,iv) and visualized by immunofluorescence confocal microscopy, overlaid with a transmitted light DIC image to enable the vascular structure of the plug to be observed. For CD34 (red), staining is most intense among endothelial cells lining the neovessels (arrows). The pan-leukocyte CD45 antibody (purple) stains isolated cells of the inflammatory infiltrate within the Matrigel matrix and leukocytes inside the lumen of the neovessels (arrows); however. CD45+ cells are also found lining the neovessels, as previously reported.34 Cell nuclei were counterstained with Sytox Green. Inset: IgG-negative controls. Objective lens used was Plan Neofluar 20×/0.5 (Carl Zeiss). Scale bar represents 50 μm. (E) Immunohistochemistry staining with an anti-Erg Ab in the Matrigel plug sections. (i) Endothelial cells lining neovessels express Erg in Luc-siRNA–treated plugs (arrows). (ii) Plugs treated with Erg siRNA show reduced Erg staining. Insert: Control IgG. Original magnification 20×/0.4 Plan N objective lens (Olympus). Scale bar represents 50 μm. (F) (i) Rare apoptotic cells, as shown by anti-Active caspase 3 staining (arrow), are found in Luc-siRNA–treated Matrigel plug samples. (ii) Active Caspase 3–positive ECs are found in Erg siRNA-treated Matrigel plug samples and appear detached from the vessel wall (arrows). *P < .05. Values are means plus or minus SEM n = 3 Erg and n = 5 control. Scale bar represents 50 μm.
Discussion
The identification of pathways that regulate endothelial apoptosis and angiogenesis and the possibility of manipulating such pathways in vivo with small molecules such as siRNA is of great interest for possible therapeutic applications.35,36 In this study, we identify the ETS family member Erg as a regulator of endothelial homeostasis and angiogenesis, and we define a new pathway that involves the junctional adhesion molecule VE-cadherin. Moreover, we demonstrate in vivo inhibition of angiogenesis using anti-Erg siRNA.
In this study we show that Erg is required for constitutive endothelial expression of VE-cadherin, and we demonstrate for the first time that Erg interacts with the human VE-cadherin promoter and enhances promoter activity. Several members of the ETS family of transcription factors are expressed in endothelial cells under basal or activated conditions.6 Another ETS family member, Ets-1, was shown to bind to and activate the mouse VE-cadherin promoter.21 However, Ets-1 is not constitutively expressed in resting ECs, whereas Erg is the most strongly expressed ETS factor in HUVECs37 and is also found in human ECs in vivo.7 Therefore, it is likely that Erg, rather than Ets-1, is involved in constitutive VE-cadherin expression. These results are in line with the observation that different ETS factors may drive transcription of the same gene, possibly depending on the cellular background or activation state or both.6 Several transcriptionally active regulatory elements were identified in the VE-cadherin promoter, including an Sp-1/Sp-3 binding site22,30 and more recently an E box associated with a GATA site.38 Thus, Erg may act in concert with other transcription factors to regulate VE-cadherin expression. For example, Erg was shown to interact with transcription factors of the ETS or other families to form multiple protein complexes, such as the one regulating the eNOS enhancer in endothelial cells, involving Erg, other ETS factors, AP-2, Sp-1–related factor, and MZF-like factors.10 Recently, Deleuze et al38 showed that VE-cadherin expression can also be regulated by the transcription factor TAL-1/SCL, a key regulator of hematopoiesis. Interestingly, although inhibition of TAL/SCL with siRNA in HUVECs decreased VE-cadherin levels, the phenotype observed was not as marked as that of Erg-deficient cells reported here. We suggest that Erg is essential for constitutive expression of VE-cadherin in endothelial cells; fluctuation in the levels of Erg and other transcription factors will contribute to the precise regulation of VE-cadherin during vascular development and angiogenesis. Studies to identify potential partners of Erg in regulating VE-cadherin are under way.
Inhibition of Erg expression in HUVECs also resulted in increased cell death, indicating that Erg regulates endothelial survival. We show that VE-cadherin mediates at least part of the Erg-dependent survival effect, suggesting a new antiapoptotic pathway involving VE-cadherin. Previous studies have highlighted the role that VE-cadherin plays in VEGF-mediated survival19 ; further studies will be required to determine whether Erg is involved in the VEGF–VE-cadherin survival pathway. This is the first evidence for a role of Erg in survival of ECs. ETS transcription factors were shown to be involved in apoptosis in various cell types, generally as protective factors. For example, Ets-2 inhibits apoptosis in macrophages by up-regulating Bcl-xL,39 whereas conflicting reports exist on the role of Ets-1 in endothelial apoptosis.40,41 Erg and Fli-1 protect fibroblasts from apoptosis induced by various agents,42 and Fli-1 was shown to drive both human and chicken Bcl-2 expression.43,44 Preliminary data from our laboratory suggest that Erg may also play a role in the regulation of Bcl-2. In HUVECs, Bcl-2 expression levels were decreased after inhibition of Erg by antisense (GB), and ChIP showed that Erg interacts with at least one of the EBS in the human Bcl-2 promoter (G.M.B., A.M.R., et al, unpublished data, July 2007). Interestingly, Erg and Bcl-2 are both implicated in the pathogenesis of prostate cancer. Erg was identified as the most highly overexpressed protein in malignant prostate epithelial cells,45 and Bcl-2 overexpression is associated with the development of androgen-independent prostate carcinomas.46 Studies to define the role of Erg in driving Bcl-2 expression in more detail are under way. It is important to point out that the survival effect mediated by Erg is also likely to be linked to other pathways, because Erg drives expression of other endothelial molecules involved in survival, such as eNOS10 and ICAM-2.7,47
Apoptosis of ECs was shown to play a crucial role in angiogenesis.48 The temporal and spatial balance between proapoptotic and antiapoptotic factors seems to be critical for effective angiogenesis. For example, Pollman et al49 showed that during the early stages of HUVEC tube formation in Matrigel, apoptosis is required to remove cells that fail to become incorporated within the capillary tube structure; network stability was associated with antiapoptotic signals and up-regulation of Bcl-2 expression. These observations are corroborated by Segura et al,50 who showed that apoptosis occurs early during capillary tube formation in Matrigel both in vitro and in vivo, through activation of a caspase-mediated pathway; overexpression of Bcl-2 disrupted the process. In light of these data, the results from our study suggest a model for a dual mechanism for the role of Erg in angiogenesis. We propose that, during the early stages of angiogenesis, Erg expression, activity, or both is down-regulated to allow disruption of EC cell-cell junctions and consequent increase in apoptosis. After formation of the neovessel, Erg expression, activity, or both increases and drives expression of VE-cadherin to provide vessel stability and survival signals. Studies on the regulation of Erg levels and activity during angiogenesis are under way.
Finally, in this study we used a recently described, innovative approach to study in vivo molecular mechanisms of angiogenesis.33 Using a combination of siRNA and the Matrigel plug mouse model of angiogenesis, we showed that Erg inhibition results in a dramatic decrease in neovascularization of the plugs and an increase in apoptotic ECs, indicating that the mechanism described in vitro also occurs in vivo. This approach may be extended to investigate other mechanisms of in vivo angiogenesis.
In conclusion, we have identified a new transcriptional pathway regulating angiogenesis, which involves the transcription factor Erg and the junctional adhesion molecule VE-cadherin, and we have established an in vivo model to inhibit angiogenesis by regulating transcription via siRNA. With the potential therapeutic application of siRNA,35 these results may open the way to a new targeted approach to inhibit angiogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Joseph Boyle and Dr Sarah De Val for helpful advice and discussions; Ms Jenny Lucas and Ms Karine Enesa for technical advice; Ms Lorraine Lawrence for Matrigel plug sectioning and processing. We acknowledge the generous support of Prof Ken Taylor.
This work was supported by grants from the British Heart Foundation (G.M.B., N.H.D., V.A., D.O.H., and A.M.R.).
National Institutes of Health
Authorship
Contribution: G.M.B. performed research, generated reagents, analyzed data, and contributed to the design of the research and writing the paper; N.H.D. and V.A. performed research and contributed to data analysis and interpretation; F.G. provided key tools and contributed to data analysis and interpretation; K.S. performed research; D.O.H. contributed to design of research, data analysis, and interpretation; E.D. provided reagents and contributed to data analysis and interpretation; J.C.M. performed research and contributed to design of research, data analysis, and interpretation; A.M.R. had the original idea, designed research, contributed keys tools and reagents, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: One of the authors (F.G.) is employed by a company (Silence Therapeutics AG, Berlin, Germany) whose product was studied in the present work. The other authors declare no competing financial interests.
Correspondence: Anna M. Randi, NHLI Cardiovascular Sciences Unit, Hammersmith Hospital, Du Cane Road, London W12 0NN, United Kingdom; e-mail: a.randi@imperial.ac.uk.

![Figure 4. VE-cadherin protects HUVECs from apoptosis. (A) HUVECs (105) grown in 6-well plates were transduced with adenovirus (VE-cadherin [VEC]–-GFP and GFP). Four days later, GFP expression levels were determined by flow cytometry, and GFP autofluorescence was visualized using confocal microscopy. Objective lens used was Plan-Neofluar 40×/1.3 oil (Carl Zeiss). VE-cadherin–GFP localizes to the endothelial cell junctions. (B) To quantify apoptosis, cells were fixed in ethanol and stained for propidium iodide. Sub-G1 apoptotic nuclei were quantified by flow cytometry. Expression of VE-cadherin–GFP significantly reduces cell death, *P < .05. Values are means plus or minus SEM, n = 7. (C) HUVECs (105) grown in 6-well dish were transduced with adenovirus (VEC-GFP and GFP). After 48 hours, cells were transfected with either control GB or Erg-specific GB (100 nM). After a further 48 hours, cells were fixed in ethanol and stained with propidium iodide to determine apoptosis. Apoptosis induced by Erg inhibition was significantly decreased in the VE-cadherin overexpressing cells, indicating that the survival pathway regulated by Erg involves VE-cadherin. *P < .05, **P < .01. Values are means plus or minus SEM, n = 7.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/7/10.1182_blood-2007-08-105346/5/m_zh80060816910004.jpeg?Expires=1769092395&Signature=rQ1upEzABFGx0HahpMLiBdEW3I6VGCc~sJ7GKIQOycSDSFbNebX9QzMKE~4Nf6Eco4YLNmIdIirZAHxFR3Pss7gLzuKz8B-fa9414-oV6MkybXtXNP4HeQPzVyYthrJ6wIbKf0ZdNW0wHzA82CtBPY4-q~NeZOwSPrWkd1zxOS1QW2oBJJSGhPj1br0IQeHCEJKA~4LqTDVAWc78J6OclTeHuUcUROG720JjG3y-Yv-l2FLkjsqJ2EInQ~GQkrZSgfHhbFnFHTY~FoCx8jDhWwGOESX8MN65MScRw2Tk1lDkrqTDLkmgzW0hmhH4Zn~0XA1RLEFNWcPxv5Z33at7UQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal