Abstract
Mantle cell lymphoma (MCL) is a non-Hodgkin lymphoma with poor prognosis. Its hallmark is the translocation t(11:14)q (13;32), leading to overexpression of cyclin D1, a positive regulator of the cell cycle. As cyclin D1 up-regulation is not sufficient for inducing malignant transformation, we combined DNA microarray and RNA interference (RNAi) approaches to identify novel deregulated genes involved in the progression of MCL. DNA microarray analysis identified 46 genes specifically up-regulated in MCL compared with normal B cells; 20 of these were chosen for further studies based on their cellular functions, such as growth and proliferation. The Granta 519 cell line was selected as an MCL in vitro model, to set up the RNAi protocol. To confirm the functionality of overexpression of the 20 disease-associated genes, they were knocked down using small interfering RNAs (siRNAs). In particular, knockdown of 3 genes, encoding the hepatoma-derived growth factor related protein 3 (HDGFRP3), the frizzled homolog 2 (FZD2), and the dual specificity phosphatase 5 (DUSP5), induced proliferative arrest in Granta 519 MCL cells. These genes emerged as functionally associated in MCL, in relation to growth and survival, and interfering with their function would increase insight into lymphoma growth regulation, potentially leading to novel clinical intervention modalities.
Introduction
Mantle cell lymphoma (MCL) is one of the most aggressive forms of non-Hodgkin lymphomas (NHLs) and is incurable, with a 5-year overall survival of less than 30%.1 Its cytogenetic profile is typically defined by the t(11:14)(q13;q32) translocation, hallmark of this disease, which places the gene encoding cyclin D1 (CCND1) under control of the immunoglobulin heavy chain enhancer.2 CCND1 interacts with cyclin-dependent kinase (CDK) 4 and CDK6, and its overexpression is involved in an increased cell cycling.3 However, additional factors are necessary for MCL tumor development, demonstrated by the fact that CCND1 transgenic mice only show small changes in cell-cycle behavior and do not spontaneously form lymphoid tumors.4 Tumor suppressor genes may be involved in the pathogenesis of MCL, since patients show recurrent deletions of certain chromosomal regions.5 Indeed, one such gene was recently identified as the ataxia telangiectasia mutated (ATM), which when inhibited, might play a role in the initiation and progression of MCL.6 In addition, by global transcription profiling, we have previously reported several up-regulated genes in MCL, such as IL10R, IL18, Bcl-2, c-mer proto-oncogene tyrosine kinase (MERTK), 4-1BB ligand, and CCL-5.7,8 As DNA microarray analysis usually results in the definition of a large number of dysfunctional genes, it is necessary to corroborate these results. Consequently, overexpression of several of identified genes has been verified on the protein level using flow cytometry and immunohistochemistry, and the results demonstrate their implication in MCL pathogenesis.8 However, until recently, techniques that allow high-throughput functional screening of deregulated genes has been lacking, but recent developments in RNA interference (RNAi) offer such possibilities.
RNAi causes sequence-specific degradation of endogenous mRNA, originally found in Caenorhabditis elegans as an antiviral mechanism.9 Knockdown of a specific gene transcript is mediated by binding of a short double-stranded RNA of 19 to 21 nt in length, called small interfering RNA (siRNA) and by engagement of the RNA-induced silencing complex (RISC).10 The role of RNAi as a tool in functional genomics became evident when Elbashir and coworkers demonstrated that mRNA in cultured mammalian cells could be targeted by this approach.11 As a result, large loss-of-function screenings have been carried out to find new components in signaling pathways and tumor suppressor genes12,13 . However, such screenings require the construction of a vector-based RNAi library comprising thousands of gene targets, resulting in a somewhat cumbersome and time-consuming approach.
In this study, we have instead used transcriptional profiling to zoom in on a limited number of target genes potentially of interest for the manipulation of tumor cell growth and proliferation. Consequently, loss-of-function RNAi screenings were carried out for 20 overexpressed genes, leading to the identification of 3 genes that, when silenced, caused proliferative arrest. These genes have previously not been shown to be involved in the growth regulation of MCL tumor cells and might open up new possibilities for immune intervention.
Methods
Cell culture
The cell lines used in this study were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany) and were cultured in RPMI 1640 medium, supplemented with 10% fetal calf serum (FCS) and 2 mM L-glutamine (all from Invitrogen, Carlsbad, CA) unless otherwise stated. JVM-2 (ACC 12), Jeko-1 (ACC 553; grown in 20% FCS), Rec-1 (ACC 584), Granta 51914,15 (ACC 342), SP-53,16 NCEB-1,17 and Z-138 (kindly provided by Prof Martin J. S. Dyer, Leicester University, United Kingdom) were all free of mycoplasma and cultured in a humidified atmosphere at 37°C using a 5% CO2 atmosphere. The cells were maintained between 2 × 105 and 106 cells/mL. For gene chip analysis, the cells were grown for 72 hours at exponential growth phase, followed by washing in phosphate-buffered saline (PBS) and resuspension in TRIzol (Invitrogen).
Purification and preparation of MCL tumors
The characteristics and purification of the MCL samples have previously been described.18 Briefly, fresh tumors were sliced into small pieces and suspended in RPMI-1640, supplemented with 10% FCS. Tissue debris was removed from the cell suspension by filtration through a cell strainer (BD Labware, Franklin Lakes, NJ). Lymphocytes were purified by gradient centrifugation, using Ficoll-Isopaque (Amersham Pharmacia Biotech, Uppsala, Sweden) followed by T-cell depletion using CD3+ Dynabeads (Dynal, Oslo, Norway). The CD5+/CD19+ MCL tumor cells, with more than 95% purity, were resuspended in TRIzol for mRNA extraction. Frozen tissue from 19 MCL tumors were homogenized (2 × 15 seconds) directly into TRIzol using an Ultra Turrax knife homogenizer (IKA-WERK; Tamro Med Lab, Mölndal, Sweden).
Isolation of normal B-cell populations
B-cell populations were purified from fresh pediatric tonsils (Malmö University Hospital, Malmö, Sweden), as previously described.7 Following Ficoll-Isopaque centrifugation, the different B-cell populations were sorted on a FACSVantageSE (BD Immunocytometry Systems, San Jose, CA) based on the following phenotypes: naive B cells (IgD+/CD23−), ligand-selected B cells (IgD+/CD23+), centroblasts (IgD−/CD38+/CD77+), centrocytes (IgD−/CD38+/CD77−), and memory B cells (post–germinal center [GC] B cells; CD38−/IgD−). The purified cell subpopulations were lyzed in TRIzol.
Sample preparation and microarray analysis
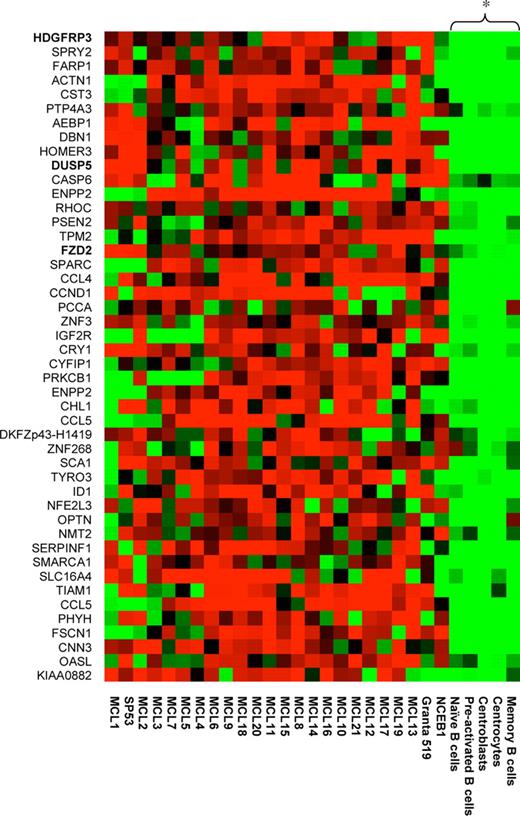
The preparation of mRNA, in vitro transcription, and hybridization of labeled cRNA to the U95Av2 arrays were performed according to standard protocols (Affymetrix, Santa Clara, CA) as previously described.7 The expression level for each probe set is given as a signal value (Micro Array Suite 5.0; Affymetrix). The signal values were normalized by scaling the values against a median target value of 500 on each array. This was performed to enable comparison between different arrays. A total of 19 MCL samples were compared with 11 primary B-cell samples, generating 209 comparison chip files. The genes in the 209 comparative chip files were then filtered for differential expression using Micro Array Suite 5.0. A total of 2 different algorithms were used to determine the qualitative change between the MCL and B-cell samples: detection algorithm and the change algorithm. Quantitative change is calculated using the signal log ratio algorithm. The statistical significance of the algorithms has been described previously.19 Thus, for each sample, a present or absent call is determined. In addition, for each comparison file (the comparison between 2 different samples), a change call and signal log ratio are determined. After generating the comparison files, all data were subsequently imported into Gene Spring 5.0 (Silicon Genetics, Redwood City, CA) for further data analysis and filtering. To allow for some variation in signal between the samples, a 50% or 30% cut-off was chosen. Thus, only genes that had (1) greater than 2-fold change and an increase call in at least 66 of 209 comparison files and a present call in at least 7 of 21 MCL samples or (2) greater than 1.5-fold change and an increase call in at least 105 of 209 comparison files and a present call in 10 of 21 MCL samples were chosen. To select for targets that were expressed in the tumor cells and also that were possible to study in our in vitro models, additional restrictions were added: the gene had to be present in (1) 1 of 2 purified MCL samples and (2) in at least 2 of 3 MCL cell lines. Using the described criteria, a list of 46 genes was generated that are specifically up-regulated in the MCL patient samples compared with the normal B-cell populations and that also were present in most of the available in vitro models (Figure 1). For hierarchic clustering, a distance measure was used, and bootstrapping (n = 100) was performed to validate the robustness of the generated tree.
Gene expression profile of MCL-associated genes. A total of 46 MCL-associated genes were identified by filtering the transcriptionally profiled genomes. The relative expression of selected genes is shown for the 21 MCL samples, 5 B-cell populations, and the 3 different MCL cell lines (NCEB1, Granta 519, and SP53). Hierarchic clustering using bootstrapping (n = 100) was performed, and the 5 B-cell populations were shown to be significantly (P < .05) separated from the MCL samples and the tumor cell lines.
Gene expression profile of MCL-associated genes. A total of 46 MCL-associated genes were identified by filtering the transcriptionally profiled genomes. The relative expression of selected genes is shown for the 21 MCL samples, 5 B-cell populations, and the 3 different MCL cell lines (NCEB1, Granta 519, and SP53). Hierarchic clustering using bootstrapping (n = 100) was performed, and the 5 B-cell populations were shown to be significantly (P < .05) separated from the MCL samples and the tumor cell lines.
siRNA design and transfection
Granta 519 cells were chosen as a MCL in vitro model system for RNAi. A total of 2 unique siRNAs were designed for targeting every gene in the study using an online tool by Ambion (Austin, TX; www.ambion.com). The siRNAs were produced and purified by Ambion. Freeze-dried siRNAs were suspended in nuclease-free water (Ambion) in a final concentration of 20 μM with the exception of Eg5 and the negative control, which were provided in a final concentration of 50 μM by the manufacturer (Ambion), and transfections were carried out on an Amaxa Nucleofection Device (Amaxa Biosystems, Cologne, Germany) using the O-17 program and Cell Line Nucleofector Solution T. For every transfection, 2 × 106 cells were electroporated with 100 pmol siRNA. The different siRNA sequences used are shown in Table 1.
siRNAs sequences used for functional screening
| Gene . | Sequence 1 (5′ → 3′) . | Sequence 2 (5′ → 3′) . | ||
|---|---|---|---|---|
| Sense | Antisense | Sense | Antisense | |
| HDGFRP3 | CCAGUGAAGGGACCUAACUtt | AGUUAGGUCCCUUCACUGGtt | GGCCAUGUGUAAAGUUUAAtt | UUAAACUUUACACAUGGCCtc |
| CST3 | CCUUGUCGAAAUCCACCUGtt | CAGGUGGAUUUCGACAAGGtc | GCCACACAUCUGAAAAGGAAAtt | UUUCCUUUUCAGAUGUGGCtg |
| PTP4A3 | CCUGUUCUCGGCACCUUAAtt | UUAAGGUGCCGAGAACAGGtc | CCCGAAGGCAAUAAAACAGtt | CUGUUUUAUUGCCUUCGGGTtg |
| HOMER3 | GGGAGAAAUCUCAGGAUGGtt | CCAUCCUGAGAUUUCUCCCtg | GGACAGCAACAACAAGCUGtt | CAGCUUGUUGUUGCUGUCCtg |
| DUSP5 | GGUAGUUGGUUGAAGUAGCtt | GCUACUUCAACCAACUACCtc | GCAAUAGACUUCAUUGACUtt | AGUCAAUGAAGUCUAUUGCtt |
| CASP6 | CCCAAGUGCAAUUGGAAAGtt | CUUUCCAAUUGCACUUGGGtc | GGAAUUAAAUCAUCCCUUAtt | UAAGGGAUGAUUUAAUUCCtg |
| RHOC | GGUGUUUGAGAUGGCCACUtt | AGUGGCCAUCUCAAACACCtc | GCCUGGAAAACAUUCCUGAtt | UCAGGAAUGUUUUCCAGGCtg |
| FZD2 | GCCCUCAUGAACAAGUUCGtt | CGAACUUGUUCAUGAGGGCtt | CGUACUUGGUAGACAUGCAtt | UGCAUGUCUACCAAGUACGtg |
| CCND1 | GGGUUAUCUUAGAUGUUUCtt | GAAACAUCUAAGAUAACCCtt | GGAGGGGUUGUGCUACAGAUtt | AUCUGUAGCACAACCCUCCtc |
| CRY1 | GGAUGAAACAGAUCUAUCAtt | UGAUAGAUCUGUUUCAUCCtt | GGUGGCGAUUUUUGCUUCAtt | UGAAGCAAAAUCGCCACCtg |
| CYFIP1 | GCGGCUUCGAGAUAAAAGUtt | ACUUUUAUGUCGAAGCCGCtc | CGGUUACGAUCAGAAUGCAtt | UGCAUUCUGAUCGUAACCGtt |
| ENPP2 | GGAUAUCCAUUUAUUGGUGtt | CACCAAUAAAUGGAUAUCCtc | GGGCGAGAGAAAUUUAAUCtt | GAUUAAAUUUCUCUCGCCCtc |
| CCL5 | GCUAGGAUGGAGAGUCCUUtt | AAGGACUCUCCAUCCUAGCtc | GGGCCCAGAUUCUACCACAtt | UGUGGUAGAAUCUGGGCCCtt |
| SCA1 | CCUUGUCAUUAGAGUUACAtt | UGUAACUCUAAUGACAAGGtt | GCUUGGUUUACAACAGGGAtt | UCCCUGUUGUAAACCAAGCtc |
| TYRO3 | GGUGUGCCAUUUUUCACAGtt | CUGUGAAAAAUGGCACACCTT | GGUCUGAAUCUUCACCAUCtt | GAUGGUGAAGAUUCAGACCtg |
| SLC16A4 | CCUAAACUAAUGUCAUUGGtt | CCAAUGACAUUAGUUUAGGtt | GCUAUCGCAUUGAAUUUGGtt | CCAAAUUCAAUGCGAUAGCtc |
| PHYH | GGAUAUUUGGAUGUUUCGAtt | UCGAAACAUCCAAAUAUCCTT | GGGAUUCCGGAAGGCAAUUtt | AAUUGCCUUCCGGAAUCCCTG |
| FSCN1 | CCCUCAGGUCAACAUCUACtt | GUAGAUGUUGACCUGAGGGtg | GCAGCCUGAAGAAGAAGCAtt | UGCUUCUUCUUCAGGCUGCtg |
| OASL | GGGUACAGAUGGGACAUCGtt | CGAUGUCCCAUCUGUACCCtt | CGAGACAUCCACUUGACAGtt | CUGUCAAGUGGAUGUCUCGtg |
| KIAA0882 | GCUGUACGACUGCAUGCAAtt | UUGCAUGCAGUCGUACAGCtg | CCUCAAUGUAGGUACAUUAtt | UAAUGUACCUACAUUGAGGtg |
| Gene . | Sequence 1 (5′ → 3′) . | Sequence 2 (5′ → 3′) . | ||
|---|---|---|---|---|
| Sense | Antisense | Sense | Antisense | |
| HDGFRP3 | CCAGUGAAGGGACCUAACUtt | AGUUAGGUCCCUUCACUGGtt | GGCCAUGUGUAAAGUUUAAtt | UUAAACUUUACACAUGGCCtc |
| CST3 | CCUUGUCGAAAUCCACCUGtt | CAGGUGGAUUUCGACAAGGtc | GCCACACAUCUGAAAAGGAAAtt | UUUCCUUUUCAGAUGUGGCtg |
| PTP4A3 | CCUGUUCUCGGCACCUUAAtt | UUAAGGUGCCGAGAACAGGtc | CCCGAAGGCAAUAAAACAGtt | CUGUUUUAUUGCCUUCGGGTtg |
| HOMER3 | GGGAGAAAUCUCAGGAUGGtt | CCAUCCUGAGAUUUCUCCCtg | GGACAGCAACAACAAGCUGtt | CAGCUUGUUGUUGCUGUCCtg |
| DUSP5 | GGUAGUUGGUUGAAGUAGCtt | GCUACUUCAACCAACUACCtc | GCAAUAGACUUCAUUGACUtt | AGUCAAUGAAGUCUAUUGCtt |
| CASP6 | CCCAAGUGCAAUUGGAAAGtt | CUUUCCAAUUGCACUUGGGtc | GGAAUUAAAUCAUCCCUUAtt | UAAGGGAUGAUUUAAUUCCtg |
| RHOC | GGUGUUUGAGAUGGCCACUtt | AGUGGCCAUCUCAAACACCtc | GCCUGGAAAACAUUCCUGAtt | UCAGGAAUGUUUUCCAGGCtg |
| FZD2 | GCCCUCAUGAACAAGUUCGtt | CGAACUUGUUCAUGAGGGCtt | CGUACUUGGUAGACAUGCAtt | UGCAUGUCUACCAAGUACGtg |
| CCND1 | GGGUUAUCUUAGAUGUUUCtt | GAAACAUCUAAGAUAACCCtt | GGAGGGGUUGUGCUACAGAUtt | AUCUGUAGCACAACCCUCCtc |
| CRY1 | GGAUGAAACAGAUCUAUCAtt | UGAUAGAUCUGUUUCAUCCtt | GGUGGCGAUUUUUGCUUCAtt | UGAAGCAAAAUCGCCACCtg |
| CYFIP1 | GCGGCUUCGAGAUAAAAGUtt | ACUUUUAUGUCGAAGCCGCtc | CGGUUACGAUCAGAAUGCAtt | UGCAUUCUGAUCGUAACCGtt |
| ENPP2 | GGAUAUCCAUUUAUUGGUGtt | CACCAAUAAAUGGAUAUCCtc | GGGCGAGAGAAAUUUAAUCtt | GAUUAAAUUUCUCUCGCCCtc |
| CCL5 | GCUAGGAUGGAGAGUCCUUtt | AAGGACUCUCCAUCCUAGCtc | GGGCCCAGAUUCUACCACAtt | UGUGGUAGAAUCUGGGCCCtt |
| SCA1 | CCUUGUCAUUAGAGUUACAtt | UGUAACUCUAAUGACAAGGtt | GCUUGGUUUACAACAGGGAtt | UCCCUGUUGUAAACCAAGCtc |
| TYRO3 | GGUGUGCCAUUUUUCACAGtt | CUGUGAAAAAUGGCACACCTT | GGUCUGAAUCUUCACCAUCtt | GAUGGUGAAGAUUCAGACCtg |
| SLC16A4 | CCUAAACUAAUGUCAUUGGtt | CCAAUGACAUUAGUUUAGGtt | GCUAUCGCAUUGAAUUUGGtt | CCAAAUUCAAUGCGAUAGCtc |
| PHYH | GGAUAUUUGGAUGUUUCGAtt | UCGAAACAUCCAAAUAUCCTT | GGGAUUCCGGAAGGCAAUUtt | AAUUGCCUUCCGGAAUCCCTG |
| FSCN1 | CCCUCAGGUCAACAUCUACtt | GUAGAUGUUGACCUGAGGGtg | GCAGCCUGAAGAAGAAGCAtt | UGCUUCUUCUUCAGGCUGCtg |
| OASL | GGGUACAGAUGGGACAUCGtt | CGAUGUCCCAUCUGUACCCtt | CGAGACAUCCACUUGACAGtt | CUGUCAAGUGGAUGUCUCGtg |
| KIAA0882 | GCUGUACGACUGCAUGCAAtt | UUGCAUGCAGUCGUACAGCtg | CCUCAAUGUAGGUACAUUAtt | UAAUGUACCUACAUUGAGGtg |
Lower case letters in the primers mean overhang bases.
Flow cytometry analysis
The effect of transfection with siRNAs targeting the CD40 transcript was measured by flow cytometry. At indicated time points following nucleofection, Granta 519 cells were washed in PBS and incubated with a FITC-conjugated mouse monoclonal anti-CD40 antibody (BD Pharmingen, San Diego, CA). The binding of the antibody was determined on a FACScan (BD Pharmingen).
Cell-cycle analysis
Eg5 (Ambion) was used as positive siRNA control. This siRNA inhibits proliferation of transfected cells by silencing the Eg5 motor protein, which is necessary for cell mitosis. Granta 519 cells were electroporated, and 48 hours after transfection, cells were fixed in cold 70% ethanol for 30 minutes at 4°C. Cells were washed twice in cold PBS and treated with ribonuclease A (Sigma-Aldrich, St Louis, MO) at 100 μg/mL, followed by 50 μg/mL propidium iodide. The DNA content of the cells was determined by FACScan, and the different cell-cycle phases were analyzed using Flow Jo software (Tree Star, Ashland, OR).
Proliferation assay
The proliferation of transfected Granta 519 cells was assessed by a [3H] thymidine incorporation assay. At 3 hours after nucleofection, which allowed recovery of membrane integrity, cells were transferred to a 96-well plate (approximately 150 000 cells/well in triplicates). After 64 hours, cells were pulsed by 0.0185 MBq/well (0.5 μCi/well) of [3H] thymidine (Amersham Pharmacia Biotech, Uppsala, Sweden) and incubated for 8 hours. After 72 hours, cells were harvested, and the incorporation of [3H] thymidine was determined by a 1450 Micro Beta Liquid Scintillation Counter (Amersham Pharmacia Biotech).
RNA isolation and real-time RT-PCR
RNA isolation was carried out in accordance with the TRIzol Reagent (Invitrogen) method, 48 hours after transfection. Briefly, cells were washed twice in cold PBS and centrifuged at 311g for 5 minutes at 4°C between each wash, the resultant pellets were resuspended in 400 μL of TRIzol solution, and 1 μL of linear polyacrylamide (LPA; 10 μg/μL, RNAse-free) was added to enable better visualization of RNA pellet at later stages of RNA preparation. A volume of 200 μL of chloroform was added, and the solution was mixed thoroughly. Mixtures were placed on ice for 10 minutes followed by centrifugation at 16000g for 30 minutes. The aqueous phase was placed into fresh tubes and mixed with 80 μL of ice-cold isopropanol and placed at −20°C overnight. The RNA precipitates were pelleted via centrifuged at 16000g for 30 minutes, and the supernatant was removed. The remaining pellets were washed with 500 μL of ice-cold ethanol and centrifuged at 16000g for 30 minutes, after which the supernatants were removed and the pellets were allowed to dry at room temperature for 1 to 2 minutes. RNA samples were finally redissolved in 5 to 10 μL RNAse-free water and heated to 65°C for 5 minutes.
Quantification of RNA samples were carried out on the NanoDrop (NanoDrop Technologies, Wilmington, DE) set for RNA measurement (A260/A280 ratio). Complementary DNA (cDNA) were generated from the RNA isolates in accordance to the procedure out lined in the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas Life Science, Burlington, ON). RNA quantities of 100 ng were used in conjunction with random hexamer primer (0.2 μg/μL). The resultant generated 20 μL of cDNA was made up to 100 μL with RNAse-free water, and 5 μL of the dilution was used for each real-time reverse transcription–polymerase chain reaction (RT-PCR) run on the Bio-Rad iQ5 cycler (Hercules, CA).
Human FZD2, HDGFRP3, DUSP5, and MYC together with G3PD expression were analyzed using iQ SYBR Green Supermix (Bio-Rad). The primers were as follows: FZD2, 5′-GGTGCCATCCTATCTCAG-3′ and 5′-TGGTGACAGTGAAGAAGG-3′; DUSP5, 5′-GTCCTCACCTCGCTACTC-3′ and 5′- CATCCACGCAACACTCAG-3′; HDGFRP3, 5′-GGACCTAACTACCATAATGAATGC-3′ and 5′-CCCGAAACACAACAGAGAGG-3′; MYC, 5′-ACACATCAGCACAACTACG-3′ and 5′-CGCCTCTTGACATTCTCC-3′; and G3PD, 5′- TGGTATCGTGGAAGGACTC-3′ and 5′-AGTAGAGGCAGGGATGATG-3′. Samples were denatured at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds and annealing/amplification at 55°C for 30 seconds.
Data collection and real-time analysis were enabled and carried out with denaturation at 95°C for 1 minute, followed by annealing/amplification at 55°C for 1 minute. The temperature was increased from 55°C to 95°C in 0.5°C increments after the second cycle for a total of 81 cycles in order to obtain data for a melting curve analysis. Data were analyzed with the iCycler iQ Optical System software (Bio-Rad). The relative expression in each sample was calculated adjusting for the real-time RT-PCR efficiencies. All experiments were performed in triplicate, and the expression of the targets and MYC was normalized to the expression of G3PD.
Results
The rationale of using up-regulated genes for identification of functionally associated targets by means of RNAi is based on the fact that tumor cells normally acquire a growth/survival advantage with this approach. Microarray technology provides a powerful means of establishing global transcriptional profiles of a given cell type.19 If the genomic signature is compared with that of another cell type, for example, tumor versus healthy cells, differentially expressed genes will be identified. In this study, we used DNA microarrays to create 209 comparison chip files based on samples from 17 frozen MCL tumors, 2 purified MCL tumors, and 11 primary B-cell samples from healthy individuals. Each comparison file contains a vast amount of transcriptional information, which needs to be condensed by restricting the data set. We focused on overexpressed genes in MCL compared with normal B-cell subpopulations, which could identify disease-associated genes as well as potential candidates for therapeutic interventions.20,21 Thus, a list of 46 genes was generated that contained target genes specifically up-regulated in the MCL patient samples compared with normal B-cell populations, and in addition were present in our in vitro cell models (Table 2). Figure 1 shows the results from a hierarchic clustering of these 46 genes. The normal B-cell populations formed a separate cluster, which differed significantly (P < .05) from all the tumor samples. From this list of 46 significantly differently expressed genes, we selected 20 that were potentially functionally associated due to their involvement in cellular growth and proliferation.
Overexpressed genes in MCL
| Name . | Systematic* . | GenBank accession no. . | Average fold change . |
|---|---|---|---|
| HDGFRP3 | 34673_r_at | AL109689 | 5.0 |
| SPRY2 | 33700_at | AF039843 | 3.6 |
| FARP1 | 32148_at | AI701049 | 5.5 |
| ACTN1 | 39329_at | X15804 | 10.8 |
| CST3 | 39689_at | AI362017 | 11.0 |
| PTP4A3 | 36008_at | AF041434 | 2.0 |
| AEBP1 | 39069_at | AF053944 | 8.1 |
| DBN1 | 37981_at | U00802 | 6.6 |
| HOMER3 | 38233_at | AF093265 | 6.5 |
| DUSP5 | 529_at | U15932 | 5.3 |
| CASP6 | 35662_at | U20536 | 2.3 |
| ENPP2 | 41124_r_at | L35594 | 22.0 |
| RHOC | 1395_at | L25081 | 3.1 |
| PSEN2 | 41349_at | L43964 | 2.4 |
| TPM2 | 32314_g_at | M75165 | 9.2 |
| FZD2 | 628_at | L37882 | 2.7 |
| SPARC | 671_at | J03040 | 24.2 |
| CCL4 | 36674_at | J04130 | 12.4 |
| CCND1 | 2020_at | M73554 | 19.7 |
| PCCA | 1348_s_at | S79219 | 2.1 |
| ZNF3 | 40118_at | X07290 | 2.5 |
| IGF2R | 160027_s_at | Y00285 | 7.8 |
| CRY1 | 38148_at | D83702 | 2.9 |
| CYFIP1 | 37306_at | D38549 | 11.2 |
| PRKCB1 | 160029_at | X07109 | 8.2 |
| ENPP2 | 41123_s_at | L35594 | 23.3 |
| CHL1 | 34193_at | AF002246 | 6.1 |
| CCL5 | 1405_i_at | M21121 | 23.8 |
| DKFZp434I | 41810_at | AA203545 | 2.0 |
| ZNF268 | 35866_at | X78926 | 1.8 |
| SCA1 | 36142_at | X79204 | 4.2 |
| TYRO3 | 2086_s_at | D17517 | 3.6 |
| ID1 | 36618_g_at | X77956 | 3.8 |
| NFE2L3 | 34282_at | AB010812 | 4.8 |
| OPTN | 41744_at | AF070533 | 2.4 |
| NMT2 | 41656_at | AF043325 | 2.0 |
| SERPINF1 | 40856_at | U29953 | 5.4 |
| SMARCA1 | 40213_at | M88163 | 2.9 |
| SLC16A4 | 39260_at | U59185 | 15.9 |
| TIAM1 | 38370_at | U90902 | 6.1 |
| CCL5 | 1403_s_at | M21121 | 11.8 |
| PHYH | 32724_at | AF023462 | 4.6 |
| FSCN1 | 39070_at | U03057 | 9.0 |
| CNN3 | 40953_at | S80562 | 12.1 |
| OASL | 38389_at | X04371 | 2.3 |
| KIAA0882 | 38254_at | AB020689 | 3.9 |
| Name . | Systematic* . | GenBank accession no. . | Average fold change . |
|---|---|---|---|
| HDGFRP3 | 34673_r_at | AL109689 | 5.0 |
| SPRY2 | 33700_at | AF039843 | 3.6 |
| FARP1 | 32148_at | AI701049 | 5.5 |
| ACTN1 | 39329_at | X15804 | 10.8 |
| CST3 | 39689_at | AI362017 | 11.0 |
| PTP4A3 | 36008_at | AF041434 | 2.0 |
| AEBP1 | 39069_at | AF053944 | 8.1 |
| DBN1 | 37981_at | U00802 | 6.6 |
| HOMER3 | 38233_at | AF093265 | 6.5 |
| DUSP5 | 529_at | U15932 | 5.3 |
| CASP6 | 35662_at | U20536 | 2.3 |
| ENPP2 | 41124_r_at | L35594 | 22.0 |
| RHOC | 1395_at | L25081 | 3.1 |
| PSEN2 | 41349_at | L43964 | 2.4 |
| TPM2 | 32314_g_at | M75165 | 9.2 |
| FZD2 | 628_at | L37882 | 2.7 |
| SPARC | 671_at | J03040 | 24.2 |
| CCL4 | 36674_at | J04130 | 12.4 |
| CCND1 | 2020_at | M73554 | 19.7 |
| PCCA | 1348_s_at | S79219 | 2.1 |
| ZNF3 | 40118_at | X07290 | 2.5 |
| IGF2R | 160027_s_at | Y00285 | 7.8 |
| CRY1 | 38148_at | D83702 | 2.9 |
| CYFIP1 | 37306_at | D38549 | 11.2 |
| PRKCB1 | 160029_at | X07109 | 8.2 |
| ENPP2 | 41123_s_at | L35594 | 23.3 |
| CHL1 | 34193_at | AF002246 | 6.1 |
| CCL5 | 1405_i_at | M21121 | 23.8 |
| DKFZp434I | 41810_at | AA203545 | 2.0 |
| ZNF268 | 35866_at | X78926 | 1.8 |
| SCA1 | 36142_at | X79204 | 4.2 |
| TYRO3 | 2086_s_at | D17517 | 3.6 |
| ID1 | 36618_g_at | X77956 | 3.8 |
| NFE2L3 | 34282_at | AB010812 | 4.8 |
| OPTN | 41744_at | AF070533 | 2.4 |
| NMT2 | 41656_at | AF043325 | 2.0 |
| SERPINF1 | 40856_at | U29953 | 5.4 |
| SMARCA1 | 40213_at | M88163 | 2.9 |
| SLC16A4 | 39260_at | U59185 | 15.9 |
| TIAM1 | 38370_at | U90902 | 6.1 |
| CCL5 | 1403_s_at | M21121 | 11.8 |
| PHYH | 32724_at | AF023462 | 4.6 |
| FSCN1 | 39070_at | U03057 | 9.0 |
| CNN3 | 40953_at | S80562 | 12.1 |
| OASL | 38389_at | X04371 | 2.3 |
| KIAA0882 | 38254_at | AB020689 | 3.9 |
Systematic Probe set name of Affimetrix-microarray U95Av2.
Establishment of RNAi in an MCL in vitro model
First, siRNAs against the CD40 cell-surface receptor were designed and electroporated into Granta 519. The silencing of CD40 could clearly be monitored through staining with fluorescently labeled antibody and flow cytometric analysis. At 3 days after transfection, the expression of CD40 receptor had decreased to less than 50% compared with cells transfected with a negative control siRNA (Figure 2A). An additional 2 siRNAs against CD40 were designed, and gave rise to similar silencing effects (data not shown). Second, to test whether silencing of genes in Granta 519 cells could induce functional effects, in particular inhibition of proliferation, an siRNA targeting the Eg5/KIF11 was used. Eg5 is necessary for mitosis, and its knockdown causes cell-cycle arrest. Granta 519 cells were transfected with 100 pmol of Eg5 siRNA, and the functional effects were determined by [3H] thymidine incorporation and cell-cycle analysis. Specifically, cells transfected with Eg5 siRNA showed more than a 40% decrease in proliferation compared with cells transfected with a scrambled siRNA sequence (negative control siRNA; Figure 2B). In addition, the cells in the G2 phase of the cell cycle increased from 10% to 18% after the introduction of Eg5 siRNA (Figure 2C).
Experimental Controls. (A) The mantle cell lymphoma cell line Granta 519 was electroporated with 100 pmol of siRNA targeting the CD40 receptor (■) or a nonexisting transcript (negative control siRNA; □). At indicated time points, the cells were harvested and the binding of a commercial anti-CD40 antibody was assessed by means of flow cytometry. The expression of CD40 (as determined by the mean fluorescence intensity [MFI] of the viable population) on cells electroporated in the absence of siRNA was set to 1. (B) To assess the RNAi knockdown on a functional level, the proliferation of transfected cells was investigated. Granta 519 cells were electroporated with 100 pmol siRNAs and incubated for 72 hours, where after the proliferative effects of the different siRNAs was determined by thymidine incorporation. Bars represent plus or minus SD. (C) Similar to panel A, Granta cells were electroporated and 48 hours later, cells were fixed and stained with propidium iodide. The DNA content of the cells was determined in the FL-2 channel in a FACScan. Cell cycle phases were analyzed using Deau-Jett-FCX model enclosed in FlowJo software (TreeStar, Ashland, OR): green lines and black picks correspond to the different phases of the cell cycle. Black horizontal lines define the amplitude of the different picks, therefore, the percentage of the different cell cycle phases.
Experimental Controls. (A) The mantle cell lymphoma cell line Granta 519 was electroporated with 100 pmol of siRNA targeting the CD40 receptor (■) or a nonexisting transcript (negative control siRNA; □). At indicated time points, the cells were harvested and the binding of a commercial anti-CD40 antibody was assessed by means of flow cytometry. The expression of CD40 (as determined by the mean fluorescence intensity [MFI] of the viable population) on cells electroporated in the absence of siRNA was set to 1. (B) To assess the RNAi knockdown on a functional level, the proliferation of transfected cells was investigated. Granta 519 cells were electroporated with 100 pmol siRNAs and incubated for 72 hours, where after the proliferative effects of the different siRNAs was determined by thymidine incorporation. Bars represent plus or minus SD. (C) Similar to panel A, Granta cells were electroporated and 48 hours later, cells were fixed and stained with propidium iodide. The DNA content of the cells was determined in the FL-2 channel in a FACScan. Cell cycle phases were analyzed using Deau-Jett-FCX model enclosed in FlowJo software (TreeStar, Ashland, OR): green lines and black picks correspond to the different phases of the cell cycle. Black horizontal lines define the amplitude of the different picks, therefore, the percentage of the different cell cycle phases.
To further substantiate the fidelity of the system, the effect of silencing MYC in Granta 519 was also investigated. The decrease in proliferation, after silencing this gene, was 35% after 72 hours, which was comparable to the effect caused by the knockdown of Eg5 (data not shown).
These experiments demonstrate that Granta 519 cells can be used as an in vitro model system to assess RNAi-mediated knockdown.
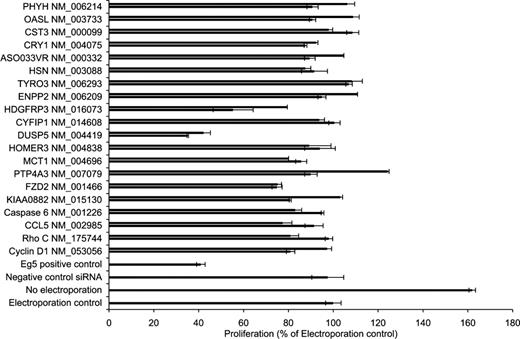
Identification of functionally associated tumor genes
The effect on cell proliferation for each siRNA was determined in Granta 519 cells 72 hours after transfection (Figure 3). A total of 3 genes were identified whose silencing by their corresponding siRNA resulted in an inhibition of proliferation of more than 30%. These genes included (1) FZD2, a member of the frizzled gene family for Wnt signaling proteins, (2) DUSP5, a dual-specific phosphatase that dephosphorylates proteins on both tyrosine and serine/threonine residues, and (3) HDGFRP3, a molecule that belongs to the hepatoma-derived growth factor family.
Assessment of proliferation after silencing of selected targets. A total of 2 unique siRNA sequences were designed and synthesized against each of 20 selected transcripts derived from the DNA microarray study. Granta 519 cells were nucleofected with the siRNAs and appropriate control siRNAs, and the proliferative effect of the sequence-specific knockdown was assessed. Electroporation control (ie, electroporation without any siRNA) was set to 100%. Bars represent plus or minus SD of triplicate samples.
Assessment of proliferation after silencing of selected targets. A total of 2 unique siRNA sequences were designed and synthesized against each of 20 selected transcripts derived from the DNA microarray study. Granta 519 cells were nucleofected with the siRNAs and appropriate control siRNAs, and the proliferative effect of the sequence-specific knockdown was assessed. Electroporation control (ie, electroporation without any siRNA) was set to 100%. Bars represent plus or minus SD of triplicate samples.
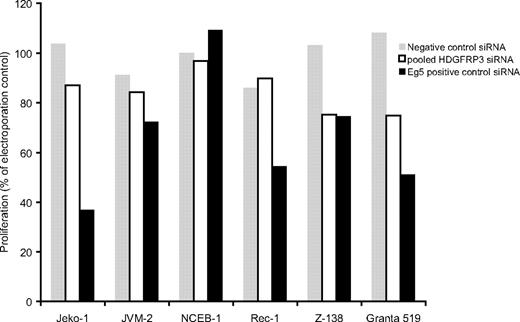
Furthermore, to substantiate the effect in other cell lines, pooled siRNAs targeting HDGFRP3 were transfected into 6 different MCL cell lines. The effect on proliferation was most pronounced in Granta 519, but 3 other cell lines (Jeko-1, JVM-2, and Z-138) showed an siRNA-induced decrease in proliferation (Figure 4).
Effect of HDGFRP3 siRNAs on different MCL cell lines. A total of 6 MCL cell lines were electroporated with 100 pmol of a pool of siRNAs targeting the HDGFRP3 (□), negative control siRNA (▩), or Eg5+ control (■). At 72 hours after transfection, the proliferation of each cell line was determined.
Effect of HDGFRP3 siRNAs on different MCL cell lines. A total of 6 MCL cell lines were electroporated with 100 pmol of a pool of siRNAs targeting the HDGFRP3 (□), negative control siRNA (▩), or Eg5+ control (■). At 72 hours after transfection, the proliferation of each cell line was determined.
Validation of selected functionally associated tumor genes by real-time RT-PCR
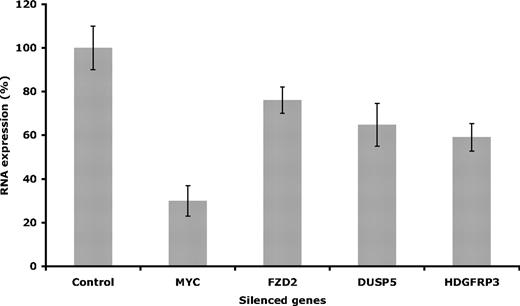
To confirm the siRNA-induced silencing and functional effect on proliferation, the selected target genes were evaluated on the mRNA level using real-time RT-PCR. Once the siRNA procedure was completed, as described in “Methods,” the efficiency of the knockdowns was determined. The effect on mRNA level varied depending on the specific target, where the highest reduction was found after knocking down MYC (around 70%), compared with 35% mRNA reduction for DUSP5, 40% to HDGFRP3, and 24% for FZD2 (Figure 5).
Evaluation of gene expression. Gene expression was evaluated after silencing the selected targets in Granta 519. Evaluation of mRNA levels was performed at 48 hours after electroporation. Error bars are plus or minus standard deviation of 3 independent experiments.
Evaluation of gene expression. Gene expression was evaluated after silencing the selected targets in Granta 519. Evaluation of mRNA levels was performed at 48 hours after electroporation. Error bars are plus or minus standard deviation of 3 independent experiments.
Discussion
New therapeutic approaches for MCL are constantly being evaluated in clinical trials, including, for example, CDK inhibitors and proteasome inhibitors.22-24 Specifically, interfering with cellular pathways deregulated in MCL could induce a therapeutic beneficial response, as demonstrated by single-agent administration of temsirolimus, a regulator of CCND1 levels.25 Aiming for a better understanding of MCL pathogenesis and definition of potential therapeutic targets, the present study combines the power of DNA microarrays and RNAi to identify possible candidate genes aberrantly expressed in this disease.
We established an assay based on RNA interference to screen the overexpressed genes for functional association, where the introduction of synthetic siRNAs will cause sequence-specific knockdown. The effect was screened by a proliferation assay. RNAi has been shown to be highly efficient in several adherently growing cell lines, but studies on suspension cell lines, such as B- and T-cell lymphomas, have been lacking mainly because of the problem of transfecting hematopoietic cell lines.26 However, a novel transfection technique has been introduced by Amaxa Biosystems using a nucleofection device that combines electroporation programs with cell type–specific transfection solutions. This approach has proven useful in difficult-to-transfect cells, such as primary cells, T cells, dendritic cells, and B cells,27,28 although to date, very few studies of RNAi in MCL cell lines have been published using this technology.29-31
We have previously demonstrated that the Granta 519 cell line is a good in vitro model for the study of MCL.18 To determine the suitability of this cell line for siRNA screening of overexpressed genes, a series of control experiments were carried out. The RNAi effect is usually demonstrated by the decrease of mRNA expression after siRNA transfection. Although changes in mRNA levels do not necessarily correlate with alteration in protein concentration due to, for example, posttranscriptional mechanisms, different turnovers of specific proteins and their mRNAs, intracellular location, and protein-protein interactions,32 levels of all selected targets, as well as of Eg5 and MYC, were assessed.
To screen for functionally associated genes, we selected 20 of the most promising and designed the appropriate siRNAs. To avoid false positives due to “off-target” effects,33,34 we required that the 2 siRNAs for each gene exercised a similar effect on cell proliferation. Since proliferation is one of the final measures of cellular status, the assessment of a proliferative change will allow us to identify relevant genes. Eg5-siRNA was included in the screening as a positive control, as well as MYC. Following this strategy, we found 3 targets (FZD2, HDGFRP3, and DUSP5) affecting the proliferation of Granta 519. The efficiency of the knockdown was assessed by real-time RT-PCR and varied depending on the target, showing up to 70% efficacy for MYC using an siRNA validated by the manufacturer. No effect on proliferation was observed after the transient silencing of CCDN1. This is supported by the observations of Pscherer et al,31 who had to use a stable and inducible RNAi plasmid to knock down CCND1. Even so, the effect on proliferation in that study was not visible before day 4.
FZD2 belongs to the frizzleds (FZDs), a family of 7 transmembrane (7TM) receptors that upon binding of Wnt ligands induce downstream signaling events, ultimately causing changes in cell proliferation and morphology.35 The signaling pathway is divided into 3 branches, the Wnt/β catenin, Wnt/Ca2+, and Wnt polarity pathways. Specifically, FZD2 induces signaling along the Wnt/β catenin pathway,36 which activates the phosphorylation of Dishevelled (Dvl) and the binding of axin and glycogen synthase kinase 3β (GSK3β). In the absence of Wnt signaling, β-catenin, the key mediator of the pathway, is phosphorylated by GSK3β and targeted for proteasomal degradation. However, the FZD2/Dvl/Axin/GSK3β complex formed upon Wnt binding prevents phosphorylation of β-catenin and consequently, β-catenin translocates to the nucleus and induces target gene expression.37 Interestingly, one of the target genes for β-catenin is CCND1, the hallmark of MCL.38-40 Furthermore, the Wnt pathway is deregulated in other types of cancers,41,42 and therapeutic intervention of the pathway has proven useful in, for example, melanoma, as demonstrated by blocking antibodies toward both frizzled receptor and Wnt ligands.43,44 Thus, the overexpression of FZD2 in MCL together with its functional knockdown by RNAi suggest that interfering with the Wnt signaling pathway could be a potential therapeutic approach in MCL.
HDGFRP3 is a member of the hepatoma-derived growth factor (HDGF) family. The HDGFs all share a common sequence motif in their N-terminal domain, called the HATH (homologous to the amino terminus of HDGF) region.45 The HDGF has only been reported to stimulate growth of fibroblasts and some hepatoma cells.46 However, high levels of HDGF are found in a number of different cancers, suggesting a potential role in cancer development.47 In contrast to HDGF, which is expressed in a wide range of tissues, for example, heart, brain, lung, liver, muscle, kidney, and pancreas,48 HDGFRP3 has so far only been found in brain and testis.49 This is the first study that reports the overexpression of HDGFRP3 in MCL, and that its down-regulation inhibits proliferation.
Finally, we also report the overexpression of the nuclear phosphatase DUSP5 in MCL. When siRNAs against the DUSP5 transcript were transfected into Granta 519 cells, the proliferation of the MCL cells was inhibited by more than 50% (Figure 3). DUSP5 has previously been reported to be involved in the inactivation of extracellular signal–regulated kinase (ERK) 2.50 The ERK mitogen-activated protein (MAP) kinase signaling pathway mediates cell proliferation, differentiation, transformation, and survival. Furthermore, Ueda and colleagues reported that overexpression of DUSP5 suppressed the proliferation of transformed cells due to dephosphorylation of ERK1/2.51 Hence, our finding that knockdown of DUSP5 in MCL induced proliferative arrest indicates that this gene product might be involved in yet another pathway, previously not functionally described.
In summary, we describe an efficient approach for identifying genes functionally involved in tumor cell proliferation by combining genomics and RNAi. The use of DNA microarrays allowed an initial rapid filtering of overexpressed and potentially disease-related genes. The subsequent screening for siRNA-mediated inhibition of proliferation leads to the identification of FZD2, HDGFRP3, and DUSP5, previously not identified as functionally associated genes in MCL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Ann-Charlott Olsson for her technical support and Ole E. Sørensen and Kanu Patel for their help with the real-time RT-PCR experiments.
This study was supported by grant no. 6085-06 from the Leukemia and Lymphoma Society and the Strategic Center for Translational Cancer Research (www.createhealth.lth.se).
Authorship
Contribution: E.O. and J.F. designed and performed the research and wrote the paper; S.E. analyzed DNA microarray data; and C.A.K.B. supervised the research and the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carl A. K. Borrebaeck, Department of Immunotechnology, Lund University, BMC D13, SE-221 84 Lund, Sweden; e-mail: carl.borrebaeck@immun.lth.se.

![Figure 2. Experimental Controls. (A) The mantle cell lymphoma cell line Granta 519 was electroporated with 100 pmol of siRNA targeting the CD40 receptor (■) or a nonexisting transcript (negative control siRNA; □). At indicated time points, the cells were harvested and the binding of a commercial anti-CD40 antibody was assessed by means of flow cytometry. The expression of CD40 (as determined by the mean fluorescence intensity [MFI] of the viable population) on cells electroporated in the absence of siRNA was set to 1. (B) To assess the RNAi knockdown on a functional level, the proliferation of transfected cells was investigated. Granta 519 cells were electroporated with 100 pmol siRNAs and incubated for 72 hours, where after the proliferative effects of the different siRNAs was determined by thymidine incorporation. Bars represent plus or minus SD. (C) Similar to panel A, Granta cells were electroporated and 48 hours later, cells were fixed and stained with propidium iodide. The DNA content of the cells was determined in the FL-2 channel in a FACScan. Cell cycle phases were analyzed using Deau-Jett-FCX model enclosed in FlowJo software (TreeStar, Ashland, OR): green lines and black picks correspond to the different phases of the cell cycle. Black horizontal lines define the amplitude of the different picks, therefore, the percentage of the different cell cycle phases.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/3/10.1182_blood-2007-02-068791/6/m_zh80030814150002.jpeg?Expires=1769089562&Signature=v8sydBg0I~uNUQXFttn4qBc18UJ2qvVNsSbqrUI-jbLI6sQ~8nVVUZbyb6f0zBvNPnWryb4InD0dAPHiTwlB4chlGmiybTSi7C-V4sHmTn4~zldPV8hHTh-adI4I4gzyi~jimp9CuoaiNBh2AYI9kV4TtV8hR1CMcjBJM98H~L8PELxYRgt5-6i7UykPcjTTxOX938OXWfHu4h9WIEoowVzs2jLfNkA~QWDoScw8wv3VKY3LlS0tUGpLJuuttthZV8LUlB1DsgG74bO5JEgFcAvauWoY0brYP-fEUOxk3kPrZaw~PidvOLiNGlG1LanE3oEWvI2RsEzKbPSbHG7hzw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal