To the editor:
Flotho et al1 identified within a cohort of 286 childhood acute lymphoblastic leukemia (ALL) patients a simple set of 14 genes (15 Affymetrix probes) with expression in diagnostic bone marrow aspirates that was shown to be an independent predictor for patient outcome. It was asked by the authors “how could the genes showing independent prognostic significance in this study be effectively incorporated into existing systems of risk classification?”
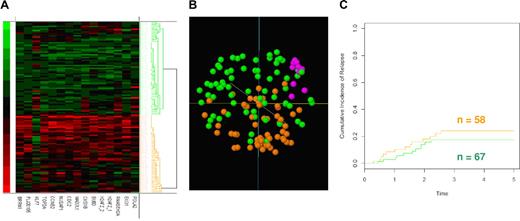
We examined the same 15 probe-sets from Affymetrix HU133A–derived gene expression data from 127 ALL patients at diagnosis generated within The Children's Hospital at Westmead. As per the original report, hierarchical clustering (Figure 1A) illustrates that the 15 probe-set classifier distinguished 2 groups of patients in the same manner as the original paper. Principal component analysis (Figure 1B) indicated that the 2 subpopulations of patients, while distinguishable, were considerably more intermingled than reported by Flotho et al.1 We included the data from 10 bone marrow aspirates from nonmalignant healthy controls, which clustered together within the proximity of both the low and high expression subgroups. Hence, while our results support the classifier as identifying 2 patient populations, the cumulative incidence of relapse show no statistical difference (P = .35) between the 2 groups (Figure 1C).
Assessment of Affymetrix data. (A) Hierarchical clustering using a simple matching similarity metric with average linkage of 127 childhood ALL patients using expression values of the 15 probe-set (14 genes). (B) Principal component analysis. Clusters indicated by green and orange correspond to hierarchical cluster dendrogram.(C) Cumulative incidence of relapse among the 127 patients for the 2 patient groups. Time is in years. Lines indicated by green and orange correspond to the clusters identified by the hierarchical dendrogram.
Assessment of Affymetrix data. (A) Hierarchical clustering using a simple matching similarity metric with average linkage of 127 childhood ALL patients using expression values of the 15 probe-set (14 genes). (B) Principal component analysis. Clusters indicated by green and orange correspond to hierarchical cluster dendrogram.(C) Cumulative incidence of relapse among the 127 patients for the 2 patient groups. Time is in years. Lines indicated by green and orange correspond to the clusters identified by the hierarchical dendrogram.
Given the similarity of the 2 studies, it is unlikely that neither technical nor platform deficiencies form the basis of this discrepancy. Unlike previous studies where gene expression signatures identified as distinguishing ALL subtypes showed no differential gene expression in our patient cohort,2 the expression of the genes in this study fall into 2 distinct subgroups. While the original study indicated that gene expression signatures can be used to identify patients at risk of relapse, it must be remembered that relapse can only be interpreted in the context of the chemotherapy protocol and treatment strategy applied. All patients in the original study were treated on the St Jude Total XIII, XIV, XV protocols, while the patients from The Children's Hospital at Westmead, Australia, were treated on the Berlin-Frankfurt-Munster (BFM) 95 protocol3 and on a complimentary protocol, Australian and New Zealand Children's Haematology and Oncology Group (ANZCHOG) VIII.4
Within this context, a “low” gene expression profile, which correlated with a poor outcome in the reported study with approximately 50% relapse, had only a 20% relapse rate in our cohort, indicating that patients with this expression profile may have a better response when treated on the BFM95/ANZCHOG VIII protocols. Conversely, the patients with high expression of the 15 genes, which corresponded to less than 10% failure rate in the original patient group, did not respond well to the German nor Australian treatment protocols. This highlights the biologic diversity of ALL patients, such that the 14 “proliferation” genes identified that distinguish response to Total XIII may not be central to the response to other chemotherapeutic protocols.
This validation study demonstrates that the power of gene expression signatures to the clinical management of pediatric ALL may not be related to their diagnosis, subclassification, or indeed prognostication. Rather, our data supports the tenets of the final line in the Flotho et al4 report; that gene expression profiles may facilitate the “selection of appropriate therapy.” With personalized therapeutics being highly sought after, the value of interpretation of gene expression data with selecting appropriate treatment protocols for patients should not be underestimated.
Authorship
This work was supported by a grant from the Australian Rotary Health Research Fund along with funds generously donated by Kayaking for Kemo Kids and the Oncology Children's Foundation. D.R.C. is a research fellow of the University of Sydney and an Adjunct Professor of the University of Technology, Sydney, Australia.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel Catchpoole, The Tumour Bank, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, 2145, NSW, Australia; e-mail: danielc@chw.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal