Abstract
The precise mechanisms by which Abl oncogenes transform hematopoietic cells are unknown. We have examined the role of Pim kinases in v-Abl–mediated transformation. In v-Abl transformants, expression of Pim-1 and Pim-2, but not Pim-3, is dependent on Abl kinase activity. Transformation assays demonstrate that v-Abl cannot efficiently transform bone marrow cells derived from Pim-1−/−/Pim-2−/− mice. Ectopic expression of either Pim-1 or Pim-2 in Pim-1−/−/Pim-2−/− cells restores transformation by v-Abl, strongly suggesting that either Pim-1 or Pim-2 is required for v-Abl–mediated tumorigenesis. Interestingly, the combined deficiency of Pim-1, Pim-2, and Suppressor of Cytokine Signalling (SOCS)-1 resulted in partial restoration of v-Abl transformation efficiency. In addition, Pim kinases are involved in modification of SOCS-1 and in regulating SOCS-1 protein levels in v-Abl–transformed cells. Furthermore, Pim kinases regulate the proapoptotic proteins Bcl-XS and BAD. Pim kinases inhibit the expression of Bcl-XS. Pim deficiency decreases the phosphorylation levels of BAD, whereas ectopic expression of Pim-1 increases the amount of phospho-BAD. This correlates with an increased protection from apoptosis in Abl transformants expressing Pim kinases. Together, these data suggest that Pim kinases play a key role in the v-Abl transformation, possibly via participating in modulation of SOCS-1 and via regulating the apoptotic signaling.
Introduction
Malignant transformation of hematopoietic cells involves the dysregulation of genes that are normally involved in regulating proliferation and survival. The mechanism of how such dysregulation can summate to transform cells is still unclear. The Pim-1 and Pim-2 oncogenes have been implicated in the transformation of both T and B lymphocytes,1,2 but the mechanism by which dysregulated expression of Pim kinases promotes the development of hematopoietic tumors has not been defined.
The Pim-1 gene was originally identified as a proviral insertion site of the Moloney murine leukemia virus (MoMuLV) in lymphomas in mice.1 Subsequently, Pim-2 and Pim-3 were cloned, and these 3 proteins constitute a distinct family of serine/threonine kinases. Increased expression of Pim kinases has been demonstrated in several different types of human tumors, including lymphomas, prostate cancer, and oral cancer.3-7 A high frequency of mutation in the Pim-1 gene has been observed in diffuse large B-cell lymphoma cases.2,8 Importantly, experiments using transgenic mouse models have demonstrated that overexpression of Pim-1 or Pim-2 predisposes the mice to the development of lymphoid tumors.9,10 In addition, the Pim family of kinases has been implicated in the development of other types of human hematopoietic malignancies. Overexpression of Pim-1 has been observed in acute myeloid and erythroid leukemias.6 These data suggest that dysregulation of Pim expression may be a common occurrence during transformation of a variety of cell types.
How then do Pim kinases propagate signals that promote tumorigenesis, and why are elevated levels of Pim kinases important for the formation of hematopoietic malignancies? Pim-1, Pim-2, and Pim-1/Pim-2–deficient mice all show only minor phenotypic aberrations.11,12 Mice deficient for all 3 Pim kinases display reduced body size and impaired responses to hematopoietic growth factors.13 Little is known about the functional role of the Pim kinases in vivo, although a number of observations have pointed to a potential role for Pim-1 in signal transduction. Pim kinases also have been demonstrated to act on several cellular substrates including BAD, NFATc, Cdc25A, and Suppressor of Cytokine Signalling (SOCS)-1.14-17 In addition, recent data suggests that Pim can phosphorylate Cot, a regulator of I-κB function.18 This appears to be important in the increased activation of NF-κB in some transformed lymphocytes.18 However, the precise role of Pim-mediated modification of these substrates in cellular transformation is not fully understood.
Interestingly, recent studies have shown that the expression of Pim-1 is induced by BCR-Abl–activated STAT5.19,20 Up-regulation of Pim expression may play an important role in cellular transformation mediated by BCR-Abl.19,21 However, a study from another group demonstrated that although Pim-1 was markedly up-regulated following activation of STAT5, the expression of Pim-1 is dispensable for transformation by BCR-Abl.22 Because these studies use growth factor–dependent cell lines such as Ba/F3 and FDCP1, the importance of Pim kinases for Abl-mediated transformation remains to be further elucidated.
To better understand the role of Pim kinases in Abl-mediated transformation, we have carried out studies of their expression and functional involvement in pre–B-cell tumorigenesis by using wild-type and Pim-deficient cells. These studies demonstrate that Pim deficiency has profound effects on the v-Abl transformation. Pim kinases are involved in multiple pathways that affect v-Abl–mediated transformation. First, Pim kinases are involved in SOCS-1 modification and in regulation of SOCS-1 protein levels in v-Abl transformants. Second, Pim kinases function as negative regulators of the expression of the proapoptotic protein Bcl-XS and the activation of BAD, and thereby may prevent BAD- and Bcl-XS–dependent apoptosis. Based on these findings, we propose a model for how Pim kinases promote the development of Abl oncogene-induced tumors.
Methods
v-Abl–mediated transformation and cell culture
Bone marrow or fetal liver transformation was performed, and v-Abl–transformed pre–B-cell lines were generated and cultured as described previously.23 Briefly, cells were infected with free viruses by spin infection. Infected cells were seeded in 96-well plates, and transformation efficiency was scored by counting the number of wells that displayed cytokine-independent growth 2 weeks after infection.
Plasmids and retroviral vectors
The bicistronic retroviral vector containing p120v-Abl and green fluorescence protein (GFP) was a gift from Dr Naomi Rosenberg (School of Medicine, Tufts University). The GFP sequence was excised from the construct, and either Pim-1, Pim-2, or Pim-3 coding DNAs were subcloned into the BsaB I/Mfe I sites to generate p120v-Abl-IRES-Pim1, p120v-Abl-IRES-Pim2, and p120v-Abl-IRES-Pim3. Either wild-type or kinase-dead mutation (K67M) of human Pim-1 were subcloned into pMSCV-IRES-EGFP (pMIG) to generate pMIGPim-1 and pMIGPim-1KD. The bicistronic retroviral vector expressing p120v-Abl and SOCS-1 has been previously described.23
Antibodies
The following antibodies were used for Western blotting or immunoprecipitation: anti–Pim-1, anti–Pim-2, and anti–Bcl-XS/L (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Abl AB-2, and AB-3 (CalBiochem, San Diego, CA), anti–X-press (Invitrogen, Carlsbad, CA), anti-BAD and anti-phospho-BAD (Ser112; Cell Signaling, Beverly, MA), anti-actin, anti-flag, and anti-hemagglutinin (HA; Sigma-Aldrich, St Louis, MO). All other anti-bodies were obtained as described previously.17,23
Cytokine and inhibitor treatments of cells
Interferon (IFN)-γ was purchased from BD Biosciences (Palo Alto, CA) and used at a concentration of 50 ng/mL for cell stimulation. For cycloheximide (Sigma-Aldrich) time course experiments, the final concentration of cycloheximide was 100 μg/mL. For LLNL (N-acetyl-leu-leu-norleucinal, Sigma-Aldrich) treatments of cells, LLNL was used at concentrations of 1 μM or 5 μM as indicated. Imatinib was a generous gift from Novartis (Cambridge, MA). Pre–B-cell lines were treated with 1 μM imatinib for 4 hours, unless otherwise indicated.
Cell extracts, immunoprecipitation, and Western blotting
Flow cytometry and apoptosis assay
Cells were washed extensively in medium and cultured with imatinib for the indicated times. For double staining with propidium iodide (PI; Sigma-Aldrich) and annexin V (Molecular Probes, Eugene, OR), cells were washed extensively with ice-cold phosphate buffered saline (PBS), and the samples were resuspended in PBS containing 2.5 μg/mL annexin V, 1 μg/mL PI, 1 mM MgCl2, and 1.8 mM CaCl2. Then the samples were analyzed by fluorescence-activated cell sorter. In some cases, cell viability was also assessed by counting the trypan blue–negative cells serially.
Metabolic radiolabeling of phosphoprotein
v-Abl–transformed pre-B cells stably expressing SOCS-1 were starved of phosphate for 3 hours, and phosphoamino acid(s) was then metabolically labeled by adding 500 μCi (18.5 MBq) of H332PO4. Using anti-Flag antibody32P-labeled SOCS-1 protein was isolated by immuno affinity, and phosphorylation was detected by autoradiography. SOCS-1 expression levels were examined by Western blot.
Results
Pim-1 and Pim-2 are expressed in v-Abl–transformed pre-B cells in an Abl kinase-dependent manner
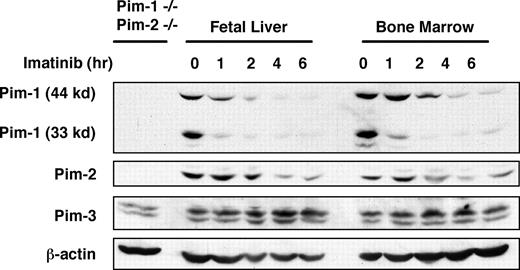
To explore the precise role of Pim kinases in Abl-mediated transformation, we generated several v-Abl–transformed pre–B-cell lines by infecting bone marrow or fetal liver cells with Abelson murine leukemia virus (A-MuLV). Although Pim-1 expression was reported to be expressed downstream of BCR-Abl signaling,19 it is still unclear if Pim kinases are expressed downstream of v-Abl. To address this issue, v-Abl–transformed bone marrow or fetal liver pre-B cells were treated in a time course with imatinib, an Abl kinase inhibitor that cannot inhibit Pim-dependent phosphorylation of SOCS-1 (Figure S1). Whole cell extracts from these samples were prepared, and Pim-1, Pim-2, and Pim-3 protein expression was examined by Western blotting. The results showed that all 3 Pim kinases are expressed in v-Abl transformants (Figure 1). However, only the levels of Pim-1 and Pim-2 consistently decreased after treatment with imatinib, indicating that expression of these 2 proteins, but not Pim-3, is Abl kinase dependent.
The expression of Pim-1, Pim-2, and Pim-3 proteins in A-MuLV–transformed pre-B-cell lines derived from fetal liver and bone marrow. v-Abl–transformed cells were treated in a time course with imatinib, an Abl kinase inhibitor. A Western blot of cell lysates was probed with indicated antibodies. Lysate of Pim-1−/−/Pim-2−/− cells was used as a control.
The expression of Pim-1, Pim-2, and Pim-3 proteins in A-MuLV–transformed pre-B-cell lines derived from fetal liver and bone marrow. v-Abl–transformed cells were treated in a time course with imatinib, an Abl kinase inhibitor. A Western blot of cell lysates was probed with indicated antibodies. Lysate of Pim-1−/−/Pim-2−/− cells was used as a control.
Pim deficiency has profound effects on the v-Abl–mediated pre–B-cell transformation
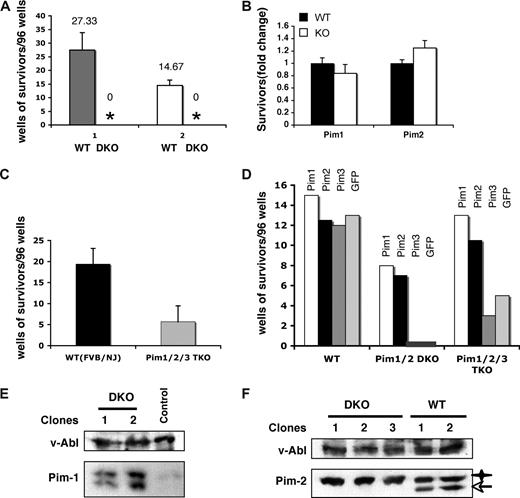
To determine whether Pim-1 and/or Pim-2 are required for v-Abl–mediated cellular transformation, bone marrow cells from Pim-1/Pim-2 double-deficient mice (Pim-1−/−/Pim-2−/−) and control wild-type mice with the same genetic background were infected with A-MuLV. In addition, another group of age-matched, 129 strain mice that have a genetic background that makes them less susceptible to A-MuLV than the mixed background of the Pim-1−/−/Pim-2−/− mice, was also included as a control. Infected cells were seeded on cytokine-free media in 96-well plates. As shown in Figure 2A, cells from every control mouse displayed significant transformation. However, none of the plates seeded with infected Pim-1−/−/Pim-2−/− cells showed growth in any wells (Figure 2A), whereas cells from either Pim-1 or Pim-2 single-deficient mice displayed wild-type levels of transformation (Figure 2B). Together, our results strongly suggest that either Pim-1 or Pim-2 is required for v-Abl–mediated pre–B-cell transformation and indicate a functional redundancy of Pim-1 and Pim-2 in this particular event.
Pim deficiency has profound effects on the v-Abl–mediated pre–B-cell transformation. (A) Well numbers per 96-well plate showing cytokine-independent growth of v-Abl–transformed wild-type (WT) and Pim-1−/−/Pim-2−/− double knockout (DKO) cells. Plotted are the average well numbers. The error bars represent the SEM (n = 3). (1) Bone marrow cells from DKO and wild-type mice with the same genetic background (mixed background) were used. (2) Mice of 129 strain, which have a genetic background that makes them less susceptible to A-MuLV than the background of the DKO mice, were also included as a control. (B) Bone marrow cells from Pim-1−/− and Pim-2−/− mice and their wild-type littermates were infected with A-MuLV. Cells were plated on soft agar medium and transformation efficiency was scored by counting foci generated 2 weeks after infection. Results are shown as fold-difference in the number of foci in the knockout with respect to wild-type mice. (C) Experiments were done as described in panel A. Pim-1−/−/Pim-2−/−/Pim-3−/− (Pim1,2,3−/−) and wild-type (WT) mice with the same genetic background (FVB/NJ) were used. Plotted are the average well numbers. The error bars represent the SEM (n = 3). (D) Bone marrow cells from Pim-1−/−/Pim-2−/−, Pim-1−/−/Pim-2−/−/Pim-3−/−, or wild-type mice were infected with bicistronic retroviruses encoding the p120 form of v-Abl and either Pim-1, Pim-2, Pim-3, or GFP. Transformation efficiency was scored as described in panel A. (E) Ectopic co-expression of Pim-1 with v-Abl allows the transformation of DKO cells by v-Abl. Shown is an immunoblot of v-Abl–transformed cell clones probed as indicated. Control is a v-Abl–transformed clone expressing Pim-2. (F) Ectopic co-expression of Pim-2 with v-Abl also allows the transformation of DKO cells by v-Abl as described in panel E. v-Abl–transformed wild-type (WT) clones were used as a control. Star, transgenic Pim-2; arrow, endogenous Pim-2.
Pim deficiency has profound effects on the v-Abl–mediated pre–B-cell transformation. (A) Well numbers per 96-well plate showing cytokine-independent growth of v-Abl–transformed wild-type (WT) and Pim-1−/−/Pim-2−/− double knockout (DKO) cells. Plotted are the average well numbers. The error bars represent the SEM (n = 3). (1) Bone marrow cells from DKO and wild-type mice with the same genetic background (mixed background) were used. (2) Mice of 129 strain, which have a genetic background that makes them less susceptible to A-MuLV than the background of the DKO mice, were also included as a control. (B) Bone marrow cells from Pim-1−/− and Pim-2−/− mice and their wild-type littermates were infected with A-MuLV. Cells were plated on soft agar medium and transformation efficiency was scored by counting foci generated 2 weeks after infection. Results are shown as fold-difference in the number of foci in the knockout with respect to wild-type mice. (C) Experiments were done as described in panel A. Pim-1−/−/Pim-2−/−/Pim-3−/− (Pim1,2,3−/−) and wild-type (WT) mice with the same genetic background (FVB/NJ) were used. Plotted are the average well numbers. The error bars represent the SEM (n = 3). (D) Bone marrow cells from Pim-1−/−/Pim-2−/−, Pim-1−/−/Pim-2−/−/Pim-3−/−, or wild-type mice were infected with bicistronic retroviruses encoding the p120 form of v-Abl and either Pim-1, Pim-2, Pim-3, or GFP. Transformation efficiency was scored as described in panel A. (E) Ectopic co-expression of Pim-1 with v-Abl allows the transformation of DKO cells by v-Abl. Shown is an immunoblot of v-Abl–transformed cell clones probed as indicated. Control is a v-Abl–transformed clone expressing Pim-2. (F) Ectopic co-expression of Pim-2 with v-Abl also allows the transformation of DKO cells by v-Abl as described in panel E. v-Abl–transformed wild-type (WT) clones were used as a control. Star, transgenic Pim-2; arrow, endogenous Pim-2.
To further confirm the functional relevance of Pim family of serine/threonine kinases in v-Abl–mediated transformation, we repeated A-MuLV infection experiment using bone marrow cells derived from mice deficient for all Pim kinases (Pim-1−/−/Pim-2−/−/Pim-3−/−) with FVB/NJ genetic background. As expected, Pim-1−/−/Pim-2−/−/Pim-3−/− cells displayed marked reduction in v-Abl transformation efficiency (Figure 2C). Interestingly, despite low efficiency, some wells showed cytokine-independent cell growth, and v-Abl–transformed pre–B-cell lines were generated. It is unclear why there existed v-Abl transformants in Pim-1−/−/Pim-2−/−/Pim-3−/− cells but not in Pim-1−/−/Pim-2−/− cells. One possibility is that the levels of pro-B and early pre-B cells (A-MuLV target cells) in Pim-1−/−/Pim-2−/−/Pim-3−/− mice are increased about 3-fold compared with wild-type mice,13 whereas no significant change was observed in the number of these cells in Pim-1−/−/Pim-2−/− animals (data not shown). Another plausible explanation may be the genetic background difference of the Pim-1−/−/Pim-2−/−and the Pim-1−/−/Pim-2−/−/Pim-3−/− mice.
To directly demonstrate the involvement of Pim kinases in pre–B-cell tumorigenesis, bone marrow cells from Pim-1−/−/Pim-2−/−, Pim-1−/−/Pim-2−/−/Pim-3−/−, or wild-type mice were infected with bicistronic retroviruses encoding the p120 form of v-Abl and either Pim-1, Pim-2, Pim-3, or GFP. As expected, GFP and Pim-3 failed to complement Pim kinase deficiency for v-Abl–mediated transformation of Pim-1−/−/Pim-2−/−cells (Figure 2D). In contrast, transformants were found in cells infected with bicistronic retroviruses encoding v-Abl and either Pim-1 or Pim-2. Subsequent Western blot analysis confirmed the ectopic expression of Pim-1 or Pim-2 in these transformants (Figure 2E,F). These data demonstrated that Pim-1 or Pim-2 is required for v-Abl–mediated pre–B-cell transformation. Based on these results, we anticipated that Pim-3 is not required for v-Abl transformation or it might have different effects on this process. Indeed, expression of Pim-3 slightly suppressed v-Abl transformation of Pim-1−/−/Pim-2−/−/Pim-3−/− cells as compared with empty vector, whereas, under same conditions, expression of either Pim-1 or Pim-2 greatly increased the v-Abl transformation efficiency (P < .01) (Figure 2D).
Pim-1 and Pim-2 interact with SOCS-1 but overcome negative regulation of SOCS-1 in v-Abl–transformed pre-B cells
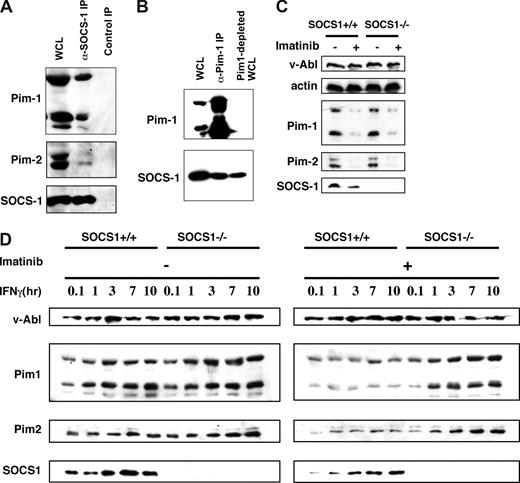
A previous study from our laboratory has shown that in 293T cells, Pim kinases interact with, and phosphorylate, SOCS-1. These phosphorylation events alter the properties of SOCS-1.17 Because the expression of Pim kinases is induced downstream of v-Abl and SOCS-1 protein is present in v-Abl–transformed pre-B cells,23 it is possible that Pim kinases interact with SOCS-1 in v-Abl transformants. To test this possibility, we performed immunoprecipitation experiments with lysates prepared from pre-B cells transformed by bicistronic retroviruses carrying v-Abl and epitope-tagged SOCS-1. Co-precipitation was observed when either anti-HA was used to precipitate SOCS-1 (Figure 3A) or anti-Pim-1 was used (Figure 3B). Furthermore, the co-immunoprecipitation experiments were quantified, and binding efficiencies were given as a percentage of the total imported protein. It was found that 10.5% of Pim-1 and 6.7% of Pim-2 were co-precipitated with SOCS-1. Co-localization of Pim-1 and SOCS-1 in the cytoplasm was further confirmed by immunofluorescence (Figure S2A). These results indicate that there are pools of Pim-1 and Pim-2 that are associated with SOCS-1 in v-Abl transformants.
There are pools of Pim-1 and Pim-2 that are associated with SOCS-1 but overcome inhibitory effects of SOCS-1 in v-Abl–transformed pre–B-cells. (A) Shown is an immunoblot of whole cell lysate (WCL) and proteins precipitated with either anti-HA that recognizes HA-tagged SOCS-1 or normal rabbit serum control. The blot was probed with indicated antibodies. (B) Immunoprecipitation using an anti–Pim-1 antibody was performed as in panel A. A blot of WCL, precipitated proteins, and Pim-1–depleted WCL was probed as indicated. (C) Shown is a Western blot of A-MuLV–transformed wild-type and SOCS-1−/− cells treated with or without imatinib and probed with indicated antibodies. (D) Effects of the expression of SOCS-1 protein on the levels of Pim-1 and Pim-2 were examined by using v-Abl–transformed SOCS-1–deficient or wild-type cell lines. The cells were treated with or without imatinib before addition of IFNγ in a time course. Western blotting shows that SOCS-1 does not inhibit v-Abl–induced expression of Pim-1 and Pim-2 but does have inhibitory effects on IFNγ-dependent expression of these kinases.
There are pools of Pim-1 and Pim-2 that are associated with SOCS-1 but overcome inhibitory effects of SOCS-1 in v-Abl–transformed pre–B-cells. (A) Shown is an immunoblot of whole cell lysate (WCL) and proteins precipitated with either anti-HA that recognizes HA-tagged SOCS-1 or normal rabbit serum control. The blot was probed with indicated antibodies. (B) Immunoprecipitation using an anti–Pim-1 antibody was performed as in panel A. A blot of WCL, precipitated proteins, and Pim-1–depleted WCL was probed as indicated. (C) Shown is a Western blot of A-MuLV–transformed wild-type and SOCS-1−/− cells treated with or without imatinib and probed with indicated antibodies. (D) Effects of the expression of SOCS-1 protein on the levels of Pim-1 and Pim-2 were examined by using v-Abl–transformed SOCS-1–deficient or wild-type cell lines. The cells were treated with or without imatinib before addition of IFNγ in a time course. Western blotting shows that SOCS-1 does not inhibit v-Abl–induced expression of Pim-1 and Pim-2 but does have inhibitory effects on IFNγ-dependent expression of these kinases.
It has been demonstrated that by binding its C-terminal SOCS box to the Elongin BC complex, SOCS-1 becomes part of an E3 enzyme and serves to target bound protein substrates such as Jak kinases to proteasomal degradation.24 Because our results suggest that Pim kinases associate with SOCS-1, we explored whether SOCS-1 has an inhibitory effect on the levels of Pim-1 and Pim-2. To this end, we used the A-MuLV–transformed pre–B-cell lines derived from SOCS-1–deficient mice and wild-type littermates. No significant differences in the protein levels of Pim-1 and Pim-2 were observed between A-MuLV–transformed wild-type and SOCS-1−/− cells treated with or without imatinib (Figure 3C). These data suggest that expression of SOCS-1 has no effect on the levels of Pim-1 and Pim-2 in v-Abl transformants. When the cells were treated with IFNγ alone, the levels of Pim-1 and Pim-2 in wild-type cells were still comparable to those found in SOCS-1−/− cells (Figure 3D). These results are consistent with our previous observations indicating that SOCS-1 function is impaired by v-Abl so that it does not inhibit JAK/STAT signaling that mediates Pim expression.25 In order to test the effect of SOCS-1 on cytokine-induced Pim expression, we inactivated Abl kinase with imatinib. As shown in Figure 3D, addition of IFNγ together with imatinib significantly decreased the amount of Pim-1 and Pim-2 in wild-type cells with respect to SOCS-1−/− cells. These experiments provide strong evidence that SOCS-1 does not inhibit v-Abl–induced expression of Pim-1 and Pim-2 but does have inhibitory effect on cytokine-dependent expression of these kinases. Thus, Pim-1 and Pim-2 are expressed at elevated levels in v-Abl–transformed cells, which may be essential for Pim functioning during Abl transformation.
Pim kinases are involved in modulating SOCS-1 protein levels and SOCS-1 phosphorylation
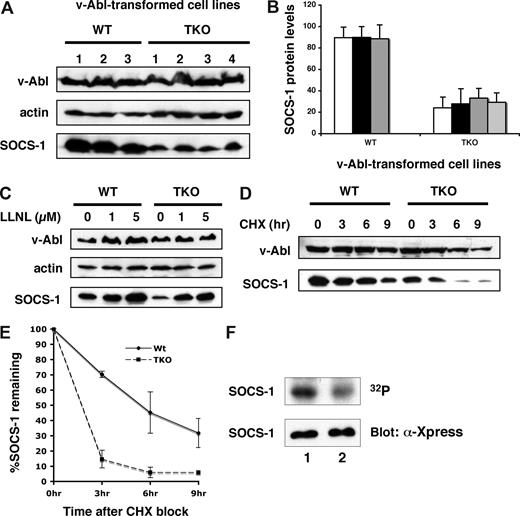
Our data reinforce previous studies that suggest a functional interaction between Pim kinases and SOCS-1.17 To further address the role Pim kinases play in v-Abl transformation, we determined whether Pim kinases had any effect on SOCS-1 protein levels and its modification. To this end, v-Abl–transformed pre–B-cell lines derived from Pim-1−/−/Pim-2−/−/Pim-3−/− or wild-type mice were generated. All cell lines showed similar levels of v-Abl expression. However, examination of these cell lines revealed that the protein levels of SOCS-1 were reduced in Pim-1−/−/Pim-2−/−/Pim-3−/− cells as compared with wild-type cells (Figure 4A,B). The effect of Pim kinase deficiency on SOCS-1 protein levels can be reversed by adding LLNL, a proteasome inhibitor, to the cells (Figure 4C). Studies using confocal microscopy showed that distribution of SOCS-1 in the microtubule organizing complex (MTOC) was increased in Pim-1−/−/Pim-2−/−/Pim-3−/− cells (Figure S2B), suggesting an increased targeting of SOCS-1 to MTOC-associated proteasome. In contrast, SOCS-1 was distributed throughout the cytoplasm in wild-type cells (Figure S2A). These observations demonstrate that a reduction in the amount of SOCS-1 protein is due to increased proteasomal degradation in the absence of Pim kinases.
Pim kinases are involved in modulating SOCS-1 protein levels and SOCS-1 phosphorylation. (A) SOCS-1 protein levels are significantly reduced in Pim-deficient v-Abl–transformed pre-B cells. The expression of SOCS-1 protein was examined in 3 wild-type and 4 Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cell lines. Shown is a Western blot probed with indicated antibodies. (B) SOCS-1 levels in panel A were quantitated by densitometry and normalized to v-Abl expression levels. In each experiment, the highest level of SOCS-1 is 100. Plotted are the average levels of SOCS-1 from 3 independent experiments. Error bars represent SEM. (C) Various concentrations of the proteasome inhibitor LLNL were added to the cells, and cells were then harvested after 14 hours of incubation. The levels of indicated proteins were examined by Western blot. (D) v-Abl–transformed wild-type or TKO pre-B cells were exposed to cycloheximide (CHX). Cells were then harvested at the indicated time points and analyzed by Western blotting. (E) Blots in panel D were quantitated as described in panel B. Plotted are the results from 3 independent experiments. Error bars represent SD. (F) v-Abl–transformed wild-type or TKO cells were metabolically labeled with 32PO4. 32P-labeled SOCS-1 was then isolated by immunoaffinity, and phosphorylation was detected by autoradiography. SOCS-1 expression levels were examined by Western blot. Lane 1, wild-type cells; lane 2, TKO cells.
Pim kinases are involved in modulating SOCS-1 protein levels and SOCS-1 phosphorylation. (A) SOCS-1 protein levels are significantly reduced in Pim-deficient v-Abl–transformed pre-B cells. The expression of SOCS-1 protein was examined in 3 wild-type and 4 Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cell lines. Shown is a Western blot probed with indicated antibodies. (B) SOCS-1 levels in panel A were quantitated by densitometry and normalized to v-Abl expression levels. In each experiment, the highest level of SOCS-1 is 100. Plotted are the average levels of SOCS-1 from 3 independent experiments. Error bars represent SEM. (C) Various concentrations of the proteasome inhibitor LLNL were added to the cells, and cells were then harvested after 14 hours of incubation. The levels of indicated proteins were examined by Western blot. (D) v-Abl–transformed wild-type or TKO pre-B cells were exposed to cycloheximide (CHX). Cells were then harvested at the indicated time points and analyzed by Western blotting. (E) Blots in panel D were quantitated as described in panel B. Plotted are the results from 3 independent experiments. Error bars represent SD. (F) v-Abl–transformed wild-type or TKO cells were metabolically labeled with 32PO4. 32P-labeled SOCS-1 was then isolated by immunoaffinity, and phosphorylation was detected by autoradiography. SOCS-1 expression levels were examined by Western blot. Lane 1, wild-type cells; lane 2, TKO cells.
Next, we estimated the relative half-life of SOCS-1 protein expressed in v-Abl–transformed Pim-1−/−/Pim-2−/−/Pim-3−/− or wild-type pre-B cells. The transformants were treated with cycloheximide and lysed after an incubation for the indicated times, and the rate of SOCS-1 remaining in the cells were given as percentages of SOCS-1 protein levels at the zero time point. As shown in Figure 4D,E, the half-life of SOCS-1 is approximately 6 hours in wild-type cells, whereas when expressed in Pim-1−/−/Pim-2−/−/Pim-3−/− cells, SOCS-1 protein appeared to be more labile as indicated by a clearly shorter half-life (less than 3 hours). The results indicate that Pim kinases are involved in regulation of SOCS-1 protein stability in v-Abl transformants.
Although SOCS-1 protein is more stable in the context of v-Abl–transformed wild-type cells, its function is disrupted by phosphorylation.23 Because Pim-dependent phosphorylation of SOCS-1 has been found in 293T cells,17 we investigated whether Pim kinases mediate this event in v-Abl transformants. Pim-1−/−/Pim-2−/−/Pim-3−/− or wild-type cells transformed with bicistronic retroviruses containing v-Abl and SOCS-1 were metabolically labeled with 32PO4. Radiolabeled SOCS-1 was then isolated by immunoprecipitation, and phosphorylation was detected by autoradiography. As shown in Figure 4F, phosphorylation levels of SOCS-1 were significantly reduced in Pim-1−/−/Pim-2−/−/Pim-3−/− cells with respect to those observed in the wild-type cells. By quantitating the autoradiography, Pim-1−/−/Pim-2−/−/Pim-3−/− cells showed 59.7% reduction in SOCS-1 phosphorylation as compared with wild-type cells. This finding suggests that Pim kinases participate in SOCS-1 phosphorylation in v-Abl transformants. In addition, the results imply that there exists other kinase(s) that are involved in SOCS-1 phosphorylation.
Because previous studies and our current data suggest that Pim kinases play a role in SOCS-1 phosphorylation that inhibits its normal function, we reasoned that the failure of transformation of Pim-1−/−/Pim-2−/− cells by v-Abl was likely due to impaired inhibition of SOCS-1 function. Thus, we anticipated that the combined triple knockout of Pim-1, Pim-2, and SOCS-1 might have positive effects on v-Abl transformation. To address this possibility, we performed v-Abl–mediated cellular transformation of fetal livers derived from Pim-1−/−/Pim-2−/−/SOCS-1−/−, Pim-1−/−/Pim-2−/−/SOCS-1−/+, and Pim-1−/−/Pim-2−/−/SOCS-1+/+ embryos because SOCS-1−/− mice die within 3 weeks of birth. Indeed, we observed that the combined loss of Pim-1, Pim-2, and SOCS-1 results in partial restoration of v-Abl transformation efficiency. Of 48 day-14 embryos derived from matings of Pim-1−/−/Pim-2−/−/SOCS-1−/+, 11 triple knockouts were obtained. Fetal livers from 4 of these triple knockouts were found to be transformed by v-Abl, whereas fetal livers from all of 28 embryos of Pim-1−/−/Pim-2−/−/SOCS-1−/+ and all of 9 embryos of Pim-1−/−/Pim-2−/−/SOCS-1+/+ did not show any viable v-Abl transformants (Table 1). Furthermore, we found that v-Abl transformants derived from these triple knockouts only survived for 5 weeks under cytokine-independent conditions. These data suggest that in addition to their role in regulating SOCS-1 function, Pim kinases play other potential roles in v-Abl transformation, likely related to apoptosis.
v-Abl–mediated fetal liver transformation
| Genotype . | Number of embryos . | Embryos displaying transformation . |
|---|---|---|
| Pim-1-/-/Pim-2-/-/SOCS-1-/- | 11 | 4 |
| Pim-1-/-/Pim-2-/-/SOCS-1-/+ | 28 | 0 |
| Pim-1-/-/Pim-2-/-/SOCS-1+/+ | 9 | 0 |
| Total | 48 | 4 |
| Genotype . | Number of embryos . | Embryos displaying transformation . |
|---|---|---|
| Pim-1-/-/Pim-2-/-/SOCS-1-/- | 11 | 4 |
| Pim-1-/-/Pim-2-/-/SOCS-1-/+ | 28 | 0 |
| Pim-1-/-/Pim-2-/-/SOCS-1+/+ | 9 | 0 |
| Total | 48 | 4 |
Fetal liver–derived pre-B cells were infected with A-MuLV. v-Abl–mediated transformation was examined 2 weeks after infection. Embryos showing at least 2 wells exhibiting cytokine-independent growth in a 96-well plate were counted as positive.
P < .02 by Fisher's exact test
Pim kinase deficiency sensitizes v-Abl–transformed cells to undergo apoptosis
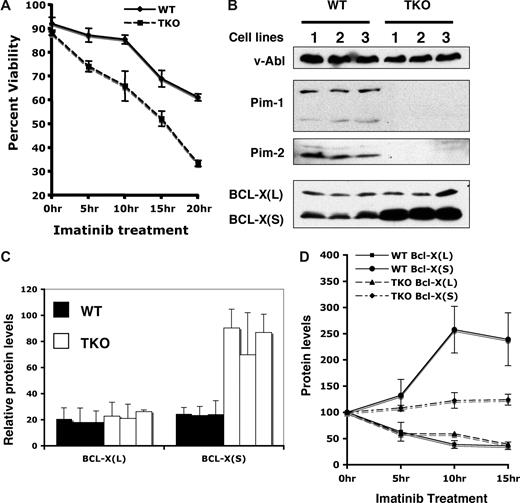
During culture of v-Abl–transformed pre-B cells in the presence of imatinib, we also observed that cells lacking all Pim kinases were highly sensitive to this inhibitor as compared with wild-type cells. Because a role for Pim kinases in antiapoptosis has been established, we investigated whether Pim deficiency sensitizes v-Abl–transformed cells to undergo apoptosis in response to imatinib treatment. As determined by the propidium iodide/Annexin V binding assay, v-Abl–transformed pre-B cells initiated apoptosis following imatinib treatment, and approximately 60% of wild-type cells remained viable after 20 hours of addition of this inhibitor into our system (Figure 5A). By contrast, only approximately 30% of Pim-1−/−/Pim-2−/−/Pim-3−/− cells were viable under the same culture conditions (Figure 5A). Similar results were obtained using staurosporine to induce apoptosis (data not shown). These experiments demonstrate that Pim kinases play a role in Abl-mediated cell survival.
Pim deficiency sensitizes v-Abl–transformed cells to undergo apoptosis. (A) v-Abl–transformed wild-type (WT) or Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells were treated with imatinib at indicated times. Apoptosis and cell survival were analyzed by the propidium iodide/Annexin V binding and measured by flow cytometry. Plotted are the results from 3 independent experiments. (B) v-Abl–transformed wild-type or TKO cell lines were analyzed by Western blotting with indicated antibodies. (C) Blots in panel B were quantitated as in Figure 4. Plotted are the average levels of Bcl-XL and Bcl-XS from 3 independent experiments. (D) v-Abl–transformed wild-type or TKO cells were treated with imatinib in a time course. Lysates from these cells were analyzed by immunoblotting. Blots were then quantitated as described above. The average levels of Bcl-XL and Bcl-XS from 3 independent experiments are plotted such that the protein level at 0 hour time point is 100%. Error bars represent SD.
Pim deficiency sensitizes v-Abl–transformed cells to undergo apoptosis. (A) v-Abl–transformed wild-type (WT) or Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells were treated with imatinib at indicated times. Apoptosis and cell survival were analyzed by the propidium iodide/Annexin V binding and measured by flow cytometry. Plotted are the results from 3 independent experiments. (B) v-Abl–transformed wild-type or TKO cell lines were analyzed by Western blotting with indicated antibodies. (C) Blots in panel B were quantitated as in Figure 4. Plotted are the average levels of Bcl-XL and Bcl-XS from 3 independent experiments. (D) v-Abl–transformed wild-type or TKO cells were treated with imatinib in a time course. Lysates from these cells were analyzed by immunoblotting. Blots were then quantitated as described above. The average levels of Bcl-XL and Bcl-XS from 3 independent experiments are plotted such that the protein level at 0 hour time point is 100%. Error bars represent SD.
A previous study from our laboratory suggests that inefficient apoptotic signaling in v-Abl transformants may be attributed to increased expression of antiapoptotic proteins such as Bcl-2 and Bcl-XL.26 The Bcl-2 family of proteins includes both anti- and proapoptotic members that are the critical regulators of the apoptosis. In an attempt to gain a better understanding of the molecular basis of the altered apoptosis profile, the expression of several members of the BCL-2 family was compared between wild-type and Pim-1−/−/Pim-2−/−/Pim-3−/− cells. Western blot analyses, using polyclonal anti–Bcl-X antiserum that recognizes both Bcl-XL, the antiapoptotic Bcl-X splice variant, and Bcl-XS, the proapoptotic Bcl-X splice variant, showed that v-Abl transformants expressed both Bcl-XL and Bcl-XS proteins (Figure 5B). Interestingly, averaged data from 3 independent experiments showed that the level of Bcl-XS increased by approximately 3-fold in Pim-1−/−/Pim-2−/−/Pim-3−/− cells as compared with wild-type cells (Figure 5B,C). In contrast, no significant changes in protein expression in Pim-1−/−/Pim-2−/−/Pim-3−/− cells were observed for Bcl-XL, Bcl-2, and Bax when compared with their levels in wild-type cells (Figure 5B,C and data not shown).
Because the results presented above indicate that Pim-1 and Pim-2 are expressed in v-Abl transformants in a v-Abl kinase-dependent manner, we determined whether treatment with imatinib had any effect on the expression of Bcl-XS protein. Analysis of protein expression was performed after treatment with 1 μM of imatinib for 0, 5, 10, and 15 hours, respectively. As expected, time-course studies showed that the levels of Pim-1 and Pim-2 in wild-type cells dramatically decreased after treatment with imatinib for 5 hours or longer (data not shown). After inhibition of the Abl kinase, the expression of Bcl-XL also was markedly down-regulated (Figure 5D), consistent with the earlier observations.26,27 Importantly, the level of Bcl-XS increased by approximately 2.5-fold in wild-type cells cultured in the presence of imatinib (Figure 5D), which is in striking contrast to the reduced Bcl-XL level. However, the BCL-XS level was only slightly increased in Pim-1−/−/Pim-2−/−/Pim-3−/− cells cultured under the same conditions (Figure 5D). Together, these results suggest a negative role for Pim kinases induced by v-Abl in the production of BCL-XS in v-Abl transformants.
Ectopic overexpression of Pim-1 increases the resistance of v-Abl–transformed cells to apoptosis induced by imatinib
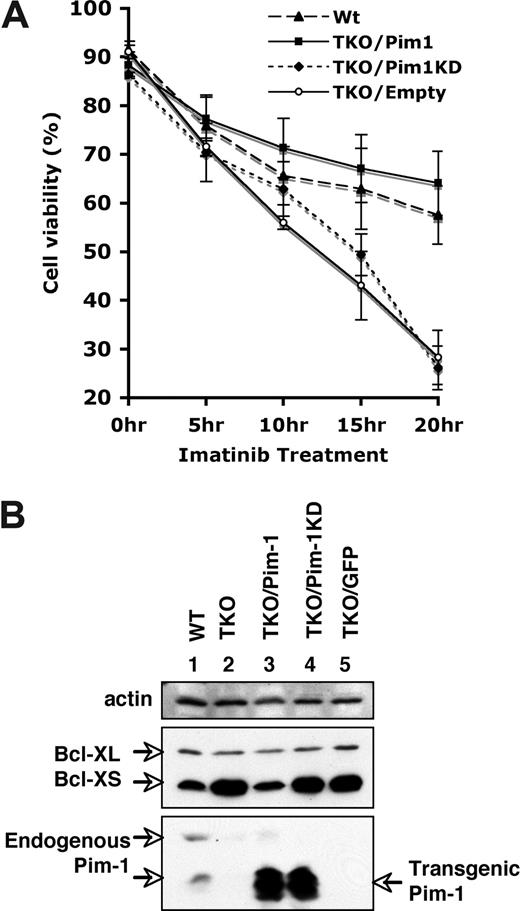
To further confirm that the alteration of cell viability and BCL-XS expression was the result of the lack of Pim kinases, we investigated whether ectopic expression of Pim-1 could complement Pim deficiency. To this end, we generated v-Abl–transformed Pim-1−/−/Pim-2−/−/Pim-3−/− cell lines stably expressing either wild-type Pim-1, kinase-dead Pim-1 (Pim-1KD) that contains a K67M point mutation in the adenosine triphosphate (ATP)–binding pocket, or GFP (empty vector) using bicistronic retroviruses. Western blotting demonstrated that the cell lines infected with the retroviruses encoding Pim-1 or Pim-1KD expressed comparable levels of these proteins (Figure 6B). Pim-1 transgenes yield 2 isoforms of approximately 32 and approximately 35 kDa that differ at their N termini but contain identical catalytic sites. Cell survival was then examined after treating these cell lines with imatinib. We observed that approximately 60% of Pim-1−/−/Pim-2−/−/Pim-3−/− cells overexpressing wild-type Pim-1 remained viable after 20 hours of imatinib treatment, which is similar to the viability of wild-type cells under the same conditions (Figure 6A). The survival-promoting effect of Pim-1 was dependent on catalytic activity, as the cells expressing kinase-dead Pim-1KD recapitulated that of the empty vector control (Figure 6A). These results indicate that when constitutively expressed in v-Abl–transformed cells, Pim-1 conferred apoptotic resistance to imatinib stimulus.
Ectopic expression of Pim-1 enhances the resistance of v-Abl–transformed cells to apoptosis induced by imatinib. (A) Apoptosis and cell survival were examined as described in Figure 5. Shown are wild-type (WT) cells and Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells ectopically expressing either wild-type Pim-1 (TKO/Pim1), kinase-death Pim1 (TKO/Pim1KD), or empty vector (TKO/Empty). Plotted are the results from 3 independent experiments. Error bars represent SD. (B) An immunoblot of v-Abl–transformed WT cells, TKO cells, and TKO cells expressing different proteins was probed with indicated antibodies.
Ectopic expression of Pim-1 enhances the resistance of v-Abl–transformed cells to apoptosis induced by imatinib. (A) Apoptosis and cell survival were examined as described in Figure 5. Shown are wild-type (WT) cells and Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells ectopically expressing either wild-type Pim-1 (TKO/Pim1), kinase-death Pim1 (TKO/Pim1KD), or empty vector (TKO/Empty). Plotted are the results from 3 independent experiments. Error bars represent SD. (B) An immunoblot of v-Abl–transformed WT cells, TKO cells, and TKO cells expressing different proteins was probed with indicated antibodies.
We next examined whether ectopic expression of Pim-1 was sufficient to regulate Bcl-XS protein expression. As shown in Figure 6B, in Pim-1−/−/Pim-2−/−/Pim-3−/− cells, constitutively expressing Pim-1 decreased the amount of Bcl-XS protein to the levels observed in wild-type cells. Expression of either Pim-1KD or empty vector had no effect on Bcl-XS levels, confirming a requirement for the kinase activity (Figure 6B). These observations, taken together, demonstrate that the alterations of cell viability and Bcl-XS expression correlated with the functioning of Pim kinases.
Pim kinases increase phosphorylation level of BAD in v-Abl–transformed cells
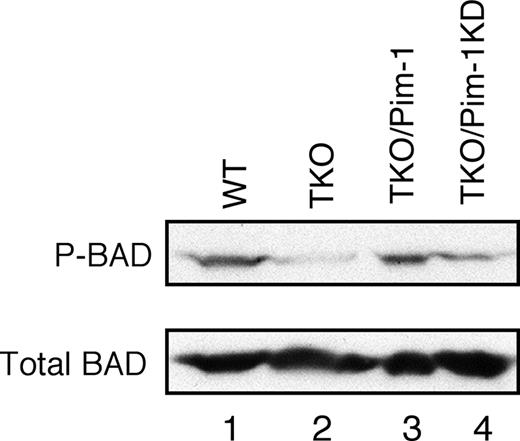
It has been documented that Pim kinases can phosphorylate Ser112 of BAD, the proapoptotic member of Bcl-2 family, and thereby block BAD-induced apoptosis.15,28,29 Hence, we explored whether Pim kinase deficiency could decrease BAD phosphorylation. Indeed, the amount of phospho-Ser112 BAD was markedly reduced in Pim-1−/−/Pim-2−/−/Pim-3−/− cells, whereas the total amount of BAD protein was not significantly affected (Figure 7 lanes 1 and 2). We then determined whether ectopic expression of Pim-1 could mediate BAD phosphorylation. As expected, expressing Pim-1 in Pim-1−/−/Pim-2−/−/Pim-3−/− cells increased BAD phosphorylation to a level comparable to that of v-Abl–transformed wild-type cells (Figure 7 lane 3), whereas the level of Bad phosphorylation was not significantly affected by overexpression of Pim-1KD (Figure 7 lane 4). These results indicate that Pim-1 is required for BAD phosphorylation in v-Abl transformants. Collectively, our findings suggest that Pim kinases promote Abl-mediated cell survival via inhibiting Bcl-XS– and BAD-dependent apoptosis.
Pim mediates phosphorylation of Ser112 of BAD in v-Abl–transformed cells. Shown is a blot probed as indicated. Lane1, wild-type cells; 2, Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells; 3, TKO cells expressing Pim-1; 4, TKO cells expressing Pim-1 kinase-dead (Pim-1KD).
Pim mediates phosphorylation of Ser112 of BAD in v-Abl–transformed cells. Shown is a blot probed as indicated. Lane1, wild-type cells; 2, Pim-1−/−/Pim-2−/−/Pim-3−/− (TKO) cells; 3, TKO cells expressing Pim-1; 4, TKO cells expressing Pim-1 kinase-dead (Pim-1KD).
Discussion
Cellular transformation by Abl oncogenes is a complicated process that requires the activation of several proliferative signaling pathways and the disruption of apoptosis. The molecular basis of how Abl can overcome the negative regulatory mechanisms to constitutively activate these signals, and how Abl-transformed cells can circumvent apoptosis is poorly understood. Our experiments demonstrate that Pim kinases are involved in these processes.
Our studies show that Pim-1 and Pim-2 are expressed at elevated levels in v-Abl–transformed cells in an Abl kinase-dependent manner, and Pim kinase deficiency has profound effects on the v-Abl transformation efficiency. Ectopic expression of either Pim-1 or Pim-2 in the Pim-1−/−/Pim-2−/− cells restores transformation by v-Abl. These data suggest that Pim-1 and Pim-2 play a key role in v-Abl–induced tumorigenesis. Although ectopic expression of Pim-1 or Pim-2 is absolutely required for v-Abl–mediated transformation of Pim-1−/−/Pim-2−/− cells, bone marrow cells deficient for all Pim kinases are not completely defective in v-Abl transformation. One possible explanation for this difference is that Pim-3 might have an opposing (inhibitory) effect on v-Abl–mediated transformation. However, we found that expression of Pim-3 only slightly suppressed v-Abl transformation of Pim-1−/−/Pim-2−/−/Pim-3−/− cells as compared with empty vector. Another possibility may be a genetic background difference between the Pim-1−/−/Pim-2−/− (mixed) and Pim-1−/−/Pim-2−/−/Pim-3−/− (FVB/NJ) mice. The number of targets for the Abelson virus differs in the bone marrow of these mouse strains. Alternatively, the levels of other kinases that compensate for the loss of Pim-1 and Pim-2 may differ in cells from different strains of mice. Interestingly, our experiments revealed that ectopic expression of either Pim-1 or Pim-2 in Pim-1−/−/Pim-2−/−/Pim-3−/− cells markedly increased the v-Abl transformation efficiency as compared with empty vector (Figure 2D). These results suggest that Pim-2−/−/Pim-3−/− and Pim-1−/−/Pim-3−/− cells could be transformed by v-Abl, and expression of either Pim-1 or Pim-2 is sufficient for Abl-mediated transformation.
Our experiments also have begun to address the mechanism by which Pim may affect Abl transformation. We observed that Pim kinases are involved in regulating protein stability and phosphorylation of SOCS-1 in v-Abl transformants. A recent study from others also demonstrates that tyrosine phosphorylation of SOCS-3 inhibits turnover of SOCS-3 protein and renders it unable to normally regulate JAK2 kinase.30 These findings suggest that alteration of SOCS function by phosphorylation may allow cells to overcome SOCS inhibition. Interestingly, the combined triple knockout of Pim-1, Pim-2, and SOCS-1 results in partial restoration of v-Abl transformation efficiency. These data support our previous model that v-Abl may induce downstream kinases that can modulate and modify SOCS-1 protein and eliminate the inhibitory effects of SOCS-1.23 Pim-1−/−/Pim-2−/−/Pim-3−/− cells showed about 60% reduction in SOCS-1 phosphorylation as compared with wild-type cells. This finding suggests that Pim kinases participate in regulating SOCS-1 phosphorylation. It is also possible that both Pim and other unknown kinase(s) induced by v-Abl can phosphorylate SOCS-1 in v-Abl–transformed cells.
Another serine/threonine kinase, Akt, shows similar substrate specificity as Pim kinases. Previous studies from our laboratory and others have uncovered that v-Abl and BCR/Abl can induce Akt activation.31,32 These observations prompted us to evaluate the effects of Pim deficiency on Akt activation. We found that Akt is phosphorylated in both wild-type and Pim-1−/−/Pim-2−/−/Pim-3−/− cells in an Abl kinase-dependent manner (data not shown). No significant differences in Akt phosphorylation levels were observed in these cells. This is consistent with a previous report indicating that Pim-2 does not affect Akt activation.33 Abl kinase may induce both Pim and Akt expression and activation, which results in activation of 2 distinct pathways that contribute to Abl transformation. However, whether activated Akt can compensate for Pim deficiency to phosphorylate SOCS-1 in v-Abl transformants remains to be determined.
It has been well documented that resistance to apoptosis is fundamentally important for the development and maintenance of malignancies. Therefore, to transform cells, Abl kinases must activate antiapoptotic pathways in the face of death-inducing stimuli. Recently, increasing evidence indicates that Pim kinases are involved in antiapoptosis through regulating the expression or activity of the Bcl-2 family of proteins.15,28,29,34 The Bcl-2 protein family is the critical regulator of caspase activation and apoptosis. In this study, we have found that the expression of Bcl-XL is Abl kinase dependent in v-Abl–transformed cells. By contrast, the expression of Bcl-XS is induced by inhibiting the Abl kinase. Interestingly, the level of Bcl-XS is markedly increased in Pim-1−/−/Pim-2−/−/Pim-3−/− cells, suggesting a negative role for the Pim kinases in the production of Bcl-XS in v-Abl transformants. Bcl-X, which is an important member of the Bcl-2 family of apoptotic genes, has several alternatively spliced forms that encode functionally distinct protein products such as Bcl-XL and Bcl-XS. Bcl-XS was identified by the Thompson group as a proapoptotic isoform of Bcl-X.35 Several studies have demonstrated that forced expression of Bcl-XS protein results in an increase in apoptosis after treatment of cells with proapoptotic stimuli.36,37 However, little is known about the factors that regulate the expression of Bcl-XS in normal and cancer cells. Our data indicate that Pim kinases function as negative regulators of the expression of Bcl-XS in v-Abl transformants. The mechanism by which Pim kinases control the production of Bcl-XS awaits further investigation.
Our experiments demonstrate for the first time that Pim kinases mediate the phosphorylation of BAD in v-Abl–transformed cells. BAD is a BH3-domain–only molecule that is associated with pro-apoptosis through its differential phosphorylation in response to extracellular survival factors. Active BAD induces apoptosis by binding to antiapoptotic Bcl-2 family members such as Bcl-2 and Bcl-XL. This allows other proapoptotic proteins such as BAX and BAK to aggregate, inducing cytochrome C release from the mitochondria and caspase activation. When phosphorylated, BAD becomes inactive, resides in the cytosol, and can be bound to 14–3-3 protein.38,39 It has been proposed that Pim-1, Pim-2, and Pim-3 can phosphorylate BAD on Ser112, which accounts in part for their ability to reverse BAD-induced cell death.15,28,29 More recently, it has been discovered that proapoptotic proteins BIM and BAD mediate imatinib-induced killing of BCR/Abl + leukemic cells.40 This suggests that BAD is an important apoptotic mediator in BCR/Abl transformants.
Based on these data, our model suggests that Pim kinases induced by v-Abl are involved in multiple pathways that control critical processes in v-Abl transformation. First, Pim kinases are involved in SOCS-1 modification and in regulation of SOCS-1 protein levels. Second, Pim kinases regulate cell survival via down-regulating Bcl-XS expression and eliminating BAD function, and thereby prevent Bcl-XS– and BAD-dependent apoptosis in v-Abl–transformed cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr M. Stamnes for comments on the manuscript, Dr D. Hilton for the monoclonal SOCS-1 antibody, and members of Rothman laboratory for helpful discussions.
Authorship
Contribution: J.-L.C. designed and performed research and wrote the paper; A.L. performed research and contributed to writing the paper; and P.B.R. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul B. Rothman, Department of Internal Medicine, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242; e-mail: paul-rothman@uiowa.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal