The FOXP1 forkhead transcription factor is targeted by recurrent chromosome translocations in several subtypes of B-cell non-Hodgkin lymphomas, where high-level FOXP1 protein expression has been linked to a poor prognosis. Western blotting studies of diffuse large B-cell lymphoma (DLBCL) cell lines unexpectedly identified the atypical high-level expression of 2 smaller, 60 to 65 kDa, FOXP1 isoforms in all 5 of those with the activated B cell (ABC)–like DLBCL subtype and in a subgroup of primary DLBCL. The anti-FOXP1 (JC12) monoclonal antibody cannot distinguish FOXP1 isoforms by immunohistochemistry, a finding that may be clinically relevant as high-level expression of the full-length FOXP1 protein was observed in some germinal center–derived DLBCLs. ABC-like DLBCL-derived cell lines were observed to express 2 novel, alternatively spliced FOXP1 mRNA isoforms, encoding N-terminally truncated proteins. These transcripts and the smaller protein isoforms were induced as a consequence of normal B-cell activation, which thus represents an additional mechanism for up-regulating FOXP1 expression in lymphomas. The expression of potentially oncogenic smaller FOXP1 isoforms may resolve the previously contradictory findings that FOXP1 represents a favorable prognostic marker in breast cancer and an adverse risk factor in B-cell lymphomas.
Introduction
Microarray-based gene expression profiling studies have played an important role in defining clinically relevant subtypes within the heterogeneous disease entity diffuse large B-cell lymphoma (DLBCL). These include an activated B cell (ABC)–like subtype whose gene expression profile is most similar to that of in vitro–activated peripheral blood B cells.1 Significantly, the average 5-year survival of patients with ABC-like DLBCL was only 16% compared with 76% for those patients with DLBCL having a gene expression profile similar to that of germinal-center B (GCB) cells. While no single marker is sufficient to define these DLBCL subtypes, immunohistochemistry with antibodies to the GCB markers CD10 and BCL6 and the activation marker MUM1/IRF4 was as effective as the gene expression profile in distinguishing high- and low-risk patients.2 However, while the gene expression profiling studies identified at least 2 distinct groups of poor-prognosis patients, ABC-like and type 3,3 these were not distinguished by the immunohistochemical subtyping method that categorized patients as either GC-like or non–GC-like.2
The ABC-like subtype of DLBCL was observed to have high-level expression of target genes regulated by nuclear factor-kappaB (NF-κB).2,3 Significantly, another independent gene expression profiling study also identified a clinically refractory group of DLBCL having both elevated protein kinase C-beta expression and NF-κB activity.4 NF-κB is a key transcription factor that regulates the survival of B cells during specific maturation stages and on B-cell activation (reviewed by Gugasyan et al5 ). Further biologic studies using 2 ABC-like DLBCL cell lines, OCI-Ly3 and OCI-Ly10, confirmed the constitutive activation of the NF-κB pathway and furthermore demonstrated that this pathway was required for their survival.6 More recently, a small molecule IkappaB kinase (IKK) inhibitor, PS-1145, has been reported to demonstrate toxicity in the ABC-like DLBCL cell lines via inhibition of the NF-κB pathway.7 The identification of ABC-like DLBCL and further elucidation of the mechanisms underlying NF-κB activation are therefore clinically relevant to assist the development of therapies targeting NF-κB in these lymphomas.
Gene expression profiling also identified increased mRNA expression of the FOXP1 forkhead transcription factor as one of the best markers of the ABC-like subtype of DLBCL.8 However, FOXP1 protein expression (positivity defined as 30% tumoral nuclear staining with the JC12 monoclonal antibody) was not found to distinguish between the GC and non-GC subtypes of DLBCL determined by immunohistochemistry with antibodies to CD10, BCL6, and MUM1, and FOXP1 expression did not have prognostic value in this series.2 However, 2 studies reported that high-level FOXP1 protein expression (one study using uniform high-level tumoral expression and the other, 30% nuclear tumoral positivity as a cutoff) was significantly associated with a poor prognosis in DLBCL patients.9,10 Subsequent studies have also reported that FOXP1 expression identifies poor-prognosis patients with cutaneous large B-cell lymphomas11,12 or mucosal tissue–associated lymphoid tissue (MALT) lymphomas.13 The current data from the study of B-cell lymphomas thus seem to suggest a role for FOXP1 as an oncogene in lymphomagenesis.
However, FOXP1 maps to a solid tumor suppressor locus on 3p14.1 and commonly exhibits a loss of nuclear protein expression in epithelial malignancies. Furthermore, the loss of nuclear FOXP1 expression is an adverse risk factor in breast cancer patients.14 The loss of Foxp1 in knockout mice leads to an embryonic lethal phenotype indicating that this molecule has an essential role in development.15 Certain Foxp1−/− cardiac tissues showed reduced apoptosis or increased cellular proliferation, findings that are consistent with the proposition that FOXP1 represents a candidate tumor suppressor gene.16 The lack of Foxp1 caused a profound defect in early B-cell development with a block at the transition from pro-B to pre-B cells.17 In this model, a small number of IgM+IgD+ B cells were detected in the spleen, suggesting that Foxp1 may also have a role at a more mature stage of B-cell development.17 Thus the existing data demonstrate key roles for Foxp1 in both B cells and nonhematopoietic cell types.
Recurrent chromosome translocations targeting the FOXP1 locus, which commonly but not exclusively involve the immunoglobulin heavy chain locus, together with trisomy chromosome 3 have been linked to increased FOXP1 expression levels in both MALT lymphomas and in DLBCL.18,,–21 Although structural and numeric aberrations at the FOXP1 locus have been implicated in up-regulating FOXP1 expression, there are a significant number of cases without these alterations that also exhibit strong FOXP1 expression. Thus other, as yet unidentified, mechanisms are also proposed to regulate FOXP1 expression.13,21,22
Here we report that B-cell activation represents an alternative mechanism up-regulating FOXP1 expression. Moreover, B-cell activation induced expression of smaller FOXP1 isoforms in normal B cells and these are specifically expressed at high levels in ABC-like DLBCL. These smaller FOXP1 isoforms, rather than the full-length protein, which may also act as a tumor suppressor in B cells, have a potentially oncogenic role in B-cell non-Hodgkin lymphoma (B-NHL).
Methods
Patient samples
All patient samples were collected with informed consent obtained in accordance with the Declaration of Helsinki, and the study was performed under local ethics committee approval from the Leeds West Regional Ethics Committee, Leeds, United Kingdom. Frozen and paraffin biopsy material, collected prior to therapy, was obtained from the Leeds General Infirmary, Leeds, United Kingdom. Blood buffy coat preparations were obtained from the National Blood Service (Bristol, United Kingdom).
Cell lines and culture conditions
The OCI-Ly3, OCI-Ly10 (ABC-derived), SUDHL6, SUDHL10, and DB (GC-derived) DLBCL cell lines were a kind gift from Dr Eric Davis (National Cancer Institute, Bethesda, MD), RIVA and HBL-1 (ABC-like DLBCL) were a kind gift from Prof Martin Dyer (Leicester University, Leicester, United Kingdom), and the LIB, MIEU, and HLY-1 DLBCL cell lines were generously provided by Dr Talal Al Saati (Purpan Hospital, Toulouse, France). Cells were maintained in RPMI 1640 media supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, and antibiotics (streptomycin [50 μg/mL] and penicillin [50 U/mL]) at 37°C and 5% CO2.
Antibodies
The JC12 antibody against FOXP1 has been previously described.16 The FOXP1 polyclonal antibody was a kind gift from Prof Ed Morrisey (University of Pennsylvania, Philadelphia, PA) and the MUM-1 antibody was a kind gift from Dr Brunangelo Falini (Institute of Hematology and Internal Medicine, Perugia, Italy). The anti–beta actin mouse monoclonal antibody (clone AC-15) was purchased from Abcam (Cambridge, United Kingdom). Hybridoma supernatants containing the additional antibodies used in the study were kindly provided by the LRF Immunodiagnostics Unit (Oxford, United Kingdom).
Immunohistochemistry
Immunostaining was performed using the EnVision kit according to the manufacturer's instructions (DAKO Cytomation, Ely, United Kingdom). Cytospins were immunostained with antibodies to FOXP1 (JC12, 1 in 10 dilution), CD25 (Act-1, 1 in 10 dilution), CD30 (Ber-H2, undiluted), CD20 (L26, undiluted), and rabbit immunoglobulin (MR12, 1 in 10 dilution). Antibodies were diluted in 1× PBS, 10% (vol/vol) human serum.
Western blotting
For adherent cell lines, proteins were extracted from one confluent 150-mm tissue culture plate. For suspension cells, proteins were extracted from 2 × 107 cells. Nuclear proteins were extracted using the Pierce NE-PER kit (Thermo Fisher Scientific, Loughborough, United Kingdom). Proteins were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred electrophoretically to Immobilon (Millipore UK, Watford, United Kingdom) by conventional semidry blotting. The membrane was incubated in blocking buffer (1× PBS, 5% [wt/vol] Marvel) for 1 hour followed by incubation with JC12 monoclonal antibody (at a 1 in 30 dilution) in blocking buffer for 90 minutes. The membrane was washed extensively for 3 times 10 minutes each in wash buffer (1× PBS, 0.05% [vol/vol] Tween). Incubation with species-specific horseradish peroxidase (HRP)–conjugated secondary antibodies (DAKO Cytomation) at a 1 in 1500 dilution was performed in blocking buffer for 90 minutes. Labeling was detected using the enhanced chemiluminescence (ECL) reagent (Amersham Biosciences, Amersham, United Kingdom). Membranes were then stripped in 100 mM 2-mercaptoethanol, 2% (wt/vol) SDS, and 62.5 mM Tris-HCl (pH 6.8) at 50°C for 30 minutes before repeating the blotting and detection procedures with anti–beta actin mouse monoclonal antibody at a dilution of 1 in 20 000 or the antinucleophosmin antibody NA24 undiluted hybridoma supernatant for 30 minutes at room temperature.
Expression analysis
Gene-expression studies were performed using the HG U133 plus 2.0 DNA chips from Affymetrix (Affymetrix, Santa Clara, CA). RNA was isolated from frozen cell pellets (107 cells) of the cell lines SUDHL6, SUDHL10, SUDHL4, LIB, DB, KARPAS 422, MIEU, HLY-1, OCI-Ly3, and OCI-Ly10 with the ALLPrep DNA/RNA Mini Extraction Kit from Qiagen (Valencia, CA) following the manufacturer's instructions. Unlabeled and internally labeled target cRNA was generated following the standard Affymetrix protocol for eukaryotic gene expression analysis. The data were normalized to 500 arbitrary units by the Gene Chip Operating Software (GCOS) from Affymetrix. For statistical analysis, the data were log2 transformed and absent signals were floored to a common baseline (50 units). CLUSTER and TREEVIEW software (http://rana.lbl.gov/EisenSoftware.htm) was used to cluster and visualize the data. The filtering options were defined by a max/min value of 3.5, thereby filtering 4284 of 54 675 probe sets.
Naive B-cell purification and activation
Peripheral blood mononuclear cells (PBMCs) were purified from a blood buffy coat by density gradient centrifugation over Ficoll Paque. Naive B cells were purified from 60 × 107 PBMCs using the naive B-cell isolation kit according to the manufacturer's instructions (Miltenyi Biotech, Bisley, United Kingdom). Naive B cells were activated as previously described.23 Essentially, purified naive B cells were cultured for 1 hour at 37°C prior to activation and then treated under the following conditions: (1) unstimulated (naive), (2) stimulated with 50 μg/mL F(ab′)2 goat antihuman IgM (Cambridge Biosciences, Cambridge, United Kingdom), or (3) stimulated with 1:20 000 wt/vol Staphylococcus aureus, Cowan (SAC) strain (Pansorbin cells; Merck Biosciences, Nottingham, United Kingdom) plus5 ng/mL recombinant interleukin-2 (rIL-2; R&D Systems, Minneapolis, MN). Cells were incubated at 37°C, 5% CO2 for 44 hours. After activation, cytospins were prepared from each sample and then total protein was extracted from the remaining cells using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 8.0], 1% [vol/vol] Triton-X-100, 0.1% [wt/vol] SDS, 0.1% [wt/vol] sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl plus protease inhibitors).
Reverse-transcription–polymerase chain reaction
mRNA was extracted from pellets of approximately 107 cells using the μMACS mRNA Isolation Kit according to the manufacturer's instructions (Miltenyi Biotech). Approximately 100 ng mRNA was used as a template for cDNA synthesis using Superscript III (Invitrogen). Polymerase chain reaction (PCR) was set up using 2 μL cDNA template, 1× buffer for KOD Hot Start Polymerase, 0.2 mM dNTPs, 1 mM MgSO4, 0.3 μM each primer, and 1 U KOD Hot Start Polymerase; and the thermocycler was programmed at 94°C for 2 minutes, followed by 35 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, and a final extension of 72°C for 10 minutes. Samples (10-15 μL) of each reaction were loaded onto a 1% agarose gel. Details concerning the PCR primers used and the expected product sizes are provided in Tables 1 and 2. Products generated from the PJB020/PJB021 and PJB045/PJB038 primer sets were A-tailed and then TA-cloned using pGEMTEasy (Promega, Madison, WI) and the inserts verified by sequencing.
Details of PCR primers used in this study
| Primer . | Sequence . | Exon . |
|---|---|---|
| PJB017 (for) | 5′-GTCGGGCGGCAGCAACCACTTACTAG-3′ | E6 |
| PJB018 (rev) | 5′-AAGGCCTTGGCGCTGCAAAGACAGGA-3′ | E10 |
| PJB019 (for) | 5′-ATCCAAAGGCAGACAGTACGGGCTCC-3′ | E6b |
| PJB020 (for) | 5′-GTAGCTAACTCAACTGTCAGAACTGC-3′ | E7c |
| PJB021 (rev) | 5′-AGGAGACACATGTCGTGGTCAGATCC-3′ | E11 |
| PJB045 (for) | 5′-CTCGGATCCGCATTTATTAACAGTGAGGCTGCTGAA-3′ | E4a |
| PJB038 (rev) | 5′-CTCGAATTCTCACTGCTGGGCGTGGGCGAGGTCAGCTGC-3′ | E6 |
| Actin (for) | 5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′ | — |
| Actin (rev) | 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′ | — |
| Primer . | Sequence . | Exon . |
|---|---|---|
| PJB017 (for) | 5′-GTCGGGCGGCAGCAACCACTTACTAG-3′ | E6 |
| PJB018 (rev) | 5′-AAGGCCTTGGCGCTGCAAAGACAGGA-3′ | E10 |
| PJB019 (for) | 5′-ATCCAAAGGCAGACAGTACGGGCTCC-3′ | E6b |
| PJB020 (for) | 5′-GTAGCTAACTCAACTGTCAGAACTGC-3′ | E7c |
| PJB021 (rev) | 5′-AGGAGACACATGTCGTGGTCAGATCC-3′ | E11 |
| PJB045 (for) | 5′-CTCGGATCCGCATTTATTAACAGTGAGGCTGCTGAA-3′ | E4a |
| PJB038 (rev) | 5′-CTCGAATTCTCACTGCTGGGCGTGGGCGAGGTCAGCTGC-3′ | E6 |
| Actin (for) | 5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′ | — |
| Actin (rev) | 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′ | — |
The non-FOXP1 sequences in primers PJB045 and PJB038 (cloning sites) are underlined.
— indicates not applicable.
Primer pairs used to PCR amplify FOXP1 isoforms and product sizes
| Primer pair . | Isoform no. and PCR product size . | Isoform no. and smaller PCR product size . |
|---|---|---|
| PJB017/PJB018 | Isoform 1 = 556 bp | Isoform 3 = 328 bp |
| PJB019/PJB018 | Isoform 4 = 483 bp | Isoform 5 = 255 bp |
| PJB020/PJB021 | Isoform 9 = 535 bp | Isoform 8 = 271 bp |
| PJB045/PJB038 | Isoform 3 = 305 bp | — |
| Actin for/rev | Actin = 837 bp | — |
| Primer pair . | Isoform no. and PCR product size . | Isoform no. and smaller PCR product size . |
|---|---|---|
| PJB017/PJB018 | Isoform 1 = 556 bp | Isoform 3 = 328 bp |
| PJB019/PJB018 | Isoform 4 = 483 bp | Isoform 5 = 255 bp |
| PJB020/PJB021 | Isoform 9 = 535 bp | Isoform 8 = 271 bp |
| PJB045/PJB038 | Isoform 3 = 305 bp | — |
| Actin for/rev | Actin = 837 bp | — |
— indicates not applicable.
Results
ABC-like DLBCL cell lines abundantly express smaller FOXP1 isoforms
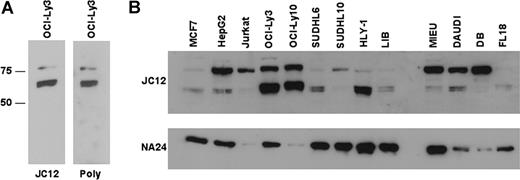
Nuclear FOXP1 protein was expressed at particularly high levels in the 2 ABC-like DLBCL cell lines OCI-Ly3 and OCI-Ly10 compared with GCB-derived cell lines SUDHL10 and SUDHL6, while the GCB-derived cell line DB and the unsubtyped HLY-1 DLBCL cell line were strongly FOXP1 positive (Figure 1, Table 3). Thus, as previously reported in DLBCL biopsies,2 FOXP1 expression determined by immunohistochemistry using the JC12 monoclonal antibody was unable to distinguish ABC-like and GCB-like DLBCL cell lines. However, Western blotting unexpectedly demonstrated that the high-level FOXP1 expression in the ABC-like DLBCL cell line OCI-Ly3 was predominantly of 2 smaller protein species (approximately 60-65 kDa) rather than the full-length protein of approximately 75 kDa (Figure 2A). Western blotting detection of these FOXP1 isoforms in the OCI-Ly3 cell line with a polyclonal antibody against the C-terminus of FOXP1 (kindly provided by Prof Ed Morrisey) confirmed their identity as FOXP1 isoforms (Figure 2A).
Immunoperoxidase labeling of DLBCL cell lines using the JC12 monoclonal antibody. The GCB-derived cell lines SUDHL6 and SUDHL10 are FOXP1 negative by immunohistochemistry, while the GCB-derived cell line DB, the ABC-like DLBCL cell lines OCI-Ly3 and OCI-Ly10, and the unsubtyped DLBCL cell line HLY-1 exhibit strong nuclear positivity. Cells were counterstained with hematoxylin Gill No. 2 (Sigma-Aldrich, St Louis, MO), mounted in Aquatex (VWR International, Poole, United Kingdom) and viewed using a bright field microscope (Axioskop; Zeiss, Welwyn Garden City, United Kingdom) fitted with a 40×/0.65 numerical aperture objective (Zeiss). The images were captured using a Micropublisher 5MP RTV camera (QImaging, Surrey, BC).
Immunoperoxidase labeling of DLBCL cell lines using the JC12 monoclonal antibody. The GCB-derived cell lines SUDHL6 and SUDHL10 are FOXP1 negative by immunohistochemistry, while the GCB-derived cell line DB, the ABC-like DLBCL cell lines OCI-Ly3 and OCI-Ly10, and the unsubtyped DLBCL cell line HLY-1 exhibit strong nuclear positivity. Cells were counterstained with hematoxylin Gill No. 2 (Sigma-Aldrich, St Louis, MO), mounted in Aquatex (VWR International, Poole, United Kingdom) and viewed using a bright field microscope (Axioskop; Zeiss, Welwyn Garden City, United Kingdom) fitted with a 40×/0.65 numerical aperture objective (Zeiss). The images were captured using a Micropublisher 5MP RTV camera (QImaging, Surrey, BC).
FOXP1 expression in DLBCL cell lines, determined by immunohistochemistry (IHC) and by Western blotting
| DLBCL cell line . | Microarray subtype . | JC12 IHC . | Smaller FOXP1 proteins . | Full-length FOXP1 protein . |
|---|---|---|---|---|
| OCI-Ly3 | ABC | ++ | ++ | + |
| OCI-Ly10 | ABC | ++ | ++ | + |
| HLY-1 | ABC | + | + | +/− |
| DB | GC | ++ | − | ++ |
| SUDHL6 | GC | − | +/− | +/− |
| SUDHL10 | GC | − | − | +/− |
| MIEU | GC | nt | +/− | + |
| LIB | Non-ABC | nt | +/− | − |
| KARPAS 422 | GC | nt | − | ++ |
| SUDHL4 | GC | nt | − | + |
| DLBCL cell line . | Microarray subtype . | JC12 IHC . | Smaller FOXP1 proteins . | Full-length FOXP1 protein . |
|---|---|---|---|---|
| OCI-Ly3 | ABC | ++ | ++ | + |
| OCI-Ly10 | ABC | ++ | ++ | + |
| HLY-1 | ABC | + | + | +/− |
| DB | GC | ++ | − | ++ |
| SUDHL6 | GC | − | +/− | +/− |
| SUDHL10 | GC | − | − | +/− |
| MIEU | GC | nt | +/− | + |
| LIB | Non-ABC | nt | +/− | − |
| KARPAS 422 | GC | nt | − | ++ |
| SUDHL4 | GC | nt | − | + |
The intensity of FOXP1 expression is indicated, with −, +/−, +, and ++ being negative, weak, moderate, and strong expression, respectively.
nt indicates not tested.
Investigation of FOXP1 isoform expression using Western blotting. (A) Western blotting of the OCI-Ly3 ABC-like DLBCL cell line with the JC12 monoclonal antibody or an anti-FOXP1 polyclonal antibody demonstrates the predominance of smaller isoforms. (B) Western blotting of a panel of cell lines using the JC12 antibody demonstrating high-level expression of the smaller isoforms in the OCI-Ly3, OCI-Ly10, and HLY-1 DLBCL cell lines.
Investigation of FOXP1 isoform expression using Western blotting. (A) Western blotting of the OCI-Ly3 ABC-like DLBCL cell line with the JC12 monoclonal antibody or an anti-FOXP1 polyclonal antibody demonstrates the predominance of smaller isoforms. (B) Western blotting of a panel of cell lines using the JC12 antibody demonstrating high-level expression of the smaller isoforms in the OCI-Ly3, OCI-Ly10, and HLY-1 DLBCL cell lines.
The ABC-like FOXP1 expression pattern is rarely observed in other cell lines
Further Western blotting studies were undertaken on 13 cell lines, including 8 derived from DLBCL, to investigate the expression of FOXP1 isoforms (Figure 2B). These data identified 3 cell lines, the 2 known ABC-like DLBCL cell lines and one unsubtyped DLBCL cell line, HLY-1, that demonstrated high-level expression of predominantly the smaller FOXP1 isoforms. The Western blotting study was extended to include a wider range of cell lines including T-cell: MOLT4, JURKAT, KARPAS 299, SUDHL1; B-cell: RAJI, KMH2, SUDHL4, KARPAS 422; neuroblastoma: SKNMC, IMR32; lung: H441; kidney: 293T, COS1 (data not shown); breast: MCF-7, MDA-MB-231, MDA-MB-468, MDA-MB-435, ZR751, T47D, SKBR3; and prostate: LNCaP, DU145, PC3 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). While the occasional cell lines such as SUDHL6, LIB (Figure 2B), and MDA-MB-231 (Figure S1) exhibited higher expression levels of the smaller FOXP1 isoforms than the full-length protein, their abundance was not comparable with the levels in the ABC-like DLBCL cell lines.
The HLY-1 cell line is a novel ABC-like DLBCL cell line
Immunophenotyping, by immunohistochemistry using the markers MUM1, BCL-6, and CD10, can distinguish GCB- and non–GC-derived DLBCL. However, this approach cannot distinguish the ABC-like subtype within the non-GC group and, in addition to this, approximately 20% of patients were differently classified by microarray gene expression profiling versus immunohistochemistry.2 Thus, although immunohistochemistry indicated that the HLY-1 cell line had a non-GC phenotype (data not shown), gene expression profiling analysis was also performed (Figure 3). The HLY-1 cell line clustered with the 2 ABC-like DLBCL cell lines indicating that this is indeed an ABC-like DLBCL cell line. There were some differences between the HLY-1 gene expression profile compared with the other 2 ABC-like cell lines, and FOXP1 was also less abundant in the HLY-1 cell line compared with the other 2 ABC-like cell lines. The remaining DLBCL cell lines clustered with the GCB-like cell lines SUDHL6 and SUDHL10, suggesting that both strongly FOXP1-positive (DB and Karpas 422) and moderately FOXP1-positive (MIEU and SUDHL4) cell lines expressing almost exclusively the full-length protein are not ABC derived. The identification of additional ABC-like DLBCL cell lines has been recently reported.24 Immunohistochemistry and Western blotting of the ABC-like DLBCL cell lines RIVA and HBL-1 using the JC12 antibody confirmed their high level nuclear FOXP1 expression and the abundance of smaller FOXP1 isoforms (data not shown).
Gene expression analysis. Hierarchic cluster (83 probe sets) highlighting gene expression differences among the cell lines OCI-Ly3, OCI-Ly10, and HLY-1 (ABC DLBCL cell lines); Karpas 422, DB, SUDHL10, SUDHL6, MIEU, and SUDHL4 (GCB DLBCL cell lines), and LIB (non-ABC cell line, which expressed the GCB markers CD10 and BCL6 together with the ABC marker IRF4).
Gene expression analysis. Hierarchic cluster (83 probe sets) highlighting gene expression differences among the cell lines OCI-Ly3, OCI-Ly10, and HLY-1 (ABC DLBCL cell lines); Karpas 422, DB, SUDHL10, SUDHL6, MIEU, and SUDHL4 (GCB DLBCL cell lines), and LIB (non-ABC cell line, which expressed the GCB markers CD10 and BCL6 together with the ABC marker IRF4).
The smaller FOXP1 isoforms are present in a subset of DLBCL biopsies
The expression of FOXP1 isoforms was further investigated by Western blotting of sections from 52 frozen DLBCL biopsies (Figure 4, Table 4). Of these, 44 expressed tumoral FOXP1 protein (2 cases, no. 10 and no. 25, exhibited exclusively cytoplasmic JC12 labeling) as determined by immunohistochemistry. By Western blotting, 15 of the FOXP1-positive cases lacked expression of FOXP1 isoforms with a molecular weight higher than approximately 60 kDa, yet only 3 FOXP1-positive cases (no. 35, no. 37, and no. 39) with adequate sample loading appeared negative for all isoforms (data not shown for all FOXP1 isoforms). The cases with cytoplasmic expression (no. 10 and no. 25) both exhibited expression of small FOXP1 proteins less than 60 kDa, but no isoforms that were unique to these cases compared with those with nuclear staining (data not shown).
Western blotting of DLBCL biopsy samples using the JC12 monoclonal antibody. The JC12 immunohistochemistry (IHC) scoring for FOXP1 expression in routinely fixed tissue from each case was scored as + for strongly positive, +/− for weak/moderate, c for cytoplasmic, or − for negative, and the subtyping as non-GC (N) or GCB (G) was performed as described previously.9 The arrowhead indicates the position of the full-length FOXP1 protein, while the arrow indicates the smaller isoforms. The blots were reprobed with antiactin as a loading control.
Western blotting of DLBCL biopsy samples using the JC12 monoclonal antibody. The JC12 immunohistochemistry (IHC) scoring for FOXP1 expression in routinely fixed tissue from each case was scored as + for strongly positive, +/− for weak/moderate, c for cytoplasmic, or − for negative, and the subtyping as non-GC (N) or GCB (G) was performed as described previously.9 The arrowhead indicates the position of the full-length FOXP1 protein, while the arrow indicates the smaller isoforms. The blots were reprobed with antiactin as a loading control.
DLBCL subtyping, FOXP1 expression determined by immunohistochemistry (IHC), and their relationship to FOXP1 isoforms identified by Western blotting
| DLBCL subtype by IHC, JC12 tumoral IHC . | Predominantly moderate to strong FOXP1 small isoforms . | Predominantly moderate to strong full-length FOXP1 . | Moderate to strong isoforms at equal levels . | Weak for either or both isoforms . | Negative for both isoforms . |
|---|---|---|---|---|---|
| NCG, n = 12 | |||||
| n = 5, + | 4 | 0 | 0 | 0 | 1 |
| n = 5, +/− | 0 | 0 | 1 | 3 | 1 |
| n = 2, − | 0 | 0 | 0 | 0 | 2 |
| n = 0, c | 0 | 0 | 0 | 0 | 0 |
| GC, n = 21 | |||||
| n = 9, + | 2 | 3 | 0 | 3 | 1 |
| n = 6, +/− | 1 | 0 | 0 | 2 | 3 |
| n = 4, − | 0 | 0 | 0 | 2 | 2 |
| n = 2, c | 0 | 0 | 0 | 0 | 2 |
| ?, n = 19 | |||||
| n = 7, + | 2 | 0 | 1 | 2 | 2 |
| n = 10, +/− | 1 | 0 | 0 | 5 | 4 |
| n = 2, − | 0 | 1 | 0 | 1 | 0 |
| n = 0, c | 0 | 0 | 0 | 0 | 0 |
| DLBCL subtype by IHC, JC12 tumoral IHC . | Predominantly moderate to strong FOXP1 small isoforms . | Predominantly moderate to strong full-length FOXP1 . | Moderate to strong isoforms at equal levels . | Weak for either or both isoforms . | Negative for both isoforms . |
|---|---|---|---|---|---|
| NCG, n = 12 | |||||
| n = 5, + | 4 | 0 | 0 | 0 | 1 |
| n = 5, +/− | 0 | 0 | 1 | 3 | 1 |
| n = 2, − | 0 | 0 | 0 | 0 | 2 |
| n = 0, c | 0 | 0 | 0 | 0 | 0 |
| GC, n = 21 | |||||
| n = 9, + | 2 | 3 | 0 | 3 | 1 |
| n = 6, +/− | 1 | 0 | 0 | 2 | 3 |
| n = 4, − | 0 | 0 | 0 | 2 | 2 |
| n = 2, c | 0 | 0 | 0 | 0 | 2 |
| ?, n = 19 | |||||
| n = 7, + | 2 | 0 | 1 | 2 | 2 |
| n = 10, +/− | 1 | 0 | 0 | 5 | 4 |
| n = 2, − | 0 | 1 | 0 | 1 | 0 |
| n = 0, c | 0 | 0 | 0 | 0 | 0 |
The intensity of tumoral FOXP1 expression was assessed by JC12 immunolabeling with −, +/−, +, and c reflecting negative, weak/moderate, strongly positive, and cytoplasmic labeling, respectively. The ? refers to cases for which subtyping data were not available.
The 60- to 65-kDa FOXP1 isoforms were detectable in 23 (56%) of the 41 cases that were FOXP1-positive by both blotting and IHC. Furthermore, the 60- to 65-kDa isoforms were highly expressed in 11 (25%) of the 41 FOXP1-positive cases, of which 9 (82%) also exhibited strong uniform FOXP1 positivity by IHC. Subtyping data were available for 7 of these cases and 4 (no. 1, no. 14, no. 18, and no. 34) were non-GC.
Seven cases (of which the 4 immunophenotyped cases were all GCB subtype) that did not exhibit high-level 60- to 65-kDa isoform expression, but also exhibited strong FOXP1 staining, had evaluable blotting data. Of these, 5 clearly expressed predominantly the full-length isoform (no. 17, no. 27, no. 30, no. 45, and no. 50). In normal tonsil, the full-length FOXP1 protein was the predominant isoform detected, although, low-level expression of the smaller of the 60- to 65-kDa isoforms was also just detectable (Figure S2). Low-level expression of the full-length FOXP1 isoform was also detectable in 4 of the cases (no. 5, no. 12, no. 29, and no. 31) that lacked tumoral FOXP1 expression (Figure 4). This is most likely to be derived from FOXP1 expressing nonmalignant cells in the microenvironment and is consistent with the full-length protein being the dominant isoform in tonsil (Figure S2).
Alternative splicing of FOXP1 occurs in the ABC-like DLBCL cell lines
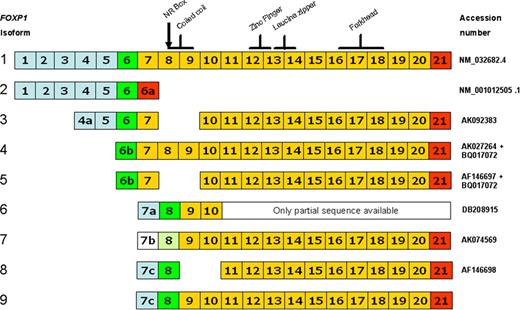
There are a variety of mechanisms that could generate smaller FOXP1 proteins. However, our previous identification of 2 alternatively spliced FOXP1 transcripts, encoding proteins with N-terminal deletions, indicated that these represented a likely source of the smaller FOXP1 proteins.16 Bioinformatics analysis of the sequence of additional FOXP1 transcripts, particularly those from the expressed sequence tag (EST) database within the human FOXP1 Unigene folder (Hs.431498), identified 9 isoforms. Their exon structure has been annotated according to the current Ensembl nomenclature in which exons 6 to 21 encode the full-length protein (Figure 5). Several of these novel FOXP1 isoforms were cloned and further validated by sequencing, including isoforms 3, 4, and 9, which were cloned from the OCI-Ly3 cell line.
Schematic diagram illustrating the exon structure of alternatively spliced FOXP1 isoforms. Blue shaded exons are 5′ noncoding; white is only potentially noncoding as the sequence lacks upstream stop codons. Green exons contain the translational start site (potential for light green shading), yellow exons are coding, while red exons contain the translational stop codon. The positions of key functional motifs in the FOXP1 protein are illustrated above isoform 1.
Schematic diagram illustrating the exon structure of alternatively spliced FOXP1 isoforms. Blue shaded exons are 5′ noncoding; white is only potentially noncoding as the sequence lacks upstream stop codons. Green exons contain the translational start site (potential for light green shading), yellow exons are coding, while red exons contain the translational stop codon. The positions of key functional motifs in the FOXP1 protein are illustrated above isoform 1.
Isoform 2 (accession no.: NM 001012505.1)25 encodes a very short FOXP1 isoform with a unique C-terminus that is not detectable using the JC12 monoclonal antibody.26 All of the remaining 8 isoforms contain the epitope recognized by the JC12 antibody (located within exons 18-21) and are thus indistinguishable by immunohistochemistry with this reagent. Isoform 1 encodes the full-length protein and has an extended 3′ UTR (accession no.: NM 032682.4)25 that was not present in the cDNA that we originally cloned.16 Isoforms 5 and 8, with internal N-terminal deletions of the exons encoding the coiled coil domain, have already been described,16 although only a partial 7c exon was present in the original cDNA clone. Isoforms 4 and 5 are predicted to initiate translation from within the 6b alternative exon and to encode FOXP1 proteins with a unique N-terminus, which in isoform 4 gives a slightly higher molecular weight full-length FOXP1 protein. In isoforms 6 to 9, the presence of upstream stop codons (not detected in the available 7b exon sequence) indicates that these are likely to encode N-terminally truncated proteins that are translated from internal methionine codons (aa's 101/102) and lack approximately the first 100 aa's of the FOXP1 protein. Unlike the other variants, a partial isoform 9 sequence was identified by cloning and sequencing an unexpectedly larger product amplified from all the 3 ABC-like DLBCL cell lines by PCR, with primers in exons 7c and 10 designed to detect isoform 8.
Reverse transcription (RT)-PCR was performed on cDNA from both GC-like and ABC-like DLBCL cell lines using the indicated primer pairs (Table 2) to investigate FOXP1 expression levels and detect the presence of specific FOXP1 splice variants (Figure 6A). PCR primer pairs designed to amplify the 5′ regions from isoform 1 (exons 6-10) and isoform 4 (exons 6b-10) indicated that both transcripts are commonly coexpressed. The size of the lower molecular weight minor product seen with each primer pair is consistent with the detection of variant transcripts lacking exons 8 and 9. Of particular interest, in relation to the abundant expression of the smaller FOXP1 protein isoforms in the ABC-like DLBCL cell lines, was the identification of 2 FOXP1 transcripts, isoforms 3 (PCR exons 4a-6) and 9 (PCR exons 7c-11) that were expressed in the ABC-like lines and not in the GC-like cell lines, including the strongly FOXP1-positive cell line DB.
Analysis of FOXP1 isoforms in DLBCL cell lines and nonmalignant B cells. (A) RT-PCR detection of FOXP1 isoforms in DLBCL cell lines. NTC indicates no template control. Actin was included as a control for cDNA quality. (B) RT-PCR detection of FOXP1 isoforms 9 and 3 in naive and activated (IgM, IL-2/SAC) peripheral blood B cells. (C) Western blotting detection of FOXP1 expression in the OCI-Ly3 cell line as a positive control and in naive and activated (IgM, IL-2/SAC) peripheral blood B cells using the JC12 monoclonal antibody. Blots were reprobed with antiactin as a loading control.
Analysis of FOXP1 isoforms in DLBCL cell lines and nonmalignant B cells. (A) RT-PCR detection of FOXP1 isoforms in DLBCL cell lines. NTC indicates no template control. Actin was included as a control for cDNA quality. (B) RT-PCR detection of FOXP1 isoforms 9 and 3 in naive and activated (IgM, IL-2/SAC) peripheral blood B cells. (C) Western blotting detection of FOXP1 expression in the OCI-Ly3 cell line as a positive control and in naive and activated (IgM, IL-2/SAC) peripheral blood B cells using the JC12 monoclonal antibody. Blots were reprobed with antiactin as a loading control.
The protein product encoded by FOXP1 isoform 9 lacks the N-terminal 100 amino acids containing a glutamine-rich region, potential phosphorylation sites, an RXXL motif that potentially binds the anaphase-promoting complex, and a cyclin-binding site. This is similar to the previously reported murine isoform Foxp1D. Isoform 3 encodes a protein with an internal N-terminal deletion of amino acids 95 to 170 including a coiled-coil domain, glutamine-rich region, and the nuclear receptor box/ LXXLL motif (aa's 126-130). Importantly, the predicted molecular weights of these isoforms, 64.8 kDa for isoform 9 and 66.4 kDa for isoform 3, are consistent with the Western blotting data.
Expression of the smaller FOXP1 isoforms is induced during nonmalignant B-cell activation
The ABC-like subtype of DLBCL is defined by a gene expression profile similar to that of nonmalignant activated B cells.8 To determine whether the high-level expression of the smaller isoforms of FOXP1 was induced in response to normal B-cell activation, the expression of FOXP1 isoforms was investigated in activated peripheral blood B cells. CD20+ peripheral blood B cells were purified and activated using IgM or IL-2 + SAC as described in the DLBCL gene expression profiling studies that reported up-regulated FOXP1 mRNA expression on B-cell activation.1,8 Immunohistochemistry using antibodies to the activation markers CD25 and CD30, together with CD20 labeling, confirmed successful B-cell purification and activation (data not shown). FOXP1 isoforms 3 and 9 were induced on nonmalignant B-cell activation (Figure 6B). Both immunohistochemistry (data not shown) and Western blotting (Figure 6C) demonstrated that B-cell activation significantly increased FOXP1 nuclear protein expression in CD20+ peripheral blood B cells. Both methods of B-cell activation induced expression of the smaller (60-65 kDa) protein isoforms in nonmalignant B cells in a comparable pattern with that observed in the ABC-like DLBCL cell lines. The RT-PCR detection of low-level isoform 9 expression in naive B cells is consistent with this transcript encoding the smallest of the two 60- to 65-kDa FOXP1 isoforms, which was also detectable at low levels in naive B cells by Western blotting.
Discussion
The FOXP1 transcription factor is widely expressed in normal tissues and has recently been shown to play an essential role in early B-cell development.17 FOXP1 is also implicated in the pathogenesis of DLBCL and MALT lymphomas, through the identification of recurrent chromosome translocations that up-regulate its expression levels18,,–21 and the correlation between high-level FOXP1 expression and poor prognosis.9,10,13 On initial examination, these data seem contrary to findings from the study of FOXP1 in epithelial malignancies, which suggest that it may represent a tumor suppressor gene.14,16 However, there are also subtypes of lymphoma that lack FOXP1 expression, for example both classical and lymphocyte predominance Hodgkin lymphomas.27 It is currently unclear as to whether the FOXP1 expression patterns in these lymphomas represent their origin from a specific stage of B-cell development that normally lacks FOXP1 expression or whether silencing of FOXP1, which might thus behave as a tumor suppressor, contributes to the disease process.
Here we report that 2 smaller 60- to 65-kDa isoforms of FOXP1, rather than the full-length protein, are preferentially expressed in DLBCL cell lines with an ABC-like gene expression profile. The expression of these smaller isoforms is up-regulated on in vitro peripheral blood B-cell activation, suggesting that their presence in ABC-like DLBCL reflects the relationship between this DLBCL subtype and activated B cells. The high-level expression of these smaller isoforms was also observed in primary DLBCL, including a proportion of those scored as strongly FOXP1 positive by immunohistochemistry. This expression pattern was not observed in the other hematologic or epithelial cell lines studied and only the smaller of these 2 FOXP1 isoforms was weakly expressed in tonsil. Notably the only 2 GC-derived DLBCL cell lines with high-level FOXP1 expression expressed the full-length protein. Variability in the expression of the full-length FOXP1 protein versus the smaller isoforms may represent an explanation as to why JC12 immunolabeling (which does not distinguish these isoforms by immunohistochemistry) does not predict survival in certain case series.2
Two activation-induced alternatively spliced FOXP1 transcripts (3 and 9) were identified that encoded proteins with N-terminal deletions upstream of the conserved leucine zipper and zinc finger motifs. Convincing evidence linking the loss of the FOXP1 N-terminus and malignancy has already been generated through the study of recurrent viral integration sites that generate avian nephroblastomas. In this model system, foxP1 was the second most frequent target of retroviral insertion, occurring in 5% of the tumor clones.28 Within the context of the current study, of particular interest were the observations that these insertions did not affect foxP1 mRNA levels and that they all clustered within the second coding exon, causing a putative NH2-terminal deletion. This led Pajer et al to propose that, “the virally altered foxP1 might interfere (in a dominant-negative fashion) with a normal function of the gene and support malignant transformation.”28 (p83) This is consistent with our data and importantly provides functional evidence that an N-terminally truncated foxP1 protein may act as an oncogene.
To date, there is little information concerning the exact location of the translocation breakpoints within the human FOXP1 gene in lymphomas. However, in one DLBCL case where the breakpoint was cloned, the translocation separated a putative upstream promoter sequence from the intact coding region.20 The finding that several of the N-terminal variant FOXP1 transcripts use novel 5′ noncoding exons raises the possibility that the expression of these isoforms may be regulated from internal promoters within the upstream introns. Consistent with this hypothesis, according to AceView29,30 there are 14 probable alternative promoters for FOXP1. Another lymphoma with a t(2;3)(q36;p13) had a breakpoint located within the region bordered by exon 5 and exon 7 (exon 6 and 8 according to Ensembl) and was thought to potentially express a fusion transcript.19 However, an alternative explanation, based on our data, would be the preferential expression of smaller N-terminally truncated FOXP1 isoforms. Sequence analysis of internal FOXP1 breakpoints will determine whether one or both possibilities occur.
Signaling events downstream of a functional B-cell receptor are key factors regulating survival of the majority of both normal and malignant B cells. An important end result of antigen-receptor stimulation in lymphocytes is gene activation via the transcription factors NFAT and NF-κB, which also play a central role in the pathogenesis of the lymphoma subtypes in which FOXP1 expression predicts poor outcome.6,31,32 Notably, the related FOXP3 transcription factor is a repressor of both NFAT and NF-κB.33 Thus, it is tempting to speculate that FOXP1 could potentially have a similar role and that the induction of smaller FOXP1 proteins may represent an important mechanism enabling NFAT and NF-κB activation in normal B cells. Targeting expression of FOXP1 isoforms 3 and 9, through the alternative internal promoter(s) that may regulate their expression, or silencing their transcription via the 5′ noncoding exons that are absent in the full-length FOXP1 isoform may potentially represent a more specific way of targeting this pathway in both normal and malignant activated B cells.
There is no novel protein sequence in the smaller isoforms, and the 6 amino acid overlap between the deleted regions is identical in the FOXP2 and FOXP4 proteins. Thus antibodies cannot be generated to specifically distinguish both these isoforms from the full-length FOXP1 protein by immunohistochemistry. As Western blotting is not practical within a routine clinical context, the potential prognostic utility of these isoforms and their expression levels in cases with chromosome translocations at the FOXP1 locus is being investigated using quantitative PCR. Further studies are also required to investigate the biologic roles of these FOXP1 isoforms and the mechanisms affecting their transcriptional regulation. As DLBCL patients with high-level FOXP1 expression have a worse outcome than those with low-level or no detectable FOXP1 expression,9,10 this may indicate that these smaller isoforms have a greater effect than simply inhibiting the function of the full-length protein. Certainly, the murine Foxp1D isoform that lacks the NH2-terminus of the protein has been found to be a significantly stronger transcriptional repressor than the full-length isoform, Foxp1A.34
In conclusion, our data address what appeared to be contradictory hypotheses that FOXP1 was a candidate tumor suppressor in epithelial malignancies and a potential oncogene in certain large B-cell lymphomas. Rather than FOXP1 having opposing actions in epithelial versus hematologic tissues, the differences are likely to reflect the unexpectedly high-level expression of smaller, potentially oncogenic isoforms, in lymphomas. Several studies have indicated that copy number gains and chromosome translocations are not the only cause of high-level FOXP1 expression in lymphomas,13,20,–22 and here we identify B-cell activation as an additional mechanism that can also up-regulate FOXP1 protein expression. However, further functional data are required: first, to confirm that FOXP1 is indeed a tumor suppressor; second, to investigate whether this role is compromised by the abundant expression of the smaller isoforms in B-cell lymphomas; and third to determine whether the recurrent chromosome translocations targeting the FOXP1 locus alter the relative abundance of FOXP1 isoforms rather than purely up-regulating expression of the full-length protein.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Paola Bignone for assistance with the Ensembl FOXP1 gene annotation.
This work was supported by the Leukemia Research Fund. E.L., C.B., and A.R. are supported by the Interdisciplinary Center for Clinical Research (IZKF) of the University of Würzburg.
Authorship
Contribution: P.J.B. and S.L.A. performed research and collected, analyzed, and interpreted data; E.L., C.B., and A.R. designed research and collected, analyzed, and interpreted microarray data; S.B., J.A.F., and A.S.J. immunophenotyped DLBCL cases and performed and scored FOXP1 immunohistochemistry; K.P. immunophenotyped the HLY-1 cell line; and A.H.B. designed research, analyzed and interpreted data, and drafted the paper.
Conflict-of-interest disclosure: A.H.B. is an inventor on a FOXP1 patent application that includes the JC12 monoclonal antibody. All other authors declare no competing financial interests.
Correspondence: Alison H. Banham, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, Level 4 Academic Block, John Radcliffe Hospital, Headington, Oxford OX3 9DU, United Kingdom; e-mail: alison.banham@ndcls.ox.ac.uk.