To the editor:
PRDM1β is a functionally impaired isoform of PRDM1,1 the master regulator of plasma cell differentiation.2 The paper by Liu et al3 suggested that PRDM1β expression correlates with chemotherapy resistance in patients with the non–germinal center B-cell (GCB)–type diffuse large B-cell lymphomas (DLBCL), which can be overcome by rituximab through down-regulation of PRDM1β. However, contrary to what is suggested by Liu et al, the use of PRDM1β mRNA as a prognostic marker in DLBCL may not have a biologic basis because of the very low abundance of the PRDM1β protein.
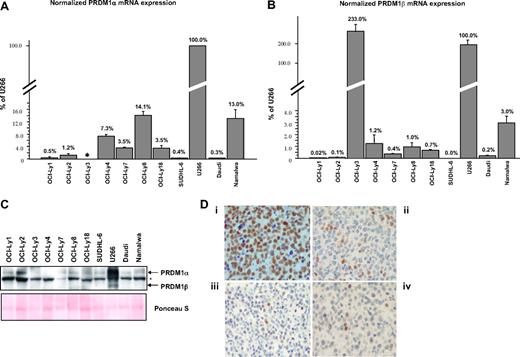
Quantitative analysis of PRDM1α and PRDM1β transcripts by real-time PCR performed on a panel of DLBCL and Burkitt lymphoma (BL) cell lines (Figure 1A,B) showed that with the exception of OCI-Ly3, PRDM1α and PRDM1β mRNA levels are generally low, ranging from less than 1% to approximately 15% and 0% to approximately 3.0% of their respective levels in U266 myeloma cells.
Low levels of PRDM1β expression in B lymphoma cell lines. (A,B) PRDM1α and PRDM1β mRNA in DLBCL cell lines (OCI-Ly1, OCI-Ly2, OCI-Ly3, OCI-Ly4, OCI-Ly7, OCI-Ly8, OCI-Ly18 and SUDHL-6) and Burkitt lymphoma cell lines (Daudi and Namalwa) were quantified by Taqman reverse-transcriptase-polymerase chain reaction, normalized with beta-glucuronidase, and expressed as a percentage relative to U266. For PRDM1α mRNA quantification: forward primer, TCCAGCACTGTGAGGTTTCA; reverse primer, TCAAACTCAGCCTCTGTCCA; probe, ATGGACATGGAGGATGCGGATATG. For PRDM1β mRNA quantification: forward primer, CCCGAACATGAAAAGACGAT; reverse primer, ATAGCGCATCCAGTTGCTTT; probe, TCCAGAGGGGAGCTTCACCACTTC. In OCI-Ly3, PRDM1α mRNA is not detectable by the primers shown above because of a chromosomal inversion breakpoint at intron 2 of the PRDM1 gene. Error bars indicate SE. (C) Western blotting of total protein extracts using the ROS monoclonal anti-PRDM1 antibody. The positions for PRDM1α and PRDM1β are indicated. Asterisk marks the nonspecific band. Ponceau S staining of the membrane is shown for protein loading control. (D) Examples of immunoperoxidase staining on paraffin tissue sections for PRDM1 in U266 cells (i) and representative DLBCL cases (ii-iv). U266 cells show uniform strong staining, while all or most of the tumor cells in DLBCL are negative for or weakly express PRDM1. A few scattered PRDM1+ cells serve as internal controls for the DLBCL cases. The DLBCL cases shown have the immunohistochemical profile of non-GCB-type DLBCL. Micrographs were acquired with a Nikon Microphot SA microscope (Nikon Instruments, Melville, NY) and a SPOT Insight Color Mosaic QE 4.2 camera and image acquisition software system (Diagnostic Instruments, Sterling Heights, MI).
Low levels of PRDM1β expression in B lymphoma cell lines. (A,B) PRDM1α and PRDM1β mRNA in DLBCL cell lines (OCI-Ly1, OCI-Ly2, OCI-Ly3, OCI-Ly4, OCI-Ly7, OCI-Ly8, OCI-Ly18 and SUDHL-6) and Burkitt lymphoma cell lines (Daudi and Namalwa) were quantified by Taqman reverse-transcriptase-polymerase chain reaction, normalized with beta-glucuronidase, and expressed as a percentage relative to U266. For PRDM1α mRNA quantification: forward primer, TCCAGCACTGTGAGGTTTCA; reverse primer, TCAAACTCAGCCTCTGTCCA; probe, ATGGACATGGAGGATGCGGATATG. For PRDM1β mRNA quantification: forward primer, CCCGAACATGAAAAGACGAT; reverse primer, ATAGCGCATCCAGTTGCTTT; probe, TCCAGAGGGGAGCTTCACCACTTC. In OCI-Ly3, PRDM1α mRNA is not detectable by the primers shown above because of a chromosomal inversion breakpoint at intron 2 of the PRDM1 gene. Error bars indicate SE. (C) Western blotting of total protein extracts using the ROS monoclonal anti-PRDM1 antibody. The positions for PRDM1α and PRDM1β are indicated. Asterisk marks the nonspecific band. Ponceau S staining of the membrane is shown for protein loading control. (D) Examples of immunoperoxidase staining on paraffin tissue sections for PRDM1 in U266 cells (i) and representative DLBCL cases (ii-iv). U266 cells show uniform strong staining, while all or most of the tumor cells in DLBCL are negative for or weakly express PRDM1. A few scattered PRDM1+ cells serve as internal controls for the DLBCL cases. The DLBCL cases shown have the immunohistochemical profile of non-GCB-type DLBCL. Micrographs were acquired with a Nikon Microphot SA microscope (Nikon Instruments, Melville, NY) and a SPOT Insight Color Mosaic QE 4.2 camera and image acquisition software system (Diagnostic Instruments, Sterling Heights, MI).
Western blots using the monoclonal ROS anti-PRDM1 antibody4 demonstrated that, despite the relatively high levels of PRDM1β mRNA in U266, PRDM1β is only weakly expressed, suggesting that it is not translated efficiently. PRDM1β in the other cell lines is virtually undetectable (Figure 1C), in agreement with their relative transcript levels, and in the case of OCI-Ly3, consistent with the presence of gene alterations that abrogate the synthesis of both full-length PRDM1α and PRDM1β.5,6 Liu et al have found more readily detectable levels of PRDM1β expression in B lymphoma cell lines. We believe the discrepancy is mainly due to differences in identification and interpretation of the PRDM1β signals in Western blots.
The band indicated as PRDM1β by Liu et al (Figures 2C,D and 3C3 ) may be nonspecific because (1) it has a different size in U266;(2) its intensities in Daudi and Namalwa relative to U266 correlate poorly with PRDM1β transcript levels as determined by our qRT-PCR assays3 ; and (3) the intensity of this band relative to PRDM1α in Namalwa is inconsistent between assays. Nonspecific bands can be detected by anti-PRDM1 antibodies, especially when total cell extracts are used. Our Western blots identified such a band below PRDM1α that is detectable in all cell lines (Figure 1C), including OCI-Ly3 (see above).
PRDM1β mRNA expression in primary DLBCL cases was also relatively low, averaging 16.4% (± 2.2%, mean ± SE) that of U266. In addition, regardless of the levels of PRDM1β mRNA, PRDM1 expression in DLBCL tends to be absent or very weak in most of the lymphoma cells (Figure 1D). Thus, PRDM1β may not be present in DLBCL cells in a quantity high enough to have a biologically significant effect on chemotherapy resistance. In addition, because of the likelihood of a misinterpretation of the PRDM1β signal (see above), the data on the down-regulation of PRDM1β by rituximab or NF-κB inhibitor becomes questionable. Nevertheless, PRDM1β mRNA may serve as a prognostic marker, possibly as a reflection of some signaling pathways. Alternatively, PRDM1β expression may simply be a surrogate marker for PRDM1α expression. The authors have stated that there is no correlation between PRDM1α expression and survival in non-GCB DLBCL; however, PRDM1α expression was assessed in an all-or-none fashion and not in a continuous quantitative manner. At this moment, the value and significance of PRDM1β expression, if any, as a prognostic marker in DLBCL needs to be interpreted with caution and requires further investigation.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wayne Tam, MD, PhD, Department of Pathology and Laboratory Medicine, Weill Medical College of Cornell University, Starr 711A, 525 East 68th Street, New York, NY 10021; e-mail: wtam@med.cornell.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal