Abstract
The inversion of chromosome 16 in the inv(16)(p13q22) is one of the most frequent cytogenetic abnormalities observed in acute myeloid leukemia (AML). The inv(16) fuses the core binding factor (CBF) beta subunit with the coiled-coil rod domain of smooth muscle myosin heavy chain (SMMHC). Expression of CBFβ-SMMHC in mice does not promote AML in the absence of secondary mutations. Patient samples with the inv(16) also possess mutually exclusive activating mutations in either N-RAS, K-RAS, or the receptor tyrosine kinases, c-KIT and FLT3, in almost 70% of cases. To test whether an activating mutation of FLT3 (FLT3-ITD) would cooperate with CBFβ-SMMHC to promote AML, we coexpressed both mutations in hematopoietic progenitor cells used to reconstitute lethally irradiated mice. Analysis of transplanted animals showed strong selection for CBFβ-SMMHC/FLT3-ITD–expressing cells in bone marrow and peripheral blood. Compared with animals transplanted with only CBFβ-SMMHC–expressing cells, FLT3-ITD further restricted early myeloid differentiation and promoted peripheralization of primitive myeloblasts as early as 2.5 weeks after transplantation. FLT3-ITD also accelerated disease progression in all CBFβ-SMMHC/FLT3-ITD–reconstituted animals, which died of a highly aggressive and transplantable AML within 3 to 5 months. These results indicate that FLT3-activating mutations can cooperate with CBFβ-SMMHC in an animal model of inv(16)-associated AML.
Introduction
Chromosomal translocations involving genes encoding the 2 subunits of core-binding factor (CBF) represent the most common cytogenetic abnormalities found in acute myeloid leukemia (AML).1-3 CBF is a transcription factor that consists of a DNA-binding subunit, RUNX1 (AML1, CBFα2, PEBP2b), and a non-DNA-binding subunit, CBFβ, which stabilizes binding of RUNX1 to DNA.4,5 CBFβ also increases the stability of RUNX1 by protecting it from proteosome-mediated degradation.6,7 Loss-of-function mutations in either Runx1 or Cbfβ result in an early embryonic lethality that is associated with intracranial hemorrhaging and the complete absence of definitive hematopoietic cells.8-12 These observations demonstrate the requirement of both RUNX1 and CBFβ for normal CBF activity.
The major translocation involving CBFβ (inv(16)(p13q22)) results from a chromosomal inversion that fuses the N-terminus of CBFβ (amino acids 1-165 in 85% of cases) to the coiled-coil domain of smooth muscle myosin heavy chain (SMMHC), which is encoded by the MYH11 gene.13,14 Mice with a knock-in of CBFβ-MYH11 into the Cbfβ locus exhibit a similar early embryonic lethal phenotype as the Runx1 or Cbfβ knockout mice, which indicates that the inv(16) encodes a dominant inhibitory protein to wild-type CBF.15 CBFβ-SMMHC binds RUNX1 with a much higher affinity and with altered stoichiometry compared with wild-type CBFβ.7,16 It can also recruit the nuclear corepressor molecules mSin3A and HDAC8 via a repression domain mapped to the C-terminus of SMMHC,17,18 which may suggest a mechanism for the inhibitory activity of the protein against CBF-regulated gene expression. CBFβ-SMMHC may also be inhibiting CBF function by sequestering RUNX1 to the cytoplasm, where it is associated with the actin cytoskeleton, or to rod-like inclusion bodies within the nucleus.19-22 Chimeric mice generated with ES cells containing a knock-in of CBFβ-MYH11 into the Cbfβ locus did not develop leukemia within the first year of life unless animals were exposed to the DNA alkylating agent, N-ethyl-N-nitrosourea.23 Leukemia was also noted in chimeric animals that were neonatally injected with the replication-competent 4070A retrovirus as a means of identifying cooperating mutations with the inv(16) by insertion site analysis.24 These results and other animal model studies25-28 demonstrated that CBFβ-SMMHC functions in association with additional mutations in the development of AML.
In humans with the inv(16), activating mutations of the receptor tyrosine kinases, FLT3 and c-KIT, and activating mutations of the N-RAS or K-RAS proto-oncogenes are observed in 60% to 70% of all cases.29,30 The observation that these mutations are mutually exclusive suggests that each may be contributing a similar cooperating function for leukemic progression associated with the inv(16). However, it is of interest that these mutations are not randomly distributed with the inv(16) or with other chromosomal translocations found in AML. For instance, activating mutations of FLT3 are relatively rare with the inv(16) (approximately 3%-8% of cases) but are observed at high frequencies in patients with the t(15;17) in acute promyelocytic leukemia and in AML patient samples with a normal karyotype (occurring in approximately 30%-40% and 30%-35% of cases, respectively).31-36 These observations suggest that there may be a reasonable degree of specificity associated with the class of mutations that can cooperate with the inv(16).
To test whether an activating allele of FLT3 (an internal tandem duplication of the juxtamembrane portion of the receptor, FLT3-ITD) is sufficient to promote AML in the presence of the inv(16), we have used retroviral vectors containing 2 spectrally distinct green fluorescent protein (GFP) variants to coexpress CBFβ-SMMHC and FLT3-ITD in hematopoietic stem/progenitor cells used to reconstitute lethally irradiated mice.37 Analysis of reconstituted animals showed that FLT3-ITD accelerated disease progression and death, with all CBFβ-SMMHC/FLT3-ITD–expressing animals succumbing to a lethal and transplantable AML by 3 to 5 months after transplant. Moribund animals exhibited splenomegaly and had extensive infiltration of CBFβ-SMMHC+/FLT3-ITD+ myeloid blasts within the interstitial areas of the liver, lungs, and kidney. Myeloid blasts were detected in the peripheral blood as early as 2.5 weeks after transplant, which suggests that CBFβ-SMMHC and FLT3-ITD are sufficient to promote peripheralization of abnormal myeloid cells in the absence of additional mutations. These results demonstrate that CBFβ-SMMHC and FLT3-ITD can cooperate in the pathogenesis of AML and suggest that these mutations have functional relevance in the context of human patient samples where they are observed.
Methods
All research described in this manuscript was performed under an IACUC-approved protocol for the use and care of animals and was done under the supervision of ALAC-accredited veterinary staff and animal housing facilities.
Bone marrow harvest, retroviral transduction, and transplantation
These experiments were done exactly as described.38 For transduction of fluorescent activated cell sorter (FACS)-purified c-Kit+Lin−Sca-1+Flt3− (KLSF) cells, bone marrow cells isolated from C57BL/6-Ly-5.2 mice were first stained with an Fc receptor blocking antibody (2.4G2) followed by staining with a biotinylated antibody to c-Kit (3C11). Cells were then incubated with streptavidin microbeads (Miltenyi Biotech, Auburn, CA) according to the manufacturer's instructions and then enriched by binding to magnetic columns (Miltenyi Biotech). Bound cells were eluted with phosphate-buffered saline (PBS) and then stained with antibodies to detect c-Kit (2B8), Sca-1, Flt3, and a panel of antigens present on mature blood cell lineages (Mac-1, Gr-1, Ter119, B220, CD3, CD4, CD5, and CD8). Stained cells were sorted once and then resorted to a purity of greater than 98% using a MoFlo cell sorter (Dako Cytomation, Fort Collins, CO). KLSF cells were pre-stimulated in stem cell factor (SCF, 50 ng/mL, R&D Systems, Minneapolis, MN) and IL-6 (5 ng/mL, R&D Systems) for 24 hours. Cells were then transferred to a 96-well plate that had been coated with retronectin according to the manufacturer's instructions (Takara, Shiga, Japan). SCF, IL-6, 4 μg/mL polybrene, 100 μL FLT3-ITD retroviral supernatant, 30 μL of CBFβ-SMMHC supernatant, and fresh Dulbecco modified Eagle medium were added to make up a total volume of 200 μL. After 6 hours, 150 μL of the volume was exchanged with fresh media to dilute the polybrene, and cells were then incubated for an additional 18 hours before reanalysis of transduction efficiencies and transplantation. Initial titers of the FLT3-ITD virus were approximately 106/mL and 5 × 106/mL for CBFβ-SMMHC. Approximately 2000 KLSF cells (both transduced and nontransduced) from the well were then transplanted into each lethally irradiated C57BL/6-Ly-5.1 recipient animal.
Western blot analysis
A total of 2 × 106 Bex+/Vex+ bone marrow cells were FACS-sorted from 2-month posttransplantation control (lane 1) or CBFβ-SMMHC/FLT3-ITD animals (lane 2), lysed in Laemmli buffer, and run on a gel for Western blotting. A polyclonal antibody to detect CBFβ (Santa Cruz Biotechnology, Santa Cruz, CA) and FLT3 (Santa Cruz Biotechnology) was used to detect CBFβ-SMMHC or the FLT3-ITD. Protein was detected using a secondary IgG antibody conjugated with HRP (Santa Cruz Biotechnology) and ECL (Amersham-Pharmacia, Piscataway, NJ).
Peripheral blood and bone marrow FACS analysis
Mice were bled beginning at 2 to 3 weeks after transplant. Red blood cells (RBCs) were removed by sedimentation in 2% dextran/PBS for 20 minutes at 37°C. Nonsedimented leukocytes and remaining RBCs were transferred to a new tube, spun, and treated with ACK to lyse the remaining RBCs. After diluting the ACK with a 10-fold volume of cold PBS, cells were washed and then stained with following antibodies: anti-B220APC-Cy7 (RA3-6B2), anti-Mac-1APC (M1/70), anti-Gr-1PE (RB6-8C5), anti-CD3PE-CY7 (KT31.1). Cells were analyzed on an LSRII flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (Tree Star, Ashland, OR). Bone marrow cells were collected from tibias and femurs to analyze KLS (c-Kit+Lin−Sca-1+) cells. Cells were stained for 25 minutes at 4°C with the following antibodies: c-KitAPC (2B8), Sca-1Texas Red (E13-161.7), and LinPE. The Lin cocktail included PE-conjugated antibodies to CD3 (145-2C11), CD4 (GK1.5), CD5 (53-7.3), CD8 (53-6.7), Gr-1 (8C5), CD11b (Mac-1), B220 (6B2), and Ter119. In addition, thymus was stained with anti-CD4PE and anti-CD8APC antibodies. Analysis was performed on a triple-laser MoFlo cell sorter (Dako Cytomation) configured with argon, krypton, and rhodamine dye lasers. Dead cells were excluded from the analysis by propidium iodide staining. All antibodies were purchased from BD-Pharmingen unless otherwise noted.
Cytospins
For cytospin preparations, 20 000 Bex+Vex+ myeloid scatter-gated bone marrow or peripheral blood cells were FACS-sorted into PBS supplemented with 12% fetal calf serum and then centrifuged onto glass slides. Cells were dried and stained with Wright-Giemsa.
Preparation of tissue and histopathology
Moribund animals were killed and full necropsies were performed. Weight of spleen was measured and recorded, and peripheral blood smears were prepared and stained with Wright-Giemsa. Spleen, liver, lung, and kidney were fixed in 10% neutral buffered formalin, dehydrated in ethanol, cleared in xylene, and embedded in paraffin block. For bone and spinal column sections, moribund mice were anesthetized with Avertin and then perfused with 10% neutral buffered saline. Femurs and spinal columns were fixed in 10% neutral buffered formalin and decalcified for one day after fixation. Paraffin sections were prepared for histologic staining with hematoxylin and eosin.
Southern blot analysis
Genomic DNA was prepared from splenocytes of moribund CBFβ-SMMHC/FLT3-ITD animals; 10μg of genomic DNA was digested with EcoRI, separated on a 0.8% agarose gels, and then transferred to Hybond membrane. Blots were independently hybridized with α-32P[dCTP]-labeled probes that were complementary to FLT3 (471 bp probe generated by PCR from the central portion of the FLT3-ITD cDNA) or CBFβ (495 bp probe generated by PCR from the 5′-end of the CBFβ-MYH11 cDNA) sequences. A unique EcoRI site within the cloned cDNA sequences was localized downstream of each probe so only single bands for unique proviral integrants would be detected. Because murine sequences were highly homologous to human sequences used for the probes, detection of endogenous Flt3 and Cbfβ was apparent in control (wild-type C57BL/6 splenocytes) lanes in both cases. An EcoRI site located within an intron between Flt3 coding sequences that were complementary to the probe resulted in the detection of 2 endogenous bands in the FLT3 control sample.
Microscopy
Images were acquired on an Olympus (Center Valley, PA) BH2 microscope equipped with a Nikon (Melville, NY) 5M camera. Plan apo objectives 10×/0.4 NA, 20×/0.7 NA, 40×/0.85 NA, and 100×/1.3 oil objectives are used in photography. Photoshop Bridge and Photoshop 9.0 imaging software were used to capture images.
Results
CBFβ-SMMHC and FLT3-ITD cooperate to induce AML in mice
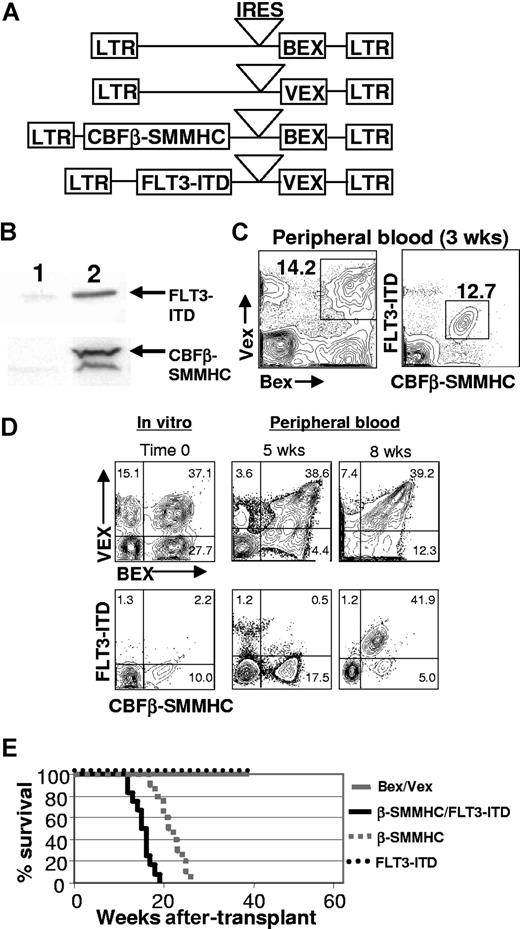
To test whether an activated allele of FLT3 (FLT3-ITD) would cooperate with CBFβ-SMMHC to promote progression to AML, we coexpressed both mutant proteins in hematopoietic progenitor cells isolated from the bone marrow of 5-fluorouracil–treated C57BL/6-Ly-5.2 animals using retroviral vectors that contained spectrally distinct GFP reporter genes (blue-excited GFP, Bex; violet-excited GFP, Vex; Figure 1A,B). Transduced cells were transplanted into lethally irradiated, C57BL/6-Ly-5.1 congenic recipient mice and analyzed at various times after transplant. One observation that was immediately apparent was the strong selection for double-expressing (CBFβ-SMMHC+/FLT3-ITD+) cells in the peripheral blood of almost all CBFβ-SMMHC/FLT3-ITD animals (n = 24) at the earliest time after transplant that analysis was done (2-3 weeks, Figure 1C), which was not evident in animals reconstituted with control Bex/Vex-expressing cells. To further characterize whether cells expressing both mutations had a selective advantage in vivo or if double-transduced cells were present at higher frequencies than single-transductants before transplantation, we transduced highly purified hematopoietic stem cells of the KLSF phenotype with retroviral supernatants and then analyzed the frequencies of single- and double-transductants immediately before transplantation of transduced cells and then at subsequent time points in peripheral blood in reconstituted animals (Figure 1D, n = 5). As shown in Figure 1D, rare double-transductants rapidly expanded in vivo and became the predominant peripheral blood population by 2 months after transplantation. The frequencies of cells that only expressed the FLT3-ITD actually declined in peripheral blood over time, whereas we noted relatively stable representation of cells that only expressed CBFβ-SMMHC.
Generation of animals transplanted with CBFβ-SMMHC/FLT3-ITD-expressing cells. (A) Structure of the MSCV (murine stem cell virus) retroviral constructs. LTR indicates long terminal repeat; IRES, internal ribosome entry site, BEX, blue-excited GFP; VEX, violet-excited GFP. (B) Western blot analysis showing expression of CBFβ-SMMHC and FLT3-ITD in 2 × 106 bone marrow cells FACS–sorted from a 2-month posttransplantation CBFβ-SMMHC/FLT3-ITD animal (lane 2). An equivalent number of bone marrow cells sorted from a Bex/Vex-control animal was used for lane 1. (C) Representative FACS analysis of peripheral blood from Bex/Vex control or CBFβ-SMMHC/FLT3-ITD–reconstituted mice at 2.5 weeks after transplant. Frequencies for double-transduced cells are shown and were variable between experiments. (D) In vitro transduction efficiencies of highly purified KLSF cells used to reconstitute lethally irradiated recipient mice and changes in chimerism of doubly transduced cells in peripheral blood over time in vivo (n = 5 for each of control and CBFβ-SMMHC/FLT3-ITD–reconstituted mice). The same animal was analyzed at 5 and 8 weeks after transplant. Numbers on plots are percentages of total cells.(E) Kaplan-Meier survival curves of mice transplanted with cells expressing CBFβ-SMMHC (n = 9), FLT3-ITD (n = 5), CBFβ-SMMHC/FLT3-ITD (n = 17), or control Bex/Vex (n = 23) retroviruses.
Generation of animals transplanted with CBFβ-SMMHC/FLT3-ITD-expressing cells. (A) Structure of the MSCV (murine stem cell virus) retroviral constructs. LTR indicates long terminal repeat; IRES, internal ribosome entry site, BEX, blue-excited GFP; VEX, violet-excited GFP. (B) Western blot analysis showing expression of CBFβ-SMMHC and FLT3-ITD in 2 × 106 bone marrow cells FACS–sorted from a 2-month posttransplantation CBFβ-SMMHC/FLT3-ITD animal (lane 2). An equivalent number of bone marrow cells sorted from a Bex/Vex-control animal was used for lane 1. (C) Representative FACS analysis of peripheral blood from Bex/Vex control or CBFβ-SMMHC/FLT3-ITD–reconstituted mice at 2.5 weeks after transplant. Frequencies for double-transduced cells are shown and were variable between experiments. (D) In vitro transduction efficiencies of highly purified KLSF cells used to reconstitute lethally irradiated recipient mice and changes in chimerism of doubly transduced cells in peripheral blood over time in vivo (n = 5 for each of control and CBFβ-SMMHC/FLT3-ITD–reconstituted mice). The same animal was analyzed at 5 and 8 weeks after transplant. Numbers on plots are percentages of total cells.(E) Kaplan-Meier survival curves of mice transplanted with cells expressing CBFβ-SMMHC (n = 9), FLT3-ITD (n = 5), CBFβ-SMMHC/FLT3-ITD (n = 17), or control Bex/Vex (n = 23) retroviruses.
Further analysis of transplanted mice showed that all CBFβ-SMMHC/FLT3-ITD-expressing animals developed a lethal AML by 3 to 5 months after transplant. FLT3-ITD increased the rate of leukemic progression compared with animals transplanted with cells that only expressed CBFβ-SMMHC, which died of AML with a latency of 5 to 7 months (Figure 1E). Expression of the FLT3-ITD alone in C57BL/6 hematopoietic progenitor cells of reconstituted mice did not result in any significant myeloid abnormalities by 9 months after transplantation. This observation was mouse strain-specific in that FLT3-ITD expression in BALB/c bone marrow resulted in a lethal myeloproliferative disease both in our hands and in experiments described by others39 (data not shown). Moribund CBFβ-SMMHC/FLT3-ITD animals were highly anemic (∼4.2 × 1012 RBCs/L vs 9.7 × 1012 RBCs/L in transplant-age matched control animals) and also had greatly reduced platelet counts (270 × 109/L vs 680 × 109/L in peripheral blood of control animals; Table 1).
Hemavet counts of peripheral blood isolated from MIB/MIV (n = 4) and moribund CBFβ-SMMHC/FLT3-ITD (n = 9) mice
| . | MIB/MIV . | β-SMMHC/FLT3-ITD . |
|---|---|---|
| White cell count, 109/L | 15.2 ± 3.2 | 108 ± 43.9 |
| RBC count, 1012/L | 9.7 ± 0.9 | 4.2 ± 2.0 |
| Hematocrit, % | 47.7 ± 3.4 | 23.1 ± 9.0 |
| Platelet count, 109/L | 680.0 ± 162.7 | 269.7 ± 118.4 |
| . | MIB/MIV . | β-SMMHC/FLT3-ITD . |
|---|---|---|
| White cell count, 109/L | 15.2 ± 3.2 | 108 ± 43.9 |
| RBC count, 1012/L | 9.7 ± 0.9 | 4.2 ± 2.0 |
| Hematocrit, % | 47.7 ± 3.4 | 23.1 ± 9.0 |
| Platelet count, 109/L | 680.0 ± 162.7 | 269.7 ± 118.4 |
Data are means plus or minus SD.
RBC indicates red blood cell.
Histologic analysis of control and moribund CBFβ-SMMHC/FLT3-ITD mice
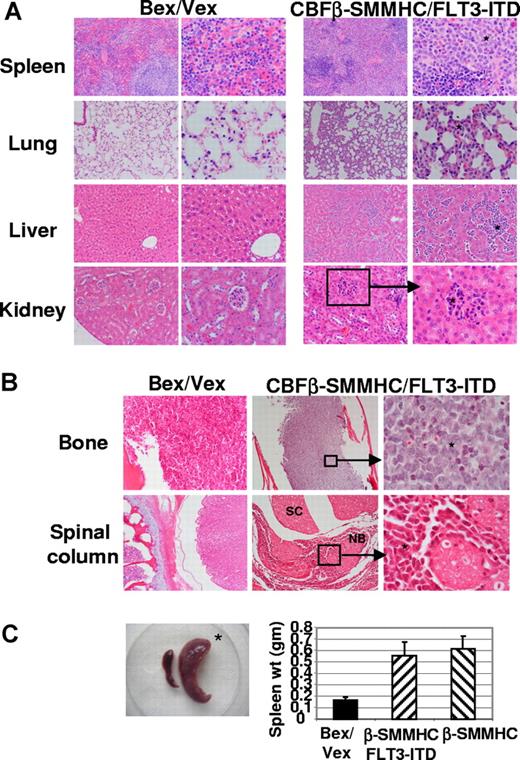
Histologic examination of moribund CBFβ-SMMHC/FLT3-ITD animals showed extensive infiltration of primitive myeloid blasts in multiple tissues. Splenic red pulp was markedly expanded and contained aggregates of leukemic cells. In some areas, residual lymphoid stroma (white pulp) was present and contained small lymphoid cells (Figure 2A). All moribund CBFβ-SMMHC and CBFβ-SMMHC/FLT3-ITD animals exhibited splenomegaly (Figure 2C). In the lung, alveolar septae were thickened (2-4 times the normal) because of mononuclear (leukemic) infiltrate inside the capillaries and also in the extravascular spaces (Figure 2A). In the liver, leukemic infiltrate was observed in the sinusoids with resultant distortion of liver lobular architecture and loss of hepatocytes. Portal spaces also showed similar neoplastic infiltrates. In the kidney, leukemic infiltrate was most concentrated in the glomerular capillaries with extension into the renal interstitium. Renal tubules were well preserved and otherwise unremarkable (Figure 2A).
Dissemination of leukemic cells into peripheral organs and tissues. (A) Hematoxylin and eosin staining of tissues from Bex/Vex control and leukemic CBFβ-SMMHC/FLT3-ITD mice. Data are representative of a minimum of 5 moribund CBFβ-SMMHC/FLT3-ITD animals. Original magnifications in the left and right columns for Bex/Vex and CBFβ-SMMHC/FLT3-ITD, ×150 and ×250, respectively, except for 3 panels that were at ×100 (left panels of control and CBFβ-SMMHC/FLT3-ITD spleen and left panel of control kidney) and 1 panel at ×500 (right panel of CBFβ-SMMHC/FLT3-ITD kidney). * Leukemic infiltrates in various tissues. (B) Representative sections of femur or spinal column stained with hematoxylin and eosin (original magnifications: bone: ×150, left panel; ×50 center panel; ×500 right panel; spinal column: ×50, left; ×50 center; ×500, right panel). SC indicates spinal cord; NB, nerve bundle. (C) Splenomegaly was observed in all CBFβ-SMMHC/FLT3-ITD mice (n = 17) (*). (Bar graph) Spleen weight (grams, means ± SD) for Bex/Vex controls (n = 6), CBFβ-SMMHC (n = 6), and CBFβ-SMMHC/FLT3-ITD mice (n = 7).
Dissemination of leukemic cells into peripheral organs and tissues. (A) Hematoxylin and eosin staining of tissues from Bex/Vex control and leukemic CBFβ-SMMHC/FLT3-ITD mice. Data are representative of a minimum of 5 moribund CBFβ-SMMHC/FLT3-ITD animals. Original magnifications in the left and right columns for Bex/Vex and CBFβ-SMMHC/FLT3-ITD, ×150 and ×250, respectively, except for 3 panels that were at ×100 (left panels of control and CBFβ-SMMHC/FLT3-ITD spleen and left panel of control kidney) and 1 panel at ×500 (right panel of CBFβ-SMMHC/FLT3-ITD kidney). * Leukemic infiltrates in various tissues. (B) Representative sections of femur or spinal column stained with hematoxylin and eosin (original magnifications: bone: ×150, left panel; ×50 center panel; ×500 right panel; spinal column: ×50, left; ×50 center; ×500, right panel). SC indicates spinal cord; NB, nerve bundle. (C) Splenomegaly was observed in all CBFβ-SMMHC/FLT3-ITD mice (n = 17) (*). (Bar graph) Spleen weight (grams, means ± SD) for Bex/Vex controls (n = 6), CBFβ-SMMHC (n = 6), and CBFβ-SMMHC/FLT3-ITD mice (n = 7).
Approximately 20% of moribund CBFβ-SMMHC/FLT3-ITD animals exhibited partial paralysis of the hind limbs. In these animals, extensive leukemic infiltrate was seen in the leptomeninges surrounding the spinal cord (Figure 2B). Part of the spinal canal was markedly narrowed by the infiltrative process; however, the spinal cord itself was free of leukemia. A similar mononuclear cell infiltrate was found in the vertebral bone marrow (> 95% cellularity) and skeletal muscle fibers (Figure 2B). The bone marrow of the tibias/femurs was hypercellular (> 95%-100%) and completely replaced by leukemic blasts. Normal hematopoietic elements were essentially absent (Figure 2B). Bony trabeculae were thin and showed erosion. In some areas, infiltrate extended through the periosteum into the surrounding muscle and soft tissue (Figure 2B and data not shown).
Characterization of myelopoiesis in CBFβ-SMMHC/FLT3-ITD mice
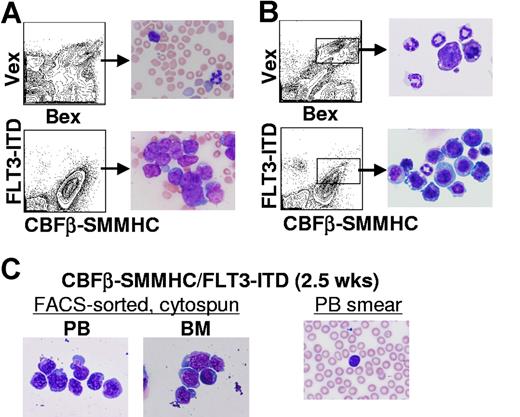
Before the development of leukemia, an extensive characterization of the hematopoietic tissues of reconstituted animals was done. As shown in Figure 3A, high percentages of myeloid blasts were evident in peripheral blood smears from CBFβ-SMMHC/FLT3-ITD animals at 2 months after transplantation (n = 6). Blasts were also evident in cytospin preparations of FACS-sorted, CBFβ-SMMHC+/FLT3-ITD+ cells isolated from bone marrow at the same time point (n = 6, Figure 3B). As early as 2 to 3 weeks after transplantation, morphologic analysis of CBFβ-SMMHC+/FLT3-ITD+ cells FACS-sorted from peripheral blood and bone marrow showed that nucleated white blood cells were highly enriched for primitive myeloid blasts that do not appear in cytospin preparations of Bex+/Vex+ cells from control animals (Figure 3B,C and data not shown). Myeloid blasts are also not detected in the periphery of animals that only express CBFβ-SMMHC until 4 to 6 months after transplantation (H.-G.K. and C.A.K., manuscript submitted). These data suggest that coexpression of CBFβ-SMMHC and FLT3-ITD is sufficient to promote rapid peripheralization of primitive myeloid cells.
Myelopoiesis in CBFβ-SMMHC/FLT3-ITD mice. (A) Representative peripheral blood smears from 2-month posttransplantation Bex/Vex (control) or CBFβ-SMMHC/FLT3-ITD mice indicating a high percentage of immature myeloid blasts in CBFβ-SMMHC/FLT3-ITD mice (original magnification ×500). (B) Wright-Giemsa staining of FACS–sorted Bex+/Vex+ cells from bone marrow of control or CBFβ-SMMHC/FLT3-ITD mice at 2 months after transplant. (C) Bex+/Vex+ cells were sorted from bone marrow or peripheral blood of CBFβ-SMMHC/FLT3-ITD mice at 2.5 weeks after transplant and cytospun onto glass slides for Wright-Giemsa staining (n = 3). A peripheral blood smear from the same animal used for isolation of FACS-sorted cells is shown at right.
Myelopoiesis in CBFβ-SMMHC/FLT3-ITD mice. (A) Representative peripheral blood smears from 2-month posttransplantation Bex/Vex (control) or CBFβ-SMMHC/FLT3-ITD mice indicating a high percentage of immature myeloid blasts in CBFβ-SMMHC/FLT3-ITD mice (original magnification ×500). (B) Wright-Giemsa staining of FACS–sorted Bex+/Vex+ cells from bone marrow of control or CBFβ-SMMHC/FLT3-ITD mice at 2 months after transplant. (C) Bex+/Vex+ cells were sorted from bone marrow or peripheral blood of CBFβ-SMMHC/FLT3-ITD mice at 2.5 weeks after transplant and cytospun onto glass slides for Wright-Giemsa staining (n = 3). A peripheral blood smear from the same animal used for isolation of FACS-sorted cells is shown at right.
Enumeration of the percentages of myeloid developmental intermediates within bone marrow of CBFβ-SMMHC/FLT3-ITD animals at 2 months after transplantation showed a strong shift in the proportion of blasts relative to mature myeloid cells (Table 2). After normalization for the percentage of Bex+/Vex+ cells in the bone marrow of each CBFβ-SMMHC/FLT3-ITD animal (range, 41%-75%), the 5 animals analyzed in Table 1 had an average of 27.8% blasts in bone marrow by 2 months after transplant (range, 11%-42%).
Differential counts of FACS-sorted Bex+/Vex+ cells isolated from bone marrow of control Bex/Vex (n = 3) or CBFβ-SMMHC/FLT3-ITD (n = 5) mice at 2 months after transplantation
| Blasts/promyelocytes . | Myelocytes/metamyelocytes . | Bands . | Eosinophils . | Monocytes . |
|---|---|---|---|---|
| Bex/Vex | ||||
| 2 | 10 | 76 | 2 | 9 |
| 3 | 21 | 66 | 2 | 8 |
| 2 | 14 | 73 | 2 | 11 |
| β-SMMHC/ FLT3-ITD | ||||
| 44 | 43 | 6 | 1 | 5 |
| 51 | 35 | 5 | 1 | 8 |
| 25 | 49 | 6 | 1 | 20 |
| 67 | 20 | 2 | 1 | 10 |
| 68 | 23 | 2 | 1 | 7 |
| Blasts/promyelocytes . | Myelocytes/metamyelocytes . | Bands . | Eosinophils . | Monocytes . |
|---|---|---|---|---|
| Bex/Vex | ||||
| 2 | 10 | 76 | 2 | 9 |
| 3 | 21 | 66 | 2 | 8 |
| 2 | 14 | 73 | 2 | 11 |
| β-SMMHC/ FLT3-ITD | ||||
| 44 | 43 | 6 | 1 | 5 |
| 51 | 35 | 5 | 1 | 8 |
| 25 | 49 | 6 | 1 | 20 |
| 67 | 20 | 2 | 1 | 10 |
| 68 | 23 | 2 | 1 | 7 |
Values represent the percentages of the indicated myeloid cell subsets as determined by typing more than 300 cells per slide.
Characterization of lymphopoiesis in CBFβ-SMMHC/FLT3-ITD mice
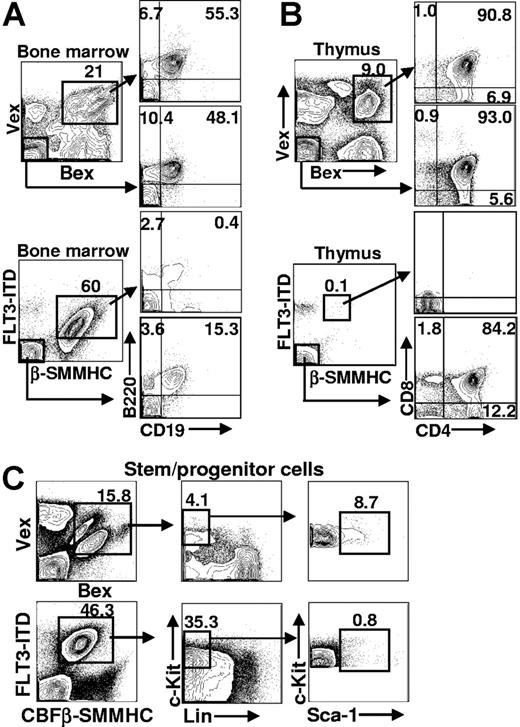
To characterize the lymphoid compartment among cells that expressed CBFβ-SMMHC/FLT3-ITD, cells isolated from bone marrow and thymus were stained with antibodies that recognized antigens on either B- or T-lineage cells. The earliest defined B-lineage intermediate expresses the B220 cell-surface marker and lacks expression of CD19 and IgM.40,41 Acquisition of CD19 expression (B220+CD19+IgM− stage) defines the earliest committed subset to the B-cell lineage. FACS analysis of CBFβ-SMMHC/FLT3-ITD-expressing bone marrow (n = 8) showed that coexpression of CBFβ-SMMHC and FLT3-ITD blocked B-cell development before CD19 expression (Figure 4A, n = 6). Analysis of the GFP− fraction of bone marrow (from the same FACS stain) indicated that B cell development proceeded normally among cells in the same animal that lacked CBFβ-SMMHC/FLT3-ITD expression. The percentage of B220+CD19+ cells (15.3%) was not statistically different from the percentage of B220+CD19+ cells in control animals (55.3% and 48.1% in Figure 4A). These results indicate that CBFβ-SMMHC/FLT3-ITD expression causes a block before, or coincident with, the earliest defined B-cell stage in bone marrow.
Representative FACS analysis of hematopoietic tissues in Bex/Vex control and CBFβ-SMMHC/FLT3-ITD–reconstituted animals. (A) Bone marrow analysis of early B-lineage development. Cells were gated for Bex/Vex expression and then analyzed for B220 and CD19 expression. Bone marrow cells that lacked Bex/Vex expression were also analyzed from the same animals. (B) FACS analysis of thymocyte development was analyzed using CD4 and CD8 staining. (C) Bone marrow cells were stained with a cocktail of antibodies to antigens on mature blood cells (Lin−) and for c-Kit and Sca-1 expression using FACS. Absolute cell numbers of c-Kit+Lin−Sca-1+ (KLS) cells that were Bex+/Vex+ was determined from total bone marrow counts and the frequency of KLS cells within the indicated gates. Numbers on plots are percentages of total cells.
Representative FACS analysis of hematopoietic tissues in Bex/Vex control and CBFβ-SMMHC/FLT3-ITD–reconstituted animals. (A) Bone marrow analysis of early B-lineage development. Cells were gated for Bex/Vex expression and then analyzed for B220 and CD19 expression. Bone marrow cells that lacked Bex/Vex expression were also analyzed from the same animals. (B) FACS analysis of thymocyte development was analyzed using CD4 and CD8 staining. (C) Bone marrow cells were stained with a cocktail of antibodies to antigens on mature blood cells (Lin−) and for c-Kit and Sca-1 expression using FACS. Absolute cell numbers of c-Kit+Lin−Sca-1+ (KLS) cells that were Bex+/Vex+ was determined from total bone marrow counts and the frequency of KLS cells within the indicated gates. Numbers on plots are percentages of total cells.
Characterization of T-cell development in the thymus of animals reconstituted with CBFβ-SMMHC/FLT3-ITD-expressing cells showed a near-complete absence of Bex/Vex-expressing cells (Figure 4B, n = 6). This was evident in animals that expressed high levels of Bex/Vex (CBFβ-SMMHC/FLT3-ITD) in the bone marrow, which indicates that early hematopoietic progenitor cells persisted in the animals (the same CBFβ-SMMHC/FLT3-ITD animal was used for stem-cell and thymic analyses in Figure 4B,C). T-cell development was also blocked at a very early stage among cells that only expressed the CBFβ-SMMHC but not by expression of the FLT3-ITD alone (Figure 4B,C).
Hematopoietic stem/progenitor cells show modest expansion as a consequence of CBFβ-SMMHC/FLT3-ITD expression
To address whether an expanded hematopoietic stem-cell population might correlate with leukemic progression observed in CBFβ-SMMHC/FLT3-ITD animals, primary transplant recipients were analyzed for the frequency and absolute number of bone marrow cells that expressed the stem/progenitor cell phenotype (defined as c-Kit+Lin−Sca-1+, “KLS,” Figure 4C). KLS cells were not expanded in absolute number among cells that only expressed CBFβ-SMMHC and were nearly absent among cells that only expressed the FLT3-ITD (data not shown). KLS cells that expressed both mutations showed a modest expansion in absolute numbers (an average of 3.7-fold at 2 months after transplantation, n = 5) compared with KLS cells from control Bex/Vex animals (n = 4). The large increase in the percentage of c-Kit+Lin−Sca-1− cells in the CBFβ-SMMHC/FLT3-ITD animals (Figure 4C, compare with same subset in Bex/Vex control) is the result of the increase in myeloblasts/promyelocytes because these cells are represented within the Sca-1− fraction of the c-Kit+Lin− subset.42
CBFβ-SMMHC/FLT3-ITD-positive cells are highly malignant on transfer into secondary hosts
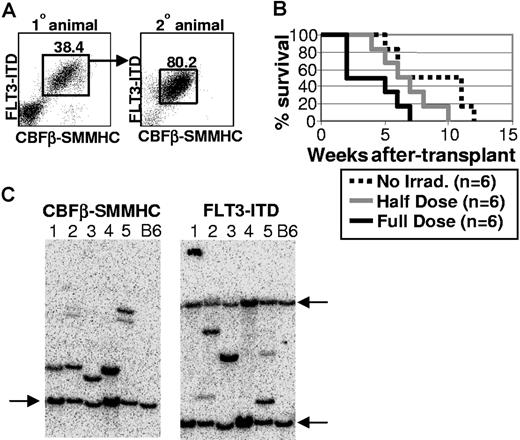
To test whether CBFβ-SMMHC/FLT3-ITD-expressing cells from primary transplant recipients would be malignant in secondary hosts, 1 million bone marrow cells were transplanted into each of 6 secondary recipients that received no irradiation, a sublethal dose of 5.0 Gy, or a lethal total exposure of 10.0 Gy given in 2 equal doses spaced by 3 hours. The Kaplan-Meier survival curve shows that all secondary transplant animals died of AML by 12 weeks after transplantation, albeit with somewhat differing kinetics depending on the irradiation dose (Figure 5B). Animals transplanted with 1 million control Bex/Vex-positive bone marrow cells were normal and continued to survive at least 4 months after transplantation, when the experiment was terminated (data not shown). These experiments were repeated with bone marrow from 2 independent primary CBFβ-SMMHC/FLT3-ITD–expressing animals with similar results. Analysis of moribund secondary animals showed that they contained expanded and highly disseminated CBFβ-SMMHC/FLT3-ITD-expressing cells that were characteristic of the bone marrow cells transplanted from the primary recipient (Figure 5A and data not shown). These results indicate that cells that coexpress CBFβ-SMMHC and FLT3-ITD are highly malignant and able to escape immune surveillance in nonirradiated, congenic secondary recipient animals.
CBFβ-SMMHC/FLT3-ITD blasts are malignant in nonirradiated secondary hosts. (A) A primary CBFβ-SMMHC/FLT3-ITD animal was killed at 3 months after transplantation, and 1 million bone marrow cells were transplanted into each of 6 secondary recipient mice that received different doses of irradiation. Representative FACS analysis of bone marrow cells from the primary donor animal and a moribund secondary recipient. Numbers on plots are percentages of total cells. (B) Kaplan-Meier survival curve for one representative experiment that was repeated using bone marrow from an independent, moribund primary CBFβ-SMMHC/FLT3-ITD animal. (C) Southern blot analysis using DNA isolated from splenocytes obtained from one of multiple secondary recipient mice transplanted at limiting dilution with bone marrow cells isolated from 5 independent, moribund primary CBFβ-SMMHC/FLT3-ITD mice. Lanes 1-5 represent one secondary recipient animal that was analyzed from each of 5 primary animals that were killed, with lane numbers representing the same DNA sample used for both blots. Blots were hybridized with radiolabeled sequences complementary to FLT3 or CBFβ sequences to detect unique proviral integrants. Arrows indicate the endogenous murine Flt3 and Cbfβ bands that were highly homologous to the radiolabeled probe derived from the human FLT3-ITD and CBFβ-SMMHC cDNA sequences. Wild-type C57BL/6 splenocytes served as control samples for each blot.
CBFβ-SMMHC/FLT3-ITD blasts are malignant in nonirradiated secondary hosts. (A) A primary CBFβ-SMMHC/FLT3-ITD animal was killed at 3 months after transplantation, and 1 million bone marrow cells were transplanted into each of 6 secondary recipient mice that received different doses of irradiation. Representative FACS analysis of bone marrow cells from the primary donor animal and a moribund secondary recipient. Numbers on plots are percentages of total cells. (B) Kaplan-Meier survival curve for one representative experiment that was repeated using bone marrow from an independent, moribund primary CBFβ-SMMHC/FLT3-ITD animal. (C) Southern blot analysis using DNA isolated from splenocytes obtained from one of multiple secondary recipient mice transplanted at limiting dilution with bone marrow cells isolated from 5 independent, moribund primary CBFβ-SMMHC/FLT3-ITD mice. Lanes 1-5 represent one secondary recipient animal that was analyzed from each of 5 primary animals that were killed, with lane numbers representing the same DNA sample used for both blots. Blots were hybridized with radiolabeled sequences complementary to FLT3 or CBFβ sequences to detect unique proviral integrants. Arrows indicate the endogenous murine Flt3 and Cbfβ bands that were highly homologous to the radiolabeled probe derived from the human FLT3-ITD and CBFβ-SMMHC cDNA sequences. Wild-type C57BL/6 splenocytes served as control samples for each blot.
To determine whether a predominant clone that expressed both CBFβ-SMMHC and FLT3-ITD was contributing to the malignant phenotype observed in primary transplant recipients, Southern blot analysis was done using genomic DNA isolated from the spleens of 5 secondary recipient animals transplanted at limiting dilution with bone marrow cells isolated from 5 independent moribund primary CBFβ-SMMHC/FLT3-ITD animals (only one secondary recipient was analyzed as a representative sample for each primary recipient (Figure 5C lanes 1-5 represent 5 independent animals). Because the 2 mutations are expressed from 2 different retroviruses that independently integrate into the genome, the presence of a single band in blots hybridized with either FLT3 or CBFβ labeled probes would be indicative of clonal hematopoiesis. As shown in Figure 5C, one primary proviral band was detected in each lane from each blot, with additional minor bands detected in lanes 2 and 5, which would represent minor contributions to hematopoiesis from an independent clone(s). These results indicate that the majority of CBFβ-SMMHC/FLT3-ITD-expressing cells in all 5 moribund primary CBFβ-SMMHC/FLT3-ITD animals were derived from a single clone that likely arose as a consequence of additional genetic changes that occurred in CBFβ-SMMHC/FLT3-ITD–expressing cells after transplantation.
Discussion
Previous studies examining human AML patient samples with the inv(16) translocation have shown a bias in the representation of specific mutations that are detected in a given patient sample. Although 60% to 70% of inv(16) patients have at least one activating mutation in N-RAS (or K-RAS), c-KIT, or FLT329,30 these mutations are not equally represented among inv(16) samples or with other chromosomal translocations that are associated with AML. Even though activating mutations in FLT3 are found in almost one third of AML cases, they are only observed in 3% to 8% of inv(16) patient samples.31-35 This suggests that there may be a reasonable degree of specificity associated with cooperativity and disease progression in AML. This specificity could result from several factors, including (1) the temporal order in which individual mutations occur with respect to each other; (2) the overall mutation rates of individual genes; (3) the degree to which given mutations cooperate with respect to differentiation arrest, deregulation of cell-cycle progression pathways, promotion of antiapoptotic pathways, peripheralization of immature cells, and escape from immunosurveillance; and (4) the cell type in which individual mutations occur. The reduced incidence of specific combinations of mutations in AML could also suggest that these combinations require additional mutations for disease progression.
In this study, we have tested whether the relatively rare combination of commonly occurring mutations (the inv(16) and FLT3-ITD) could cooperate in disease progression in a retroviral transduction/bone marrow transplant model of AML. What was perhaps most striking was the relatively potent ability of both mutations to cooperate in generating a lethal AML within 3 to 5 months after transplant and the rapid dissemination of primitive myeloid cells in the peripheral circulation within 2 to 3 weeks after transplant (Figures 1E and 3C). These observations argue that CBFβ-SMMHC and FLT3-ITD do exhibit a high degree of cooperativity when coexpressed within hematopoietic progenitor cells. This is further supported by the observation that the predominant population that expands in the reconstituted animals is expressing both mutations (Figure 1C,D). Whether coexpression of CBFβ-SMMHC and FLT3-ITD is sufficient for AML in the absence of additional mutations remains unclear, although the clonality data (Figure 5C) would argue that additional mutations have occurred by the time of significant disease onset at 3 months after transplantation. The observation that myeloblasts could readily be found in the periphery of reconstituted animals as early as 2.5 weeks after transplantation (Figure 3C) is perhaps the strongest evidence that CBFβ-SMMHC and FLT3-ITD may be sufficient for conferring a malignant phenotype on a single leukemic stem-cell clone in the absence of additional mutations, although the kinetics of disease progression (death in 3 to 5 months) is relatively slow and suggests that other mutations are required for presentation of a more aggressive AML phenotype.
Unlike CBFβ-SMMHC and FLT3-ITD, coexpression of FLT3-ITD with AML1-ETO (the other major translocation that dominantly inhibits CBF activity) does not result in selection for double-expressing cells that expand in the periphery or to lethality within the same time frame as the CBFβ-SMMHC/FLT3-ITD animals43 (Sheetal Purohit and C.A.K., unpublished observations, December 2003). Like the inv(16) and FLT3-ITD, the incidence of AML1-ETO and FLT3-ITD coexpression in human patient samples is also relatively low (on the order of 5%-10%). Our results indicate that, at least in the murine C57BL/6 model, the inv(16) is contributing a much more potent AML-promoting activity than AML1-ETO (when coexpressed with the FLT3-ITD) and that the coincident detection of specific mutations in human patient samples may not always be predictive of the outcome of functional coexpression assays in mice.
Expression of either AML1-ETO or CBFβ-SMMHC in primary bone marrow cells results in a partial block in myeloid differentiation that is measured by an increase in myeloid colony-forming cells in vitro and by a gradual increase in the percentage of myeloblasts/promyelocytes in bone marrow over time44 (H.-G.K. and C.A.K., manuscript submitted). This similarity between the AML1-ETO and CBFβ-SMMHC phenotypes suggests that both factors may be inhibiting a common downstream target (most likely, CBF) to partially arrest myeloid differentiation at the myeloblast/promyelocyte stage. Interestingly, FLT3-ITD expression with either CBFβ-SMMHC or AML1-ETO potently synergizes to further restrict myeloid differentiation at the myeloblast/promyelocyte stage, with the only difference in phenotype being the aggressive expansion of these cells in the periphery of CBFβ-SMMHC/FLT3-ITD animals. A role for FLT3 in blocking myeloid differentiation has also been noted in other studies where FLT3-ITD expression in the 32D myeloid cell line in vitro or with the PML/RARalpha translocation in vivo resulted in an enhanced myeloid differentiation block.45,46 Because sub-stoichiometric levels of CBFβ-SMMHC can completely inhibit CBF activity,7,16 it seems likely that the activated FLT3 signal is acting on another mediator of myeloblast differentiation like C/EBPα, as recently proposed.47,48
Animal modeling the coexpression of CBFβ-SMMHC and FLT3-ITD in hematopoietic progenitor cells of reconstituted mice has demonstrated that both mutations can cooperate in the pathogenesis of AML and suggests that these mutations have functional relevance in the context of human patient samples where they are observed. It remains to be determined whether coexpression of CBFβ-SMMHC with more commonly associated RTK mutations, such as activating mutations of c-KIT, will cause more rapid progression to AML than CBFβ-SMMHC/FLT3-ITD, especially if additional cooperating events are not required for AML progression. This would provide a rationale for the highly biased association of commonly occurring mutations in AML patient samples.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Scott Hiebert (Vanderbilt University) for the inv(16) cDNA and for many helpful discussions, Dr Hubert Serve (Muenster, Germany) for generously providing the FLT3-ITD clone, Dr John Kearney for use of the fluorescent microscope, Dr Larry Gartland for expert flow cytometry assistance, and Ning Wang, Rose Ko, and other members of the Klug laboratory for their help and support.
This work was supported by the National Cancer Institute (grant R01CA96798, C.A.K; and grant K01 DK067186, C.S.S.).
Authorship
Contribution: H.-G.K. performed research, designed experiments, and analyzed data; K.K., C.S.S., V.R., Y.H., and C.V.C. performed research and analyzed data; and C.A.K. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher A. Klug, University of Alabama at Birmingham, 1825 University Ave, Shelby Room 510, Birmingham, AL 35294-2182; e-mail: chrisk@uab.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal