Lipopolysaccharide binding protein (LBP) function is dependent on circulating LBP levels. Disturbance of LBP transcription regulation may influence the risk for clinical events. In a nested case-control study using a single nucleotide polymorphism haplotype tagging (tagSNP) approach, we assessed whether genetic variation in the LBP gene influences the risk for Gram-negative (GN) bacteremia after allogeneic hematopoietic cell transplantation (HCT), then validated the association in a prospective cohort by correlating genetic variation with basal serum LBP levels and mortality. Presence of the tagSNP 6878 C allele among patients was associated with a 2-fold higher risk for GN bacteremia (odds ratio = 2.15; 95% confidence interval [CI], 1.31-3.52, P = .002). TagSNP 6878 was in strong linkage disequilibrium with 3 SNPs in the LBP promoter, one of which was SNP 1683 (r2 = 0.8), located in a CAAT box that regulates LBP promoter efficiency. SNP 1683 was associated with higher median basal serum LBP levels (TT 8.07 μg/mL; TC 10.40 μg/mL; CC 17.39 μg/mL; P = .002), and a 5-fold increase in GN bacteremia related mortality after HCT (hazard ratio = 4.83; 95% CI, 1.38-16.75, P = .013). These data suggest that transcriptional regulation of the LBP gene contributes to the risk for developing GN bacteremia and death after HCT.
Introduction
The lethal effects of Gram-negative (GN) bacteria are attributable to lipopolysaccharide (LPS), a highly conserved glycolipid component of the cell wall of all GN bacteria.1,2 One of the key components of the innate immune response to LPS is lipopolysaccharide binding protein (LBP), a secretory class I acute-phase protein synthesized by hepatocytes. LBP is involved in LPS recognition and signaling.3 Circulating LBP can have both pro-inflammatory and anti-inflammatory effects on the host response to LPS. At low to normal concentrations, LBP catalyzes the transfer of disaggregated LPS to the binding site of membrane-bound and soluble forms of CD14, facilitating signaling via TLR4,4,–6 and binds directly to GN bacteria, resulting in enhanced phagocytosis and clearance from blood.7 At high concentrations, LBP can inhibit LPS-induced host cell activation, reduce LPS binding to monocytes, and attenuate the release of proinflammatory cytokines, such as tumor necrosis factor-α.8,9
The dual nature of LBP activity makes this an interesting candidate for genetic analysis. LBP's concentration-dependent immunologic function must require precise genetic regulation of gene transcription, suggesting that genetic variation in the elements controlling LBP production may affect an individual's immune response to LPS and GN bacteria. This possibility is supported by a previous detailed study of the LBP promoter region; truncation mutation experiments indicate that a region of the LBP promoter is responsible for regulating the efficiency of gene transcription.10 Based on this work, we hypothesized that genetic variation in the LBP gene may disturb LBP transcription and regulation may influence the risk for clinical events. To test this hypothesis, we performed a 2-stage genetic association study to determine whether variation in the LBP gene influences the risk for GN bacteremia in patients after allogeneic hematopoietic cell transplantation (HCT), a population in which bloodstream infections with GN bacteria remains a significant clinical problem.11,,–14
Methods
This study was performed using 2 patient populations. The first population was a retrospectively identified nested case-control population, used for identifying clinical risk factors for GN bacteremia and analysis of the association between LBP single nucleotide polymorphisms (SNP) and GN bacteremia. The second population was prospectively identified for validation of the candidate LBP SNP association with a LBP intermediate phenotype, the basal circulating LBP levels.
Nested case-control study
Patients who had their first allogeneic myeloablative HCT at the Fred Hutchinson Cancer Research Center/Seattle Cancer Care Alliance (the “Center”) between January 1, 1990, and December 31, 2000, and provided informed consent in accordance with the Declaration of Helsinki for the Fred Hutchinson Cancer Research Center institutional review board-approved genetic studies were considered for enrollment. An a priori list of GN bacterial organisms was assembled from a review of our laboratory database (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Patients were selected as a “case” if they had one or more positive blood cultures with one of these organisms before discharge from our Center. Control patients were selected at an approximate ratio of 2:1 to 3:1 (control-case) after meeting several criteria. Patients who did not have any positive blood cultures (due to any organisms) before discharge from our Center were identified as “eligible controls.” This group of “eligible controls” was further restricted by matching them to cases according to the year of transplantation plus or minus one year, then according to exposure period, defined as days to development of GN bacteremia plus or minus 10 days. Although LBP was recently found to interact with lipoteichoic acid, a cell wall component of Gram-positive bacteria, such as Staphylococcus aureus and Streptococcus pneumoniae, we did not include these as cases because of the desire to maintain a highly refined phenotype.15 For the same reason, 30 patients who had multiorganism bloodstream infections that included GN bacteria were excluded from the analysis. All cases and controls that had both patient and donor DNA available in our genetic repository were genotyped and included in the genetic analyses.16
Standard demographic, laboratory, and clinical data were extracted from a prospectively collected database. Disease risk categories were ranked according to the outcomes we have observed at our Center and have been previously described.17 Stem cell sources were classified as growth factor-mobilized blood cells, bone marrow, or other, which included cord blood or a combination of bone marrow and mobilized blood cells. Matching between the donor and recipient was determined according to donor-recipient HLA-A, HLA-B, and HLA-DR compatibility. Conditioning regimens were categorized as either total body irradiation based or not (containing no irradiation). To maintain a uniform at-risk population, patients who received a reduced intensity conditioning regimen were excluded from this analysis. Acute and chronic graft versus host disease (GVHD) was graded based on previously published clinical, histologic, and laboratory criteria.18,,–21 Acute GVHD was categorized as present (grade 2-4) or absent (grade 0-1). Chronic GVHD was categorized according to the presence or absence of clinical extensive chronic GVHD.
Neutropenia before transplantation was defined using the neutrophil count obtained closest to time of transplantation, before transplantation. Neutropenia was defined as an absolute neutrophil count (ANC) less than 500 cells/μL. After transplantation, engraftment occurred if the ANC was more than or equal to 500 cells/μL for 3 consecutive days. Neutropenia after engraftment was defined as an ANC less than 500 cells/μL after engraftment for more than or equal to one day. All patients with chemotherapy-induced neutropenia received systemic broad-spectrum prophylactic antibiotics for bacterial prophylaxis. Blood cultures were collected for evaluation of fever (core body temperature ≥ 38.3°C), and once weekly (outpatients) or twice weekly (inpatients) for patients who received systemic corticosteroids at a dose of at least 0.5 mg/kg. All patients received intermittent prophylaxis with trimethoprim/sulfamethoxazole, double-strength twice daily on Mondays and Tuesdays as first-line prophylaxis for Pneumocystis pneumonia.
Prospective cohort and LBP protein measurements
Between December 1, 2004, and January 31, 2007, we prospectively obtained consent from and enrolled 250 patients between 18 to 65 years of age scheduled to receive an allogeneic transplant at our Center. Fasting whole blood was drawn and centrifuged, and the serum was aspirated and aliquoted for storage at −80°C. LBP concentrations were measured using standard enzyme-linked immunosorbent assay techniques according to manufacturer specifications (Hycult Biotechnology, Uden, the Netherlands). A total of 234 patients who ultimately received a transplant were followed until the first episode of GN bacteremia, death, or discharge from our Center through February 9, 2007.
DNA, SNP selection, and genotyping
For the retrospective cohort, donor and recipient DNA was extracted (QIAamp DNA Blood Mini Kit; Qiagen, Valencia, CA) from B-lymphoblastoid cell lines immortalized by Epstein-Barr virus transformation.22 For the prospective cohort, DNA was isolated from citrated human whole blood using the Puregene DNA blood kit D-5500 (Gentra Systems, Minneapolis, MN).
Genetic variation data for the entire LBP gene was obtained from the Innate Immunity Program for Genomic Application (http://innateimmunity.net), a resource that contains the full LBP gene sequence, including 5000 base pairs upstream and downstream, for 23 healthy European whites. From this database, we identified 24 SNPs with a minor allele frequency (MAF) more than or equal to 10% and placed them in “bins” inferred according to the r2 linkage disequilibrium statistic (threshold ≥ 0.8).23 A maximally informative tagSNP was then selected from each bin using LDSelect (Figure 1A).24 This algorithm selects a subset of variants that efficiently describe all common patterns of variation in a gene, based on 2 primary criteria: 1) the MAF of a SNP and 2) the minimum level of association between assayed and unassayed SNPs, measured by the linkage disequilibrium statistic r2. Given these parameters, LDSelect identifies bins of SNPs such that one tagSNP per bin can be genotyped. All SNPs above the MAF threshold will either be directly genotyped or will exceed the specified level of allelic association with a SNP that is genotyped. The retrospective cohort was genotyped using the Illumina Beadarray platform.25 Data quality was assessed using random duplicate samples and gender discrimination. The prospective cohort was genotyped using the ABI Taqman Assay by Design according to manufacturer specifications (Applied Biosystems, Foster City, CA).
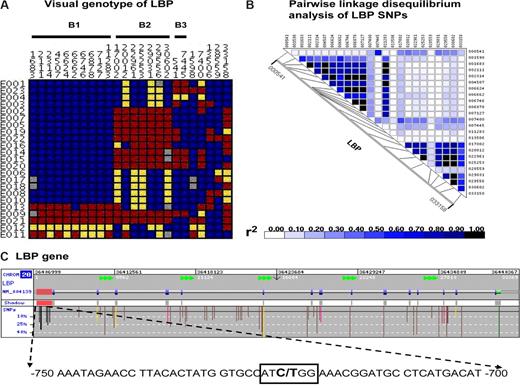
Single nucleotide polymorphisms on the lipopolysaccharide binding protein gene. (A) Visual genotype of LBP SNPs with a minor allele frequency more than or equal to 10% in 23 healthy whites. Each row represents an individual's genotype for each LBP SNP, which is represented by each column. Blue boxes represent homozygous wild-type genotypes; yellow boxes, heterozygous genotypes; red boxes, homozygous recessive genotypes; gray boxes, undetermined genotypes. Displayed data are arranged according to linkage disequilibrium bins inferred based on an r2 threshold of more than or equal to 0.8. Nineteen of 24 SNPs are in 3 linkage disequilibrium bins (B1, B2, and B3). SNP 6878 (rs2232582), SNP 17 002 (rs2232596), and SNP 541 (rs1780616) were selected as linkage disequilibrium tagging SNPs. (B) The pair-wise analysis of linkage disequilibrium, based on r2, among these LBP SNPs. These figures are based on LBP sequence data provided by the Innate Immunity Program for Genomic Application (http://innateimmunity.net), which sequenced the entire LBP gene and flanking regions in 23 CEPH European Americans from the Coriell Cell Repository. (C) Image of the entire LBP gene (provided by GeneSNPs) and SNPs identified in dbSNP with a minor allele frequency more than or equal to 10%. This includes a detailed display of SNP 1683 at −778, within a previously reported CAAT box (boxed nucleotides) in the 5′ 1.1-kb promoter region. Color codes are as follows: 5′ promoter region (red shaded area), flanking SNP (black), untranslated region (green), intronic (brown), synonymous SNPs (yellow), and nonsynonymous SNPs (pink).
Single nucleotide polymorphisms on the lipopolysaccharide binding protein gene. (A) Visual genotype of LBP SNPs with a minor allele frequency more than or equal to 10% in 23 healthy whites. Each row represents an individual's genotype for each LBP SNP, which is represented by each column. Blue boxes represent homozygous wild-type genotypes; yellow boxes, heterozygous genotypes; red boxes, homozygous recessive genotypes; gray boxes, undetermined genotypes. Displayed data are arranged according to linkage disequilibrium bins inferred based on an r2 threshold of more than or equal to 0.8. Nineteen of 24 SNPs are in 3 linkage disequilibrium bins (B1, B2, and B3). SNP 6878 (rs2232582), SNP 17 002 (rs2232596), and SNP 541 (rs1780616) were selected as linkage disequilibrium tagging SNPs. (B) The pair-wise analysis of linkage disequilibrium, based on r2, among these LBP SNPs. These figures are based on LBP sequence data provided by the Innate Immunity Program for Genomic Application (http://innateimmunity.net), which sequenced the entire LBP gene and flanking regions in 23 CEPH European Americans from the Coriell Cell Repository. (C) Image of the entire LBP gene (provided by GeneSNPs) and SNPs identified in dbSNP with a minor allele frequency more than or equal to 10%. This includes a detailed display of SNP 1683 at −778, within a previously reported CAAT box (boxed nucleotides) in the 5′ 1.1-kb promoter region. Color codes are as follows: 5′ promoter region (red shaded area), flanking SNP (black), untranslated region (green), intronic (brown), synonymous SNPs (yellow), and nonsynonymous SNPs (pink).
Statistical analysis
All statistical analyses were performed using SAS (SAS Institute, Cary, NC), R (R Foundation; http://www.r-project.org/), and STATA 8.0 (StataCorp, College Station, TX) software programs. The nested case control cohort was analyzed in 2 steps. In step 1, we identified clinical variables that may modify the genetic effects. This analysis included all cases (N = 350) and controls (N = 865) and was performed by first assessing the association between each clinical variable and GN bacteremia in univariate analysis. All clinical variables that were associated with GN bacteremia at a significance level of P less than .1 were then assessed using a forward and backward stepwise selection algorithm in conditional logistic multivariate regression analysis (Table 1). Variables with at least one statistically significant category (P < .05) in multivariate analysis were included in step 2. In step 2, we performed a genetic association analysis to determine whether LBP tagSNPs influenced the risk for developing GN bacteremia. This analysis was restricted to cases (N = 97) and controls (N = 204) that had both patient and donor DNA available in our genetic repository. We first assessed patient and donor LBP tagSNPs for deviation from Hardy-Weinberg equilibrium using a χ2 test. Each LBP tagSNP was then independently analyzed in multivariate models, which included the clinical variables previously found to be potential effect modifiers. This analysis was performed using the Hplus software, which evaluates phenotypic association with gene-based haplotypes while incorporating uncertainties due to unphased genotype data and adjustment for covariates.26
Comparison of the clinical characteristics of patients who developed Gram-negative bacteremia with patients who did not develop bacteremia
| Clinical variable . | Univariate analysis . | Multivariate analysis . | |||
|---|---|---|---|---|---|
| Controls, N = 865 . | Cases, N = 350 . | P . | Odds ratio (95% CI) . | P . | |
| Age, y | 34.0 ± 15.2 | 34.1 ± 15.9 | .95 | — | — |
| Sex match (patient/donor) | |||||
| Male/male | 305 (35) | 109 (31) | .024 | Referent | — |
| Male/female | 196 (23) | 72 (21) | — | 0.95 (0.65-1.40) | .791 |
| Female/male | 194 (22) | 108 (31) | — | 1.51 (1.06-2.16) | .023 |
| Female/female | 169 (20) | 61 (17) | — | 1.01 (0.67-1.50) | .977 |
| Race match (patient/donor) | |||||
| White/white | 765 (88) | 294 (84) | .105 | — | — |
| White/nonwhite | 7 (1) | 4 (1) | — | — | — |
| Nonwhite/white | 8 (1) | 8 (2) | — | — | — |
| Nonwhite/nonwhite | 85 (10) | 44 (13) | — | — | — |
| Disease risk | |||||
| Low | 298 (35) | 93 (26) | .015 | Referent | — |
| Moderate | 237 (27) | 97 (28) | — | 0.99 (0.68-1.45) | .974 |
| High | 330 (38) | 160 (46) | — | 1.43 (1.00-2.04) | .049 |
| Donor type | |||||
| Matched related | 452 (52) | 156 (45) | .043 | — | — |
| Mismatched related | 103 (12) | 53 (15) | — | — | — |
| Unrelated | 310 (36) | 141 (40) | — | — | — |
| Total body irradiation | |||||
| No | 335 (39) | 84 (24) | <.001 | Referent | — |
| Yes | 530 (61) | 266 (76) | — | 1.50 (1.08-2.07) | .015 |
| Stem cell source | |||||
| Bone marrow | 762 (88) | 326 (93) | .024 | — | — |
| PBSC | 96 (11) | 21 (6) | — | — | — |
| Other | 7 (1) | 3 (1) | — | — | — |
| CMV serostatus (patient/donor) | |||||
| Negative/negative | 397 (46) | 100 (29) | <.001 | Referent | — |
| Negative/positive | 152 (18) | 54 (15) | — | 1.39 (0.93-2.09) | .113 |
| Positive/negative | 125 (14) | 91 (26) | — | 2.59 (1.77-3.81) | <.001 |
| Positive/positive | 188 (22) | 104 (30) | — | 2.57 (1.78-3.69) | <.001 |
| Pretransplant neutropenia | |||||
| No | 441 (51) | 132 (38) | <.001 | Referent | — |
| Yes | 424 (49) | 218 (62) | — | 1.37 (1.01-1.87) | .045 |
| Days to engraftment | 20.5 ± 5.2 | 20.0 ± 5.8 | .127 | — | — |
| Neutropenia after initial engraftment | |||||
| No | 783 (91) | 246 (70) | <.001 | Referent | — |
| Yes | 82 (9) | 104 (30) | — | 2.77 (1.95-3.95) | <.001 |
| Acute GVHD | |||||
| No | 334 (39) | 55 (16) | <.001 | Referent | — |
| Yes | 518 (61) | 292 (84) | — | 3.03 (2.14-4.27) | <.001 |
| Chronic GVHD | |||||
| No | 574 (67) | 183 (55) | <.001 | Referent | — |
| Yes | 284 (33) | 155 (45) | — | 1.66 (1.24-2.22) | .001 |
| Clinical variable . | Univariate analysis . | Multivariate analysis . | |||
|---|---|---|---|---|---|
| Controls, N = 865 . | Cases, N = 350 . | P . | Odds ratio (95% CI) . | P . | |
| Age, y | 34.0 ± 15.2 | 34.1 ± 15.9 | .95 | — | — |
| Sex match (patient/donor) | |||||
| Male/male | 305 (35) | 109 (31) | .024 | Referent | — |
| Male/female | 196 (23) | 72 (21) | — | 0.95 (0.65-1.40) | .791 |
| Female/male | 194 (22) | 108 (31) | — | 1.51 (1.06-2.16) | .023 |
| Female/female | 169 (20) | 61 (17) | — | 1.01 (0.67-1.50) | .977 |
| Race match (patient/donor) | |||||
| White/white | 765 (88) | 294 (84) | .105 | — | — |
| White/nonwhite | 7 (1) | 4 (1) | — | — | — |
| Nonwhite/white | 8 (1) | 8 (2) | — | — | — |
| Nonwhite/nonwhite | 85 (10) | 44 (13) | — | — | — |
| Disease risk | |||||
| Low | 298 (35) | 93 (26) | .015 | Referent | — |
| Moderate | 237 (27) | 97 (28) | — | 0.99 (0.68-1.45) | .974 |
| High | 330 (38) | 160 (46) | — | 1.43 (1.00-2.04) | .049 |
| Donor type | |||||
| Matched related | 452 (52) | 156 (45) | .043 | — | — |
| Mismatched related | 103 (12) | 53 (15) | — | — | — |
| Unrelated | 310 (36) | 141 (40) | — | — | — |
| Total body irradiation | |||||
| No | 335 (39) | 84 (24) | <.001 | Referent | — |
| Yes | 530 (61) | 266 (76) | — | 1.50 (1.08-2.07) | .015 |
| Stem cell source | |||||
| Bone marrow | 762 (88) | 326 (93) | .024 | — | — |
| PBSC | 96 (11) | 21 (6) | — | — | — |
| Other | 7 (1) | 3 (1) | — | — | — |
| CMV serostatus (patient/donor) | |||||
| Negative/negative | 397 (46) | 100 (29) | <.001 | Referent | — |
| Negative/positive | 152 (18) | 54 (15) | — | 1.39 (0.93-2.09) | .113 |
| Positive/negative | 125 (14) | 91 (26) | — | 2.59 (1.77-3.81) | <.001 |
| Positive/positive | 188 (22) | 104 (30) | — | 2.57 (1.78-3.69) | <.001 |
| Pretransplant neutropenia | |||||
| No | 441 (51) | 132 (38) | <.001 | Referent | — |
| Yes | 424 (49) | 218 (62) | — | 1.37 (1.01-1.87) | .045 |
| Days to engraftment | 20.5 ± 5.2 | 20.0 ± 5.8 | .127 | — | — |
| Neutropenia after initial engraftment | |||||
| No | 783 (91) | 246 (70) | <.001 | Referent | — |
| Yes | 82 (9) | 104 (30) | — | 2.77 (1.95-3.95) | <.001 |
| Acute GVHD | |||||
| No | 334 (39) | 55 (16) | <.001 | Referent | — |
| Yes | 518 (61) | 292 (84) | — | 3.03 (2.14-4.27) | <.001 |
| Chronic GVHD | |||||
| No | 574 (67) | 183 (55) | <.001 | Referent | — |
| Yes | 284 (33) | 155 (45) | — | 1.66 (1.24-2.22) | .001 |
Data entries are numbers (%) for each item.
CI indicates confidence interval; PBSC, peripheral blood stem cell; CMV, cytomegalovirus; GVHD, graft-versus-host disease; and —, not applicable.
For the prospective cohort, one-way analysis of variance was used to assess the relationship between genotypes and log-transformed LBP serum protein levels. Multivariate Cox proportional hazard regression models were used to evaluate the relationship between the presence of the putative functional SNP and time to development of GN bacteremia and death. The mortality analysis was also stratified according to presence of GN bacteremia to assess whether the effect of the putative functional SNP on mortality was more pronounced in the presence of GN bacteremia. A stepwise selection algorithm was used as above to assess pretransplantation clinical variables (Table S2). The proportional hazards assumption was tested using the log-rank test.
Results
From 3193 HCT recipients, 350 cases and 865 controls were identified. The median time to development of GN bacteremia was 53 days (range, 1-195 days). From the univariate and multivariate analyses (Table 1), we determined that donor gender match, disease risk, tuberculosis infection status, cytomegalovirus serostatus, presence of neutropenia before transplantation and recurrence after transplantation, and the development of acute and chronic GVHD were significantly associated with GN bacteremia, and therefore, may influence the relationship between genetic variants and the risk for GN bacteremia. All of these variables were included in the subsequent LBP genetic analyses models.
Association of LBP tagSNPs and GN bacteremia
Analysis of the LBP sequence data revealed there were 24 SNPs with a minor allele frequency more than or equal to 10%, 19 of which existed in 3 linkage disequilibrium bins (Figure 1). One tagSNP from each bin was selected for genotyping: SNP 6878 (rs2232582), SNP 17 002 (rs2232596), and SNP 541 (rs1780616).
From the 350 cases and 865 controls in this epidemiologic evaluation, 97 cases and 204 controls were selected based on availability of both patient and donor DNA samples. All patient and donor genotypes were in Hardy-Weinberg equilibrium. Univariate analysis of donor genotypes revealed no association with GN bacteremia (SNP 6878, P = .104; SNP 17002, P = .907; SNP 541, P = .527), but the patient SNP 6878 genotype was significantly associated with GN bacteremia (SNP 6878, P = .002; SNP 17002, P = .079; SNP 541, P = .593). Among the cases, 7 (7%) were homozygous for the SNP 6878 C allele (minor allele) and 38 (39%) were heterozygous, versus 3 (1.5%) and 55 (27%) among the controls, respectively. Multivariate analysis revealed that patient SNP 6878 (P = .001) and SNP 17002 (P = .027) genotypes were associated with GN bacteremia (Table 2). However, in multivariate analysis restricted to whites, only the association with SNP 6878 remained significant; the presence of the SNP 6878 C allele was associated with a 2-fold higher risk for GN bacteremia (odds ratio = 2.15; 95% confidence interval [CI], 1.31-3.52, P = .002).
Association of patient LBP tagSNP genotypes with GN bacteremia
| TagSNP . | All participants (97 cases; 204 controls) . | Whites only (85 cases; 189 controls) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Allele frequencies . | Odds ratio (95% CI) . | P . | Allele frequencies . | Odds ratio (95% CI) . | P . | |||
| Case . | Control . | Case . | Control . | |||||
| 6878 (C/T) | 0.26 | 0.15 | 2.22 (1.39-3.56) | .001 | 0.27 | 0.16 | 2.15 (1.31-3.52) | .002 |
| 17002 (A/G) | 0.40 | 0.50 | 0.65 (0.44-0.95) | .027 | 0.41 | 0.49 | 0.7 (0.47-1.06) | .089 |
| 541 (C/T) | 0.34 | 0.32 | 0.93 (0.63-1.38) | .729 | 0.32 | 0.34 | 0.9 (0.6-1.35) | .602 |
| TagSNP . | All participants (97 cases; 204 controls) . | Whites only (85 cases; 189 controls) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Allele frequencies . | Odds ratio (95% CI) . | P . | Allele frequencies . | Odds ratio (95% CI) . | P . | |||
| Case . | Control . | Case . | Control . | |||||
| 6878 (C/T) | 0.26 | 0.15 | 2.22 (1.39-3.56) | .001 | 0.27 | 0.16 | 2.15 (1.31-3.52) | .002 |
| 17002 (A/G) | 0.40 | 0.50 | 0.65 (0.44-0.95) | .027 | 0.41 | 0.49 | 0.7 (0.47-1.06) | .089 |
| 541 (C/T) | 0.34 | 0.32 | 0.93 (0.63-1.38) | .729 | 0.32 | 0.34 | 0.9 (0.6-1.35) | .602 |
Each tagSNP was analyzed in independent multivariate models that included sex match, disease risk, TBI dose, CMV serostatus, presence of neutropenia pretransplant, recurrent neutropenia after engraftment, and presence of acute or chronic GVHD as covariates.
SNP 6878 tags the first LBP linkage disequilibrium bin (B1), which contains 9 other SNPs (Figure 1A). Three of these SNPs in B1, SNP 1683 (rs2232571), SNP 2111 (rs2232575), and SNP 2314 (rs2232578), map to within the 5′ 1.1-kb promoter region. SNP 6878 is in strong linkage disequilibrium with SNP 1683 (r2 = 1.0, Figure 1B). Based on previous detailed mapping of the LBP promoter region, SNP 1683 confers a C/T substitution in the CAAT box at position −778 (Figure 1C).10 In the same previous study, a promoter truncation mutation that excluded this region increased the inducibility of the promoter.10 Therefore, we hypothesized that SNP 1683 may be a functional variant that affects the efficiency of the LBP promoter.
Association of SNP 1683 with circulating LBP levels and mortality
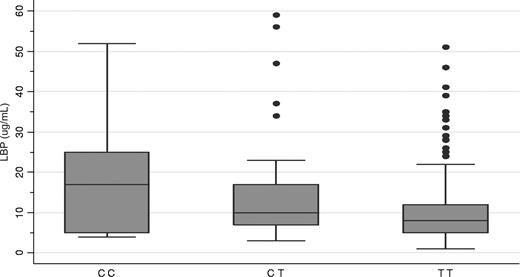
To address the above hypothesis and validate the discovery data suggesting that genetic variation in the LBP gene predisposes to GN bacteremia, we assessed whether basal LBP levels in serum collected from a prospective cohort of 250 patients being assessed for HCT correlated with the SNP 1683 genotype. SNP 1683, which was in Hardy-Weinberg equilibrium (P = .14), was found to be significantly associated with plasma LBP levels (P = .0036). The median plasma LBP levels according to SNP 1683 genotype were TT (N = 182), 8.07 μg/mL; TC (N = 59), 10.40 μg/mL; and CC (N = 9), 17.39 μg/mL (Figure 2).
Relationship between SNP 1683, circulating LBP levels, and mortality. Heterozygous and homozygous recessive patients had higher median circulating LBP levels measured before transplantation (box plot; P = .004). Box indicates 25th percentile and whiskers, 75th percentile.
Relationship between SNP 1683, circulating LBP levels, and mortality. Heterozygous and homozygous recessive patients had higher median circulating LBP levels measured before transplantation (box plot; P = .004). Box indicates 25th percentile and whiskers, 75th percentile.
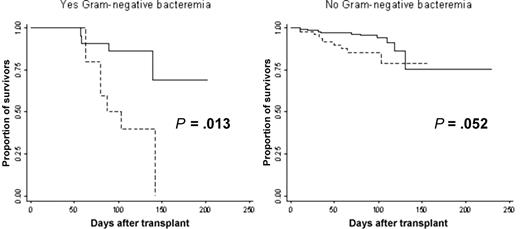
Among these patients, 234 received transplants, 32 of which developed GN bacteremia during a median follow-up time of 98 days (range, 11-230 days). The SNP 1683 C allele was significantly associated with an overall 3-fold increase in risk of death before discharge from our Center (hazard ratio [HR] = 3.30; 95% CI, 1.59-6.84, P = .001; Table 3). When this analysis was stratified according to GN bacteremia status, patients with the SNP 1683 C allele who developed GN bacteremia had a significant 5-fold increase in mortality risk (HR = 4.83; 95% CI, 1.38-16.75, P = .013; Figure 3); among patients with GN bacteremia, 64% (N = 7) of those who died had the SNP 1683 C allele, versus 14% (N = 3) among those who survived. Even among patients who did not develop GN bacteremia, patients with the SNP 1683 C allele had a borderline significant 2-fold increase in mortality risk (HR = 2.51; 95% CI, 0.99-6.37, P = .052). Only older patient age was associated with death after transplantation in univariate analysis (P = .019; Table S2). However, patient age was not a significant factor in the multivariate analysis. The SNP 1683 C allele was not significantly associated with an increase in risk for GN bacteremia, but this was expected because the size of the prospective cohort was not designed to detect an association with GN bacteremia.
Relationship between SNP 1683 and mortality after transplant in the prospective cohort
| SNP 1683 genotype . | Death, N (%) . | Total . | |
|---|---|---|---|
| No . | Yes . | ||
| TT | 158 (92) | 14 (8) | 172 |
| TC | 41 (76) | 13 (24) | 54 |
| CC | 6 (75) | 2 (25) | 8 |
| SNP 1683 genotype . | Death, N (%) . | Total . | |
|---|---|---|---|
| No . | Yes . | ||
| TT | 158 (92) | 14 (8) | 172 |
| TC | 41 (76) | 13 (24) | 54 |
| CC | 6 (75) | 2 (25) | 8 |
P = .004.
Kaplan-Meier survival curve stratified by whether Gram-negative bacteremia developed (yes vs no). As more patients with the SNP 1683 C allele ( ) died, this proportion was higher among patients who developed Gram-negative bacteremia.
) died, this proportion was higher among patients who developed Gram-negative bacteremia.
Kaplan-Meier survival curve stratified by whether Gram-negative bacteremia developed (yes vs no). As more patients with the SNP 1683 C allele ( ) died, this proportion was higher among patients who developed Gram-negative bacteremia.
) died, this proportion was higher among patients who developed Gram-negative bacteremia.
Discussion
These results demonstrate that genetic variation in the promoter region of the LBP gene is associated with the blood level of LBP and with the risk of developing GN bacteremia and GN bacteremia-related death after HCT. Transcriptional activity of the LBP gene is partly governed by the patient genotype. SNP 1683 confers a C/T substitution at position −778, which is located in one of the LBP CAAT boxes, which are transcriptional elements that regulate the efficiency of the promoter. In promoter truncation experiments that excluded this region, LBP promoter inducibility increased 3-fold compared with when the entire promoter was intact.10 Association of the patients' SNP 1683 genotype with a 2-fold higher LBP level suggests that presence of the minor SNP 1683 allele may enhance the efficiency of the promoter. Unlike previous candidate LBP SNP approaches,27,28 our study benefited from current extensive knowledge of the genetic variation across the entire LBP gene. We analyzed nearly 80% of all the common LBP SNPs, defined as SNPs with a minor allele frequency more than or equal to 10%, by genotyping for only 3 tagSNPs. In the context of a biologically relevant phenotype and a racially uniform population, this maximized our likelihood of finding a meaningful genetic association.
This finding provides novel insight into the complex biology of the innate immune response to LPS and GN infections. LBP is a secretory class I acute-phase protein, whose gene is transcriptionally activated by APRF/STAT3 and other cytokine-inducible nuclear proteins, such as AP-1 and C/EBPβ.10,29 Transcription of the LBP gene is also induced by IL-1β alone, synergistically by IL-1β and IL-6, or by tumor necrosis factor-α and dexamethasone, resulting in maximal circulating LBP concentrations within 24 to 48 hours after LPS challenge.30,31 LBP's role in modulating the innate immune response is a 2-edged sword, depending on the concentration of LBP. In its proinflammatory role, LBP binds to the amphipathic lipid A component of LPS with high affinity and catalyzes the transfer of LPS to the binding site of membrane-bound and soluble forms of CD14, facilitating the host inflammatory response. Indeed, early studies in murine models of endotoxemia found LBP−/− mice were protected against septic shock in response to intraperitoneally injected Salmonella LPS but were more susceptible to infections.32 Human studies also indicate that LBP levels are significantly higher among patients with sepsis or septic shock compared with normal control patients.33
However, more recent studies have found that high concentrations of LBP can have inhibitory effects on LPS-induced host cell activation. Murine models indicate that mice are protected by high-dose LBP after otherwise lethal intraperitoneal LPS injections or infection with GN bacteria.9 In humans, sera from patients with severe sepsis or septic shock, which contains high LBP concentrations, significantly reduce LPS binding to monocytes, reduce monocyte activation, and limit the monocyte's immunologic response to bacterial challenge.8 There are now 4 known mechanisms by which LBP anti-inflammatory activity can occur. LBP can bind apolipoprotein A- or apolipoprotein B-containing lipoproteins and transfer LPS into high- and low-density lipoproteins, very-low-density lipoproteins, or chylomicrons, resulting in the clearance of LPS from the bloodstream.34,,,–38 LBP also combines with LPS aggregates to form large LPS–LBP complexes that bind to membrane-bound CD14 and are internalized but have decreased ability to signal through MD2/TLR4.39,40 LBP can also bind LPS that has already bound to membrane-bound CD14 and attenuate cell responses.41 Finally, Gioannini et al recently found that LBP binds to LPS to expose fatty acyl chains within lipid A, which allows acyloxyacyl hydrolase and eukaryotic lipase to bind and detoxify LPS.42 Although these data suggest that high levels of circulating LBP may be beneficial for curtailing the inflammatory response to high concentrations of LPS, the resultant attenuation of cellular response to GN bacteria, especially when leukocyte counts are already low and innate immunity is even more important in host defense (as commonly found early after transplantation), may permit unchecked GN bacterial growth and ultimately result in death.
The association of SNP 1683 with a 5-fold increase in risk of death after transplantation among patients with GN bacteremia, and a borderline effect among patients without GN bacteremia, suggests several possible mechanisms by which LBP variants might influence mortality risk. This finding is consistent with the known biology of LBP and the major role it plays in modulating the host immune response to GN bacteria and LPS.43 If high levels of LBP down-regulate the innate immune response to GN bacteria, which is essentially the only immune response available during the early posttransplantation period, the outcome may be disastrous in the presence of GN bacteremia. We also suspect that the borderline association observed among patients without GN bacteremia may be related to clinically undetectable GN bacteremia. It is well known that, because of intestinal mucosal damage related to the conditioning regimen, GN bacteria commonly translocate across the intestinal mucosa during the early posttransplantation period.44 In the setting of a genetically predisposed patient whose LBP levels are high, this relatively low level of bacteremia that is undetectable by standard clinical techniques may be allowed to advance unchecked by the innate immune system, ultimately leading to increased mortality. The magnitude of the genetic attributable risk is also noteworthy. The risk for mortality associated with SNP 1683 is higher than nearly all clinical predictors of mortality we recently identified in a multistage cohort study of more than 2400 patients.45 These results suggest that use of SNP 1683 as a predictor of mortality risk may be worthy of validation in future studies in other cohorts.
The a priori intent of the prospective cohort was to confirm the clinical association with an intermediate phenotype (circulating LBP level) and a secondary clinical phenotype (mortality). Although we did not find an association between SNP 1683 and GN bacteremia in the prospective cohort, this was expected. Although the incidence of clinically detectable GN bacteremia was consistent with our retrospective data (∼10% per year), there were only 32 cases of GN bacteremia; this cohort was grossly underpowered for detection of a genetic association. We also would have preferred to demonstrate an association between SNP 1683 and LBP gene transcription activity. However, this was not possible because circulating LBP is produced by hepatocytes, which were not available from our prospective cohort.
As elegantly summarized by Mullally and Ritz in a recent publication,46 genetic variants in immune response genes can influence the outcome of HCT, independent of HLA match status. Our LBP findings add additional justification for more research in this area of HCT. However, our findings also have clinical implications beyond the HCT population. GN organisms are among the most common causes of nosocomial infections and mortality in hospitalized and critically ill patients. Future studies should focus on confirming the function of SNP 1683 in molecular assays, determining how genetic variation in LBP affects clinical outcomes, and evaluating the predictive value of this genetic biomarker for clinical outcomes in various high-risk populations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Gary Schoch for retrieval of the clinical data, Eric Mickelson for managing the research cell bank, Jeffrey Stevens and Brandon Au for the genotyping and protein measurements, Hyung Woo Kim, PhD, for performing the clinical statistical analyses, and Lue Ping Zhao, PhD, for serving as the statistical genetics consultant.
This work was supported by National Institutes of Health (grants K23HL69860, AI33484, CA15704; CA18029, and HL87690), American Lung Association of Washington Research Grant, and the Amy Strelzer Manasevit Research Award from the National Marrow Donor Program.
National Institutes of Health
Authorship
Contribution: J.W.C. provided the initial study concept and design, provided input on the study design and participated in the finalization of the design; was responsible for retrieval of the biologic samples, the genetic and protein analyses, the clinical data analyses, and the genetic analyses; and participated in the interpretation of the data and preparation of the manuscript; M.J.B. provided input on the study design and participated in the finalization of the design, was responsible for the clinical data analyses, and participated in the interpretation of the data and preparation of the manuscript; J.A.H. provided input on the study design and participated in the finalization of the design, was responsible for retrieval of the biologic samples and the genetic analyses, and participated in the interpretation of the data and preparation of the manuscript; and J.G.C. provided the initial study concept and design, provided input on the study design and participated in the finalization of the design, was responsible for the genetic and protein analyses, and participated in the interpretation of the data and preparation of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason W. Chien, Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, D5-280, Seattle, WA 98109-1024; e-mail: jchien@fhcrc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal