Abstract
We have analyzed the adhesion of human and murine platelets, and of recombinant human and murine GpVI ectodomains, to synthetic triple-helical collagen-like peptides. These included 57 peptides derived from the sequence of human type III collagen and 9 peptides derived from the cyanogen bromide fragment of bovine type III collagen, α1(III)CB4. We have identified several peptides that interact with GpVI, in particular a peptide designated III-30 with the sequence GAOGLRGGAGPOGPEGGKGAAGPOGPO. Both human and murine platelets bound to peptide III-30 in a GpVI-dependent manner. III-30 also supported binding of recombinant GpVI ectodomains. Cross-linked III-30 induced aggregation of human and murine platelets, although with a lower potency than collagen-related peptide. Modifications of the peptide sequence indicated that the hydroxyproline residues play a significant role in supporting its GpVI reactivity. However, many peptides containing OGP/GPO motifs did not support adhesion to GpVI. These data indicate that the ability of a triple-helical peptide to bind GpVI is not solely determined by the presence or spatial arrangement of these OGP/GPO motifs within the peptides.
Introduction
Platelet adhesion to collagen is a key initial step in arterial thrombosis.1 Collagen interacts directly with platelets via glycoprotein VI (GpVI), which mediates platelet activation, and integrin α2β1, which supports platelet adhesion to collagen.2
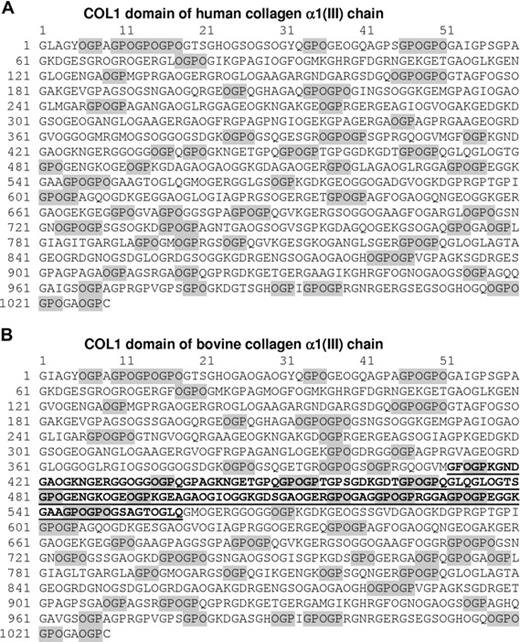
Collagen is an extracellular matrix protein containing the repeating motif (GLY-X-X′)n. Glycine (GLY, G) in every third position and a high imino acid content (proline [PRO, P]) in the X position and hydroxyproline [HYP, O] in the X′ position) promote triple-helix formation.3 Receptor interactions with collagen are dependent on this triple-helical structure4 and the primary sequence, as shown by the differential platelet reactivity of peptides prepared by cyanogen bromide fragmentation of collagen.5-8 The α1(III)CB4 fragment from bovine collagen has been identified as a platelet reactive locus (Figure 1).6
Amino acid sequence of triple-helical (COL1) domain of human and bovine type III collagen. (A) Human COL1 (triple-helical) domain from collagen α1(III) chain. OGP/GPO motifs are highlighted and comprise 20% of the full sequence. The sequence is taken from Swiss-Prot P02461 (European Bioinformatics Institute, Hinxton, United Kingdom). This is the sequence used to design type III Toolkit peptides. (B) Bovine COL1 domain from collagen α1(III) chain. OGP/GPO motifs are highlighted and comprise 21% of the full sequence. The sequence of the α1(III)CB4 fragment is underlined. The sequence is taken from Swiss-Prot Q08E14. The prolines in the X′ position are shown as the posttranslational modification hydroxyproline for both the human and bovine sequences.
Amino acid sequence of triple-helical (COL1) domain of human and bovine type III collagen. (A) Human COL1 (triple-helical) domain from collagen α1(III) chain. OGP/GPO motifs are highlighted and comprise 20% of the full sequence. The sequence is taken from Swiss-Prot P02461 (European Bioinformatics Institute, Hinxton, United Kingdom). This is the sequence used to design type III Toolkit peptides. (B) Bovine COL1 domain from collagen α1(III) chain. OGP/GPO motifs are highlighted and comprise 21% of the full sequence. The sequence of the α1(III)CB4 fragment is underlined. The sequence is taken from Swiss-Prot Q08E14. The prolines in the X′ position are shown as the posttranslational modification hydroxyproline for both the human and bovine sequences.
Subsequently, the synthetic collagen-related peptide (CRP), consisting of repeating GPO motifs, was shown to induce platelet activation9 in a GpVI-dependent manner.10,11 Platelets deficient in GpVI respond weakly to collagen10,12,13 and not at all to CRP.14 By contrast, peptides consisting of repeating GPP motifs do not support platelet binding or activation,15 indicating an important contribution of the hydroxyl group to the affinity of CRP for GpVI. Recent work using peptides comprising both GPP and GPO triplets shows that platelet and GpVI binding increases with GPO content, such that maximum interaction requires at least 4 contiguous GPO triplets.16
Although GpVI is critical in the response of platelets to collagen,17 there are no extensive stretches of OGP/GPO in collagen, unlike CRP. To explore the interaction of collagen receptors with native sequences, we synthesized a series of 57 peptides representing the 1029 residue triple-helical (COL1) domain of human type III collagen (Toolkit-III).18 Twenty percent of this COL1 domain is OGP/GPO motifs, although the longest discrete motif is GPOGPOGPO (Figure 1). Of the Toolkit-III peptides, 3 have no OGP/GPO motif and 23 contain a single HYP residue within an OGP/GPO motif. One contains 5 HYP residues and 5 contain 4 HYP residues within OGP/GPO motifs.
We have identified a major GpVI-binding locus that contains 3 HYP residues within OGP/GPO motifs. These data indicate that GpVI interacts with specific loci in collagen and that motifs other than OGP/GPO contribute to these binding sites.
Methods
Approval was obtained for these studies from the Human Biology Research Ethics Committee at the University of Cambridge. Informed consent was obtained from volunteers in accordance with the Declaration of Helsinki.
Materials
Fc receptor gamma chain–deficient (FcRγ−/−) C57/Bl6 mice were from Dr Takashi Saito19 ; wild-type (WT) C57/Bl6 mice were from Harlan (Bicester, United Kingdom); anti-GpVI antibody 10B12 and its negative control 2D4 are single-chain variable fragments (scFvs) selected from a phage display library obtained from CAT/MedImmune (Abington, United Kingdom)15 ; 6F1 was from Prof Barry Coller (Rockefeller University, New York, NY); Ha1/29 was from BD Biosciences Pharmingen (Oxford, United Kingdom); Immulon-2HB plates were from Fisher Scientific (Loughborough, United Kingdom); Horm collagen (equine tendon type I) was from Axis Shield (Dundee, United Kingdom); bovine type I collagen fibers were from Ethicon (Somerville, NJ)20 ; pepsin-digested bovine type I collagen (PDI) was previously prepared in this laboratory8 ; GR144053F was from Calbiochem (San Diego, CA); lotrafiban was from GlaxoSmithKline (King of Prussia, PA); heparin (Monoparin) was from CP Pharmaceuticals (Wrexham, United Kingdom); other reagents were from Sigma (Poole, United Kingdom).
Human platelet preparation
Citrated blood from healthy donors at the National Blood Service (Cambridge, United Kingdom) was centrifuged (240g, 15 minutes) to obtain platelet-rich plasma (PRP). PGE1 (2 μM) was added to the PRP, which was centrifuged (125g, 10 minutes) to remove residual erythrocytes. Red cell–free PRP was centrifuged (640g, 15 minutes) and the platelet pellet resuspended in 10 mL calcium-free Tyrode (CFT) (mM): NaCl, 137; NaHCO3, 11.9; NaH2PO4, 0.4; KCl, 2.7; MgCl2, 1.1; glucose, 5.6 (pH 7.4). Platelet counts were adjusted to 1.25 × 108/mL for the static adhesion assay and 2.0 × 108/mL for aggregation using a Beckman Coulter Z2 particle analyzer (High Wycombe, United Kingdom). Citrated PRP was used for aggregation assays.
Murine platelet preparation
Platelets were obtained from adult male and female WT and FcRγ−/− mice.19 FcRγ−/− mice do not express GpVI on their platelets.21 Blood was drawn under terminal anesthesia into heparin (5 IU/mL) and citrate (11 mM). PRP was obtained by centrifugation (110g, 5 minutes). PGE1 (2 μM) was added to the PRP, which was centrifuged (2000g, 6 minutes), and the platelet pellet was resuspended in CFT to a concentration of 1.25 × 108/mL for the static adhesion assay and 2.0 × 108/mL for aggregation.
Peptide synthesis, polarimetry, and cross-linking
The synthesis of Toolkit peptides and their sequences have been previously published.18 The sequences of modified peptides are shown in Table 1.
Amino acid sequences of synthetic peptides
| Peptide . | Sequence . | Tm (°C) . |
|---|---|---|
| CRP | GCO(GPO)10GCOG-NH2 | 82.3 |
| GPP | GPC(GPP)10GPC-NH2 | 4111 |
| GFOGER | GPC(GPP)5GFOGER(GPP)5GPC-NH2 | 43.927 |
| III-01 | GPC(GPP)5GLAGYOGPAGPOGPOGPOGTSGHOGSO(GPP)5GPC-NH2 | 41.918 |
| III-01a | GPC(GPP)5GLAGYOGPAGPOGPOGPOGTSGPOGPO(GPP)5GPC-NH2 | 59.0 |
| III-01b | GPC(GPP)5GLAGYOGPAGPOGPPGPPGTSGPOGPO(GPP)5GPC-NH2 | 55.0 |
| III-01c | GPC(GPP)5GPOGYOGPAGPPGPOGPOGTSGHOGSO(GPP)5GPC-NH2 | 55.1 |
| III-30 | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 42.918 |
| III-30a | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPOGPP(GPP)5GPC-NH2 | 44.7 |
| III-30b | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPPGPO(GPP)5GPC-NH2 | 45.1 |
| III-30c | GPC(GPP)5GAOGLRGGAGPPGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 43.9 |
| p1 | GPC(GPP)5GPQGLQGLOGTSGPOGEN(GPP)5GPC-NH2 | 43.5 |
| p2 | GPC(GPP)5GTSGPOGENGKOGEOGPK(GPP)5GPC-NH2 | 43.6 |
| p3 | GPC(GPP)5GKOGEOGPKGEAGAOGIO(GPP)5GPC-NH2 | 51.5 |
| p4 | GPC(GPP)5GEAGAOGIOGGKGDSGAO(GPP)5GPC-NH2 | 38.2 |
| p5 | GPC(GPP)5GGKGDSGAOGERGPOGAG(GPP)5GPC-NH2 | 34.6 |
| p6 | GPC(GPP)5GERGPOGAGGPOGPRGGA(GPP)5GPC-NH2 | 18.1 |
| p7 | GPC(GPP)5GPOGPRGGAGPOGPEGGK(GPP)5GPC-NH2 | 49.9 |
| p8 | GPC(GPP)5GPOGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 54.2 |
| p9 | GPC(GPP)5GAAGPOGPOGSAGTOGLQ(GPP)5GPC-NH2 | 50.8 |
| p8-A534 | GPC(GPP)5GPAGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 49.8 |
| p8-A546 | GPC(GPP)5GPOGPEGGKGAAGPAGPO(GPP)5GPC-NH2 | 50.9 |
| p8-A549 | GPC(GPP)5GPOGPEGGKGAAGPOGPA(GPP)5GPC-NH2 | 51.1 |
| p8-A546/549 | GPC(GPP)5GPOGPEGGKGAAGPAGPA(GPP)5GPC-NH2 | 43.8 |
| Peptide . | Sequence . | Tm (°C) . |
|---|---|---|
| CRP | GCO(GPO)10GCOG-NH2 | 82.3 |
| GPP | GPC(GPP)10GPC-NH2 | 4111 |
| GFOGER | GPC(GPP)5GFOGER(GPP)5GPC-NH2 | 43.927 |
| III-01 | GPC(GPP)5GLAGYOGPAGPOGPOGPOGTSGHOGSO(GPP)5GPC-NH2 | 41.918 |
| III-01a | GPC(GPP)5GLAGYOGPAGPOGPOGPOGTSGPOGPO(GPP)5GPC-NH2 | 59.0 |
| III-01b | GPC(GPP)5GLAGYOGPAGPOGPPGPPGTSGPOGPO(GPP)5GPC-NH2 | 55.0 |
| III-01c | GPC(GPP)5GPOGYOGPAGPPGPOGPOGTSGHOGSO(GPP)5GPC-NH2 | 55.1 |
| III-30 | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 42.918 |
| III-30a | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPOGPP(GPP)5GPC-NH2 | 44.7 |
| III-30b | GPC(GPP)5GAOGLRGGAGPOGPEGGKGAAGPPGPO(GPP)5GPC-NH2 | 45.1 |
| III-30c | GPC(GPP)5GAOGLRGGAGPPGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 43.9 |
| p1 | GPC(GPP)5GPQGLQGLOGTSGPOGEN(GPP)5GPC-NH2 | 43.5 |
| p2 | GPC(GPP)5GTSGPOGENGKOGEOGPK(GPP)5GPC-NH2 | 43.6 |
| p3 | GPC(GPP)5GKOGEOGPKGEAGAOGIO(GPP)5GPC-NH2 | 51.5 |
| p4 | GPC(GPP)5GEAGAOGIOGGKGDSGAO(GPP)5GPC-NH2 | 38.2 |
| p5 | GPC(GPP)5GGKGDSGAOGERGPOGAG(GPP)5GPC-NH2 | 34.6 |
| p6 | GPC(GPP)5GERGPOGAGGPOGPRGGA(GPP)5GPC-NH2 | 18.1 |
| p7 | GPC(GPP)5GPOGPRGGAGPOGPEGGK(GPP)5GPC-NH2 | 49.9 |
| p8 | GPC(GPP)5GPOGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 54.2 |
| p9 | GPC(GPP)5GAAGPOGPOGSAGTOGLQ(GPP)5GPC-NH2 | 50.8 |
| p8-A534 | GPC(GPP)5GPAGPEGGKGAAGPOGPO(GPP)5GPC-NH2 | 49.8 |
| p8-A546 | GPC(GPP)5GPOGPEGGKGAAGPAGPO(GPP)5GPC-NH2 | 50.9 |
| p8-A549 | GPC(GPP)5GPOGPEGGKGAAGPOGPA(GPP)5GPC-NH2 | 51.1 |
| p8-A546/549 | GPC(GPP)5GPOGPEGGKGAAGPAGPA(GPP)5GPC-NH2 | 43.8 |
Melting temperatures (Tms) were determined as previously described.18
OGP/GPO motifs are highlighted in bold, and the overlap regions between the α1(III)CB4 peptides are underlined. The 2 “GG” motifs in peptide p6 may be responsible for its low melting temperature.
Peptide melting temperatures (Tms) were determined by polarimetry to confirm triple-helical peptide conformation. Optical rotation measurements were obtained as the peptide sample was heated up. Tms were calculated by fitting the data to a theoretic equation by nonlinear regression.11
All peptides formed triple helices at room temperature except the α1(III)CB4 derivative p6, which showed a consistently low Tm (18.1°C). Data obtained using this peptide should be interpreted accordingly.
Peptides (3 mM) dissolved in 0.1 M NaHCO3 were cross-linked with 1.5 molar equivalents of SPDP (3-(2-pyridyldithio)propionic acid N-hydroxysuccinimide ester) at room temperature for 1 hour followed by dialysis against 0.01 M acetic acid for 4 hours.22 Cross-linked peptides were used only in aggregation studies.
Static platelet adhesion assay
Platelet adhesion was measured by detecting alkaline phosphatase present in platelet lysates.23 Test substrate (100 μL; 10 μg/mL in 0.01 M acetic acid) was added to 96-well plates overnight at 4°C. Excess ligand was discarded and wells were blocked with 175 μL 5% bovine serum albumin (BSA) in CFT for 1 to 2 hours, after which the wells were washed 3 times with 175 μL 0.1% BSA in CFT. Platelets (50 μL; 1.25 × 108/mL) were incubated in each well at room temperature for 1 hour. Excess platelets were discarded and the wells washed 3 times. Citrate lysis buffer (150 μL; mM: p-nitrophenyl phosphate, 3.53; trisodium citrate, 71.4; citric acid, 28.55; Triton X-100, 0.1% [vol/vol]; pH = 5.4) was added to each well for 1 hour at room temperature. Then, 100 μL 2 M NaOH was added to each well and absorbance at 405 nm was determined using a Fluostar plate reader (BMG Labtech, Aylesbury, United Kingdom).
In addition to peptides, we measured adhesion to Horm, Ethicon, and PDI collagens. CRP and the α2β1-binding peptide GFOGER were used as positive controls and GPP as a negative control (see Table 1 for sequences).
Adhesion of GpVI ectodomains (D1D2)
Receptor binding to ligands was determined using recombinant ectodomains of GpVI (D1D2) coexpressed with calmodulin. Murine and human D1D2 constructs were prepared as previously described and detected using a horseradish peroxidase (HRP)–conjugated calmodulin-binding peptide, N9A.15
The 96-well plates were coated with test substrate as described above in “Static platelet adhesion assay.” Excess ligand was discarded and wells were blocked for 2 hours at room temperature with 175 μL 5% BSA in Tris-buffered saline (TBS) (mM): NaCl, 150; TrisHCl, 50; CaCl2, 1. The wells were washed 3 times with wash buffer (0.1% BSA in TBS) and then 50 μL D1D2 (500 μg/mL in wash buffer) was added to each well for 2 hours at room temperature. Excess D1D2 was discarded and the plate washed 4 times. HRP-conjugated N9A (100 μL) was added to each well for 1 hour at room temperature. This was discarded and the wells were rewashed 4 times. HRP substrate (100 μL, KPL TMB Sure Blue Reserve; Insight Biotechnology, Wembley, United Kingdom) was added to each well and incubated for 3 minutes at room temperature. The reaction was terminated with 50 μL H2SO4 (2.5 M) and the absorbance of the wells measured at 450 nm using a Fluostar plate reader.
Aggregation
Platelet aggregation was carried out as previously described24 using Chronolog (Labmedics, Manchester, United Kingdom) and BioData PAP-4 (Alpha Laboratories, Eastleigh, United Kingdom) turbidimetric aggregometers. Aggregation of human platelets was performed in citrated PRP, and in washed platelets for murine platelets.
Data analysis
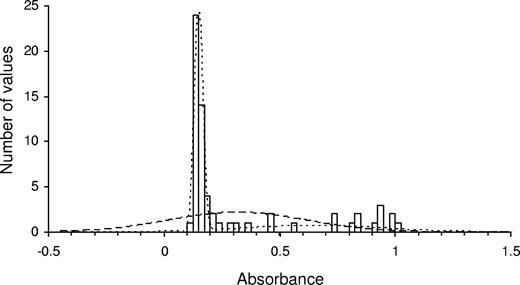
To determine which peptides supported platelet or D1D2 binding, data from each experiment was resolved into responders and nonresponders as follows. A probability density function for each dataset was calculated using 2 models: (1) a single normal distribution with 2 parameters (mean and SD of the dataset); and (2) two independent normal distributions with 5 parameters (2 means, 2 SDs, and a parameter defining the proportion of the data in each of these 2 distributions). The likelihood for each model was maximized by modifying the parameter values and a likelihood ratio test performed to determine whether model no. 1 or no. 2 best described the data.25 The test statistic is − 2ln(L1/L2), where L1 and L2 are the maximized likelihoods for model no. 1 and no. 2, and follows a chi-squared distribution. A P value was obtained using the test statistic with 3 degrees of freedom (5 minus 2 parameters). Where model no. 2 described the data best, the probabilities for each data point for each of the 2 distributions were compared and the data point was determined to belong to the distribution with the highest probability. Figure 2 shows one set of data and the representations of model no.1 and no. 2. Analysis was performed using Microsoft Excel (Redmond, WA).
Analysis of data from a single human platelet Toolkit binding experiment. The bars show the number of occurrences of any given absorbance value in the dataset (a total of 64 values, one for each adhesion substrate). The dashed line illustrates model no. 1 in which the data are represented as a single normal distribution. The dotted line illustrates model no. 2 in which the data are represented as 2 independent normal distributions, one for nonresponders (mean ± SD = 0.15 ± 0.018) and one for responders (0.65 ± 0.30). In this example, the data clearly conform to model no. 2 better than to model no. 1 (P < .001), and 21 of 64 data points were determined to be responders.
Analysis of data from a single human platelet Toolkit binding experiment. The bars show the number of occurrences of any given absorbance value in the dataset (a total of 64 values, one for each adhesion substrate). The dashed line illustrates model no. 1 in which the data are represented as a single normal distribution. The dotted line illustrates model no. 2 in which the data are represented as 2 independent normal distributions, one for nonresponders (mean ± SD = 0.15 ± 0.018) and one for responders (0.65 ± 0.30). In this example, the data clearly conform to model no. 2 better than to model no. 1 (P < .001), and 21 of 64 data points were determined to be responders.
Prior to analysis of inhibition data, equality of variances was assessed using the Levene statistic. All the raw datasets showed significant heteroscedasticity and so data were transformed as follows: y′ = ((yλ − 1)/λ) + λ, where y = raw datum, y′ = transformed datum, and λ = power of the transformation. When λ = 1, y′ = y; when λ = 0, y′ = ln(y).
λ was selected to minimize the Levene statistic. In all cases, the transformed data were no longer heteroscedastic.
Transformed data were analyzed by univariate ANOVA in which treatment groups were fixed factors and experiments random factors. Posthoc analysis was carried out using the Waller-Duncan method with a type I/type II error ratio of 100. This analysis was performed using SPSS 13.0 for Windows (Chicago, IL).
Concentration-response data were analyzed and EC50 values determined as previously described.26
Results
Adhesion of human platelets to α1(III)CB4 peptides
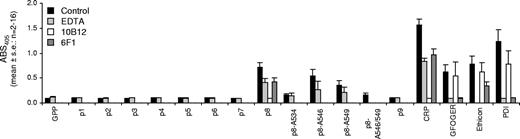
Cyanogen bromide cleavage of bovine collagen III yields fragments with differing properties. Fragment α1(III)CB4 has potent proaggregatory activity.6 We synthesized 9 peptides (Table 1) representing α1(III)CB4 to identify the reactive locus. Platelets adhered only to peptide p8. Adhesion was reduced by EDTA (2 mM) and abolished by the anti-GpVI scFv 10B12. The α2-blocking antibody 6F1 attenuated binding to p8 and to CRP, implying a nonspecific effect. Binding to GFOGER was unaffected by 10B12 but abolished by 6F1. These data suggest p8 binding is GpVI dependent (Figure 3).
Static adhesion of platelets to peptides derived from bovine α1(III)CB4 fragment. Human washed platelets adhered only to peptide p8 derived from α1(III)CB4 fragment. This binding was abolished by the anti-GpVI single chain antibody 10B12 (20 μg/mL). Data are the mean plus or minus SE (n = 2-16) for absorbance values at 405 nm.
Static adhesion of platelets to peptides derived from bovine α1(III)CB4 fragment. Human washed platelets adhered only to peptide p8 derived from α1(III)CB4 fragment. This binding was abolished by the anti-GpVI single chain antibody 10B12 (20 μg/mL). Data are the mean plus or minus SE (n = 2-16) for absorbance values at 405 nm.
HYP→ALA substitutions in p8 generated 4 new peptides (Table 1). Platelet adhesion to these peptides was attenuated. Replacing the single GPO→GPA (p8-A534) and the doublet GPOGPO→GPAGPA (p8-A546/549) abolished binding, whereas replacing either of the doublet GPO→GPA (p8-A546 and p8-A549) only attenuated binding.
Adhesion of human platelets to Toolkit peptides
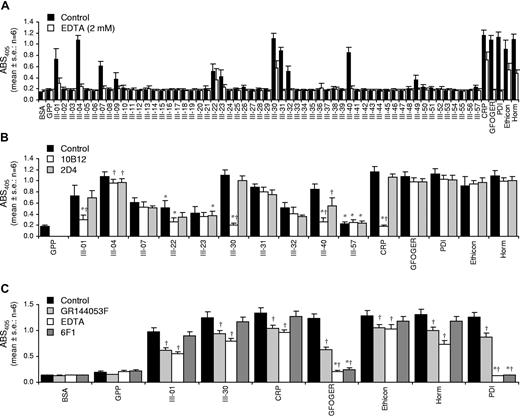
Platelet adhesion to Toolkit peptides was determined using platelets from 6 donors. Figure 4A shows adhesion of human platelets plus or minus EDTA (2 mM). Maximal adhesion was observed to CRP. Six Toolkit peptides supported binding in all 6 experiments (% control CRP binding): III-30 (94%), III-04 (92%), III-31 (72%), III-40 (69%), III-07 (45%), and III-32 (35%). Three bound in 5 of 6 experiments: III-01 (57%), III-22 (35%), and III-23 (25%). Two bound in 4 of 6 experiments: III-09 (20%) and III-49 (19%). Of the remaining 46 peptides, 34 did not bind in any experiment, 9 bound in 1 of 6 experiments, and 3 bound in 2 of 6 experiments. CRP, GFOGER, Ethicon, Horm, and PDI bound platelets in 6 of 6 experiments. GPP did not bind platelets in any.
Adhesion of human washed platelets to Toolkit-III peptides. The response is given as absorbance at 405 nm (mean ± SE: n = 6). (A) For control platelets, adhesion was observed to the following substrates (in decreasing order of magnitude): CRP (in 6 of 6 experiments); PDI (6/6); III-30 (6/6); Horm (6/6); III-04 (6/6); GFOGER (6/6); Ethicon (6/6); III-31 (6/6); III-40 (6/6); III-01 (5/6); III-07 (6/6); III-22 (5/6); III-32 (6/6); III-23 (5/6); III-09 (4/6); III-49 (4/6). Adhesion of EDTA-treated platelets was observed to the following substrates: CRP (5/6); III-30 (5/6); Ethicon (6/6); Horm (6/6); III-22 (1/6); III-01 (3/6); III-04 (3/6); III-23 (4/6). (B) Effect of GpVI-blocking antibody (10B12, 20 μg/mL) and a negative control antibody (2D4, 20 μg/mL) on static adhesion of washed human platelets to selected Toolkit peptides. *Not significantly different from adhesion to GPP. †Significantly different from control within ligand group (P < .05). (C) Effect of αIIbβ3 antagonist GR144053F (2 μM) and anti-α2β1 antibody 6F1 (5 μg/mL) on static adhesion of washed human platelets to Toolkit peptides III-01 and III-30. *Not significantly different from adhesion to GPP. †Significantly different from control within ligand group (P < .05).
Adhesion of human washed platelets to Toolkit-III peptides. The response is given as absorbance at 405 nm (mean ± SE: n = 6). (A) For control platelets, adhesion was observed to the following substrates (in decreasing order of magnitude): CRP (in 6 of 6 experiments); PDI (6/6); III-30 (6/6); Horm (6/6); III-04 (6/6); GFOGER (6/6); Ethicon (6/6); III-31 (6/6); III-40 (6/6); III-01 (5/6); III-07 (6/6); III-22 (5/6); III-32 (6/6); III-23 (5/6); III-09 (4/6); III-49 (4/6). Adhesion of EDTA-treated platelets was observed to the following substrates: CRP (5/6); III-30 (5/6); Ethicon (6/6); Horm (6/6); III-22 (1/6); III-01 (3/6); III-04 (3/6); III-23 (4/6). (B) Effect of GpVI-blocking antibody (10B12, 20 μg/mL) and a negative control antibody (2D4, 20 μg/mL) on static adhesion of washed human platelets to selected Toolkit peptides. *Not significantly different from adhesion to GPP. †Significantly different from control within ligand group (P < .05). (C) Effect of αIIbβ3 antagonist GR144053F (2 μM) and anti-α2β1 antibody 6F1 (5 μg/mL) on static adhesion of washed human platelets to Toolkit peptides III-01 and III-30. *Not significantly different from adhesion to GPP. †Significantly different from control within ligand group (P < .05).
EDTA-treated platelets bound maximally to CRP at 56% of control levels. Adhesion to CRP was observed in 5 of 6 experiments. None of the Toolkit peptides bound in all 6 experiments. III-30 bound in 5 of 6 experiments (73% of EDTA-treated platelet binding to CRP) and III-23 bound in 4 of 6 experiments (11%). III-01 (24%), III-04 (12%), and III-40 (8%) bound in 3 of 6 experiments. Of the remaining 52 peptides, 43 did not bind in any experiment, 7 bound in 1 of 6 and 2 bound in 2 of 6 experiments. EDTA-treated platelets bound to Ethicon and Horm in 6 of 6 experiments but did not bind GFOGER, PDI, and GPP in any.
Inhibition of binding by EDTA is consistent with roles for α2β1, which mediates direct attachment (eg, to GFOGER27 ), and αIIbβ3, which facilitates binding by forming platelet-platelet bonds, hence the reduced binding to CRP.
GpVI dependency of human platelet binding
The anti-GpVI scFv 10B12 abolished binding of human platelets to III-01, III-30, III-40, and CRP (P < .05). 10B12 did not inhibit adhesion to III-07, III-22, III-23, III-31, III-32, III-57, or to GFOGER, PDI, Ethicon, or Horm. There was a small but significant effect of both 10B12 and the negative control anti–HLA-A2 2D415 on adhesion to III-04. 2D4 had an incomplete inhibitory effect on binding to III-40 (Figure 4B).
Role of α2β1 and αIIbβ3 in human platelet binding
Adhesion of murine platelets to Toolkit peptides
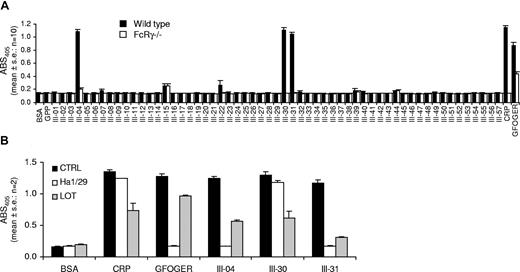
Adhesion was measured using platelets from 10 WT and 10 FcRγ−/− mice (Figure 5A). WT platelets bound maximally to CRP. The following peptides supported binding in all 10 experiments (% CRP binding): III-30 (97%), III-04 (94%), and III-31 (90%). III-15 (11.5%) bound WT platelets in 9 of 10 experiments. Of the remaining 53 peptides, 43 did not bind in any experiment, while 6 bound in 1 of 10, and 1 bound for each of 2, 3, 4, and 5 of 10 experiments. CRP and GFOGER bound WT platelets in all 10 experiments and GPP in none.
Adhesion of murine WT and FcRγ−/− washed platelets to Toolkit-III peptides. (A) The response is given as absorbance at 405 nm (mean ± SE: n = 10). For control platelets, adhesion was observed to the following substrates (in decreasing order of magnitude): CRP (in 10/10 experiments); III-30 (10/10); III-04 (10/10); III-31 (10/10); GFOGER (10/10); III-15 (9/10). (B) Effect of inhibitors of integrins α2β1 and αIIbβ3 on adhesion of murine platelets to Toolkit peptides III-04, III-30, and III-31. The anti-α2 antibody Ha1/29 (2 μg/mL) abolished binding to the α2β1 ligand GFOGER as well as III-04 and III-31, whereas it had no effect on either CRP or III-30. The αIIbβ3 antagonist lotrafiban (LOT: 10 μM) partially inhibited adhesion of platelets to all ligands, indicating a general role for platelet cross-linking by integrin in promoting adhesion (mean ± SE: n = 2).
Adhesion of murine WT and FcRγ−/− washed platelets to Toolkit-III peptides. (A) The response is given as absorbance at 405 nm (mean ± SE: n = 10). For control platelets, adhesion was observed to the following substrates (in decreasing order of magnitude): CRP (in 10/10 experiments); III-30 (10/10); III-04 (10/10); III-31 (10/10); GFOGER (10/10); III-15 (9/10). (B) Effect of inhibitors of integrins α2β1 and αIIbβ3 on adhesion of murine platelets to Toolkit peptides III-04, III-30, and III-31. The anti-α2 antibody Ha1/29 (2 μg/mL) abolished binding to the α2β1 ligand GFOGER as well as III-04 and III-31, whereas it had no effect on either CRP or III-30. The αIIbβ3 antagonist lotrafiban (LOT: 10 μM) partially inhibited adhesion of platelets to all ligands, indicating a general role for platelet cross-linking by integrin in promoting adhesion (mean ± SE: n = 2).
Maximal adhesion of FcRγ−/− platelets was to GFOGER, which was 30% of WT adhesion to CRP. III-15 (40% of binding to GFOGER) bound platelets in all 10 experiments. III-04 (23%) and III-44 (14%) bound in 9 of 10 experiments. Of the remaining 54 peptides, 46 did not bind in any experiment, while 6 bound in 1 of 10 and 1 bound for each of 2 and 4 of 10 experiments. GFOGER bound FcRγ−/− platelets in all experiments. CRP and GPP did not support any binding.
The αIIbβ3 antagonist lotrafiban (10 μM) partially inhibited binding to peptides III-04, III-30, and III-31. The anti-α2 antibody Ha1/29 (2 μg/mL) abolished binding to III-04 and III-31 but had no effect on binding to III-30 (Figure 5B).
Adhesion of recombinant GpVI ectodomains to Toolkit peptides
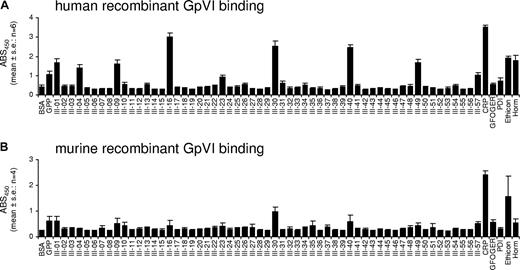
Adhesion of human GpVI ectodomains (hD1D2) to Toolkit peptides and controls was measured on 6 occasions (Figure 6A). Maximal binding was observed to CRP. Nine peptides supported binding in 6 of 6 experiments (% CRP binding): III-16 (84%), III-30 (69%), III-40 (67%), III-49 (42%), III-01 (42%), III-09 (40%), III-04 (34%), III-57 (21%), and III-23 (18%). Of the remaining 48 peptides, 34 showed no binding, 7 bound in 1; 4, in 2; 2, in 3; and 1 (III-13), in 4 experiments. hD1D2 bound to CRP, Horm, Ethicon, and GPP in 6 of 6 and to PDI in 5 of 6 experiments. Binding to GPP was unexpected and is discussed later.
Adhesion of recombinant GpVI ectodomains (D1D2-Cam) to Toolkit-III peptides. The response is given as absorbance at 450 nm (mean ± SE). (A) Human GpVI (hD1D2) bound significantly to the following peptides: CRP (6 experiments of 6); III-16 (6/6); III-30 (6/6); III-40 (6/6); Ethicon (6/6); Horm (6/6); III-49 (6/6); III-01 (6/6); III-09 (6/6); III-04 (6/6); GPP (6/6); III-57 (6/6); III-23 (6/6); PDI (5/6); III-31 (3/6); III-10 (3/6); GFOGER (3/6); III-13 (4/6). (B) Murine GpVI (mD1D2) bound in 4 of 4 experiments to CRP but in only 3 of 4 to III-30, III-01, and III-57. Overall binding of mD1D2 was clearly lower than that of hD1D2.
Adhesion of recombinant GpVI ectodomains (D1D2-Cam) to Toolkit-III peptides. The response is given as absorbance at 450 nm (mean ± SE). (A) Human GpVI (hD1D2) bound significantly to the following peptides: CRP (6 experiments of 6); III-16 (6/6); III-30 (6/6); III-40 (6/6); Ethicon (6/6); Horm (6/6); III-49 (6/6); III-01 (6/6); III-09 (6/6); III-04 (6/6); GPP (6/6); III-57 (6/6); III-23 (6/6); PDI (5/6); III-31 (3/6); III-10 (3/6); GFOGER (3/6); III-13 (4/6). (B) Murine GpVI (mD1D2) bound in 4 of 4 experiments to CRP but in only 3 of 4 to III-30, III-01, and III-57. Overall binding of mD1D2 was clearly lower than that of hD1D2.
Adhesion of murine D1D2 was measured on 4 occasions (Figure 6B). Overall, binding of mD1D2 was lower than that of hD1D2. Maximal binding was observed to CRP. No Toolkit peptide supported binding in every experiment. Three peptides supported binding in 3 of 4 experiments (% CRP binding): III-30 (33%), III-01 (16%), and III-57 (13%). Three peptides showed binding in 2 of 4 experiments: III-40 (15%), III-49 (7%), and III-26 (4%). Of the remaining 51 peptides, 37 showed no binding and 14 showed binding in 1 experiment. mD1D2 bound to CRP in 4 of 4; GFOGER and GPP, in 3 of 4; Horm and Ethicon, in 2 of 4; and PDI, in 1 of 4 experiments.
Platelet adhesion to modifications of III-01 and III-30
III-30 was the best GpVI substrate for both human and mouse platelets. It contains 3 HYP residues within OGP/GPO motifs. Peptide III-01 has the most HYP residues within such motifs, namely, 5. However, there was no detectable binding of murine platelets to III-01, and human platelet binding was heavily dependent on αIIbβ3. We synthesized modifications of these peptides to investigate the role of the HYP residues. For III-30, each HYP residue was individually replaced with a PRO. For III-01, the HIS and SER residues in the X position were replaced with PRO to increase OGP/GPO content. Subsequently, HYP residues were replaced with PRO to replicate the spatial pattern of GPO motifs found in III-30 (ie, (GPO)(GXX′)3(GPO)2) (Table 1).
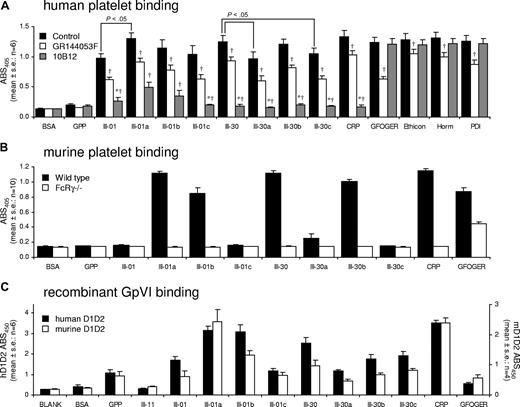
For human platelets, single HYP→PRO substitutions in III-30 had little effect on binding. III-30a and III-30c showed a small decrease in binding compared with III-30. Binding to III-30b was unchanged. 10B12 abolished platelet adhesion to III-30a, III-30b, and III-30c. Increasing OGP/GPO content in III-01a increased binding to a level comparable with CRP and III-30. Binding to III-01b and III-30c was not significantly different from that seen with III-01. Surprisingly, 10B12 did not completely abolish binding to III-01a or III-01b, compared with GPP (Figure 7A).
Adhesion of platelets and GpVI ectodomains to modifications of peptides III-01 and III-30. (A) Static adhesion of human washed platelets to modifications of Toolkit peptides III-01 and III-30 and the effects of αIIbβ3 antagonist GR144053F (2 μM) and anti-GpVI antibody 10B12 (20 μg/mL). Data are the mean plus or minus SE (n = 6) of the absorbance measured at 405 nm. *Not significantly different from adhesion to GPP; †Significantly different from control within ligand group (P < .05). (B) Static adhesion of murine WT and FcRγ−/− washed platelets to modifications of Toolkit peptides III-01 and III-30. Data are the mean plus or minus SE (n = 10) of the absorbance at 405 nm. WT platelets bound to the following peptides on 10 of 10 occasions: III-01a, III-01b, III-30, III-30b, CRP, and GFOGER. WT platelets bound to III-30a on 5 of 10 occasions. FcRγ−/− platelets bound only to GFOGER on 10 of 10 occasions. (C) Adhesion of recombinant human and murine GpVI (D1D2-Cam) to modifications of Toolkit peptides III-01 and III-30. Data are the mean plus or minus SE (n = 6 for hD1D2 and n = 4 for mD1D2) of the absorbance at 450 nm. Note the y-axes are adjusted to show equivalence of binding of hD1D2 and mD1D2.
Adhesion of platelets and GpVI ectodomains to modifications of peptides III-01 and III-30. (A) Static adhesion of human washed platelets to modifications of Toolkit peptides III-01 and III-30 and the effects of αIIbβ3 antagonist GR144053F (2 μM) and anti-GpVI antibody 10B12 (20 μg/mL). Data are the mean plus or minus SE (n = 6) of the absorbance measured at 405 nm. *Not significantly different from adhesion to GPP; †Significantly different from control within ligand group (P < .05). (B) Static adhesion of murine WT and FcRγ−/− washed platelets to modifications of Toolkit peptides III-01 and III-30. Data are the mean plus or minus SE (n = 10) of the absorbance at 405 nm. WT platelets bound to the following peptides on 10 of 10 occasions: III-01a, III-01b, III-30, III-30b, CRP, and GFOGER. WT platelets bound to III-30a on 5 of 10 occasions. FcRγ−/− platelets bound only to GFOGER on 10 of 10 occasions. (C) Adhesion of recombinant human and murine GpVI (D1D2-Cam) to modifications of Toolkit peptides III-01 and III-30. Data are the mean plus or minus SE (n = 6 for hD1D2 and n = 4 for mD1D2) of the absorbance at 450 nm. Note the y-axes are adjusted to show equivalence of binding of hD1D2 and mD1D2.
For mouse platelets, the HYP→PRO substitutions in III-30 had substantial effects. Binding to III-30c was abolished, whereas substantial binding remained to III-30b in all experiments. Adhesion to III-30a was attenuated and variable: in 5 of 10 experiments, WT platelets bound to this peptide. Binding of WT platelets was the same to III-01a and CRP. Both III-01a and III-01b bound platelets in 10 of 10 experiments, although binding was lower to III-01b. WT platelets did not bind to III-01c. FcRγ−/− platelets did not bind to any III-01 or III-30 derivatives (Figure 7B).
D1D2 binding to modifications of peptides III-01 and III-30
For III-30, the HYP→PRO substitutions caused a reduction in hD1D2 binding, especially with III-30a. III-30b and III-30c bound more than III-30a but less than III-30. A similar pattern was observed for mD1D2. For hD1D2, binding was observed to each of these modified peptides in 6 of 6 experiments. For mD1D2, III-30a bound on 2 of 4; III-30b, on 3 of 4; and III-30c, on 4 of 4 occasions.
For III-01, binding increased with III-01a, most notably with mD1D2, which bound at levels similar to CRP. The same was true for hD1D2. Binding of hD1D2 to III-01b was similar to III-01a and CRP, whereas mD1D2 binding to III-01b was reduced. Binding of hD1D2 and mD1D2 to III-01c was comparable with peptide III-01 (Figure 7C).
Platelet aggregation induced by cross-linked peptides
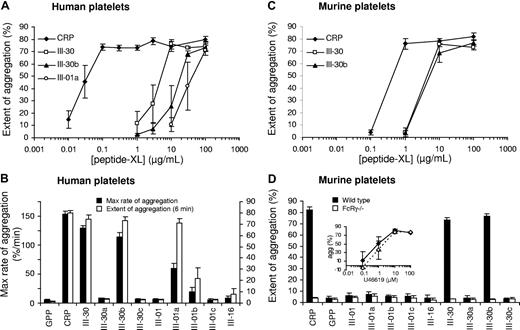
Cross-linking of peptides (-XL) is necessary for proaggregatory activity.9 Several peptides were cross-linked to determine their ability to induce aggregation (Figure 8). Non–cross-linked peptides did not induce aggregation (data not shown). At 100 μg/mL, CRP-XL, III-30-XL, III-30b-XL, and III-01a-XL induced aggregation of human platelets and CRP-XL, III-30-XL, and III-30b-XL induced aggregation of murine platelets. III-01b-XL induced weak and inconsistent aggregation of human platelets. Although III-01a-XL induced full aggregation of human platelets, the rate of aggregation was lower than for the other peptides, indicating less potent activation (Figure 8B). Concentration-response curves (Figure 8A) revealed CRP-XL as the most potent agonist (EC50 = 20 ng/mL in human platelets) compared with III-30-XL (EC50 = 2.8 μg/mL) and III-30b-XL (EC50 = 12.7 μg/mL). The difference between III-30-XL and III-30b-XL was statistically significant (P = .03). An estimate for the EC50 of III-01a-XL was not obtained as the concentration-response curve did not level off (Figure 8A). CRP-XL was less potent in murine compared with human platelets (EC50 = 321 ng/mL). However, III-30-XL and III-30b-XL were as potent in the mouse (EC50 = 1.8 and 2.3 μg/mL, respectively) as III-30-XL was in human platelets. U46619 induced aggregation in FcRγ−/− platelets; however, they did not, as expected, respond to any peptides.
Platelet aggregation induced by cross-linked peptides. Aggregation of human platelets in citrated PRP (A,B) and murine washed platelets (C,D) induced by cross-linked peptides. Peptide concentration in panels C,D was 100 μg/mL in all cases. FcRγ−/− platelets did not respond to any of the collagen peptides; however, they did respond normally to the TxA2 analog U46619 (D inset). CRP was more potent in humans than in the mouse. III-30 in human platelets and III-30 and III-30b in murine platelets showed similar levels of reactivity, whereas III-30b was less potent in the human. III-01a induced aggregation in human platelets at high concentrations. Although III-01a showed a similar extent of aggregation to the other active peptides, the response was much slower as indicated by the lower rate of aggregation. III-01a did not induce aggregation in murine platelets. Monomeric peptides induced no aggregation (data not shown). Extent of aggregation was determined 6 minutes after addition of agonist. Data shown are mean plus or minus SE (n = 3 or 4).
Platelet aggregation induced by cross-linked peptides. Aggregation of human platelets in citrated PRP (A,B) and murine washed platelets (C,D) induced by cross-linked peptides. Peptide concentration in panels C,D was 100 μg/mL in all cases. FcRγ−/− platelets did not respond to any of the collagen peptides; however, they did respond normally to the TxA2 analog U46619 (D inset). CRP was more potent in humans than in the mouse. III-30 in human platelets and III-30 and III-30b in murine platelets showed similar levels of reactivity, whereas III-30b was less potent in the human. III-01a induced aggregation in human platelets at high concentrations. Although III-01a showed a similar extent of aggregation to the other active peptides, the response was much slower as indicated by the lower rate of aggregation. III-01a did not induce aggregation in murine platelets. Monomeric peptides induced no aggregation (data not shown). Extent of aggregation was determined 6 minutes after addition of agonist. Data shown are mean plus or minus SE (n = 3 or 4).
Of the α1(III)CB4 peptides, p8-XL induced aggregation at 20 μg/mL and p7-XL at concentrations higher than 200 μg/mL. None of the others induced aggregation at concentrations higher than 200 μg/mL. The HYP→ALA-substituted α1(III)CB4 peptides did not induce aggregation at concentrations higher than 500 μg/mL (data not shown).
Discussion
Collagen has long been recognized as a platelet activator.28,29 The identification of the α1(III)CB4 fragment following cyanogen bromide cleavage of collagen as a platelet agonist was an early step toward locating platelet-binding sites within collagen. Similar studies resulted in the development of CRP,9,11 a ligand for GpVI, the major activatory collagen receptor on platelets.17 We extended this strategy by synthesizing the collagen Toolkit,18 identifying novel binding loci for α2β118 and von Willebrand factor (VWF).30 Although CRP is a selective GpVI agonist, and collagen-induced platelet activation is GpVI dependent,12 collagen itself does not contain continuous regions of GPO like CRP. We therefore used the Toolkit to continue the work begun with cyanogen bromide fragmentation to identify native GpVI-binding motifs within collagen.
Several peptides bound both human and murine platelets. Murine platelets bound substantially to III-04, III-30, and III-31. We previously reported that III-04 contains an α2β1-binding motif: GROGER.18 As expected, α2β1 blockade abolished murine platelet adhesion to III-04 and to III-31, which contains the α2β1-binding motif GMOGER,31 whereas binding to III-30 was unaffected. Murine platelet adhesion to these peptides was dependent on GpVI (Figure 5A), suggesting a functional interaction between GpVI and α2β1 in facilitating binding to III-04 and III-31, while showing that III-30 mediates adhesion via GpVI alone.
Human platelets bound to more peptides than murine platelets. In addition to III-04, III-30, and III-31, platelets bound to III-01, III-07, III-09, III-22, III-23, III-32, III-40, and III-49. EDTA attenuated binding, although it remained strongest to III-30. Platelet adhesion to III-30 was abolished by the anti-GpVI antibody 10B12, unaffected by the anti-α2 antibody 6F1, and partially inhibited by the αIIbβ3 antagonist GR144053F. αIIbβ3 facilitates binding, especially when direct adhesion to the ligand is weak, by enabling platelet-platelet interactions. In some cases (eg, III-40), GpVI-dependent adhesion was observed only because of αIIbβ3-dependent cross-linking of platelets. Therefore, III-30 supports platelet adhesion via GpVI in a divalent cation-independent manner. III-01 also showed GpVI-dependent binding. The effect of EDTA and lack of effect of 10B12 on III-04, III-07, III-31, and III-32 is consistent with their binding to α2β1.18 Weak binding to III-23 (in 4/6 experi-ments) may reflect VWF-mediated adhesion.30
We also used recombinant GpVI ectodomains (D1D2) to identify binding sites in collagen. hD1D2 bound to III-01, III-04, III-09, III-16, III-30, III-40, III-49, and III-57. The more extensive binding with hD1D2 than platelets may be because the adhesive forces required are lower for isolated GpVI than platelets. The number of peptides to which hD1D2 did not bind was striking. Surprisingly, hD1D2 bound to III-16, containing just one OGP, but not to other peptides containing OGP or GPO. III-40 contains OGPO and OGPOGP motifs and bound hD1D2, whereas III-41 contains OGPOGP, GPOGP, and OGP motifs but did not bind hD1D2.
Adhesion of mD1D2 was not as strong as hD1D2. This indicates a lower affinity of mD1D2 for CRP than hD1D2.15 mD1D2 bound best to III-30, although this was less than to CRP. mD1D2 also bound weakly to III-01 and III-57, which both have OGP/GPO motifs.
hD1D2 and mD1D2 bound noticeably to GPP, although not to many Toolkit peptides. This is surprising, since Toolkit peptides have (GPP)5 motifs at either end. Although GPP is structurally similar to GPO, and would align with the CRP-binding site on GpVI,32 the interaction is thought to be too weak to sustain binding. However, several D1D2 molecules may align and stack alongside each other, effectively increasing their overall affinity.16 That this may occur on a (GPP)10 sequence but not on shorter (GPP)5 ends of Toolkit peptides is consistent with the recent suggestion that a single GpVI molecule spans approximately 3 GXX′ triplets.16,32 Nevertheless, this interaction may be insufficient to support platelet rather than D1D2 binding to (GPP)10. Terminal “fraying” of the triple helix may also reduce the availability of GPP sequence for binding.
Peptide p8 from the α1(III)CB4 fragment6 contains most of its GpVI reactivity. This locus in p8 is equivalent to that in III-30 of the Toolkit. Furthermore, III-30-XL and p8-XL both induced aggregation. p7-XL also induced aggregation at high concentrations, although it did not support adhesion. However, p7 differs from human collagen in that it contains a GPO motif instead of GAO. This may explain its reactivity.
Peptide III-30 stands out as the best GpVI-binding substrate. Its specific sequence is GAOGLRGGAGPOGPEGGKGAAGPOGPOGP, containing 3 HYP residues within OGP/GPO motifs (bold type highlights OGP/GPO-containing portions of the peptide). However, it does not have the highest OGP/GPO content. III-01 contains 5 HYPs within GPOGPOGPO and 2 OGP motifs, and III-41 contains 4 HYPs within OGPOGP, GPOGP, and OGP motifs. Other peptides containing 4 HYP residues within OGP/GPO motifs are III-12, III-22, III-40, and III-57. The distribution of OGP/GPO motifs within these peptides does not reveal an obvious pattern for determining GpVI reactivity. The 2 longest stretches of OGP/GPO in the Toolkit are in III-01 (GPOGPOGPO) and III-09 (OGPOGPOGP). These peptides show some human but no murine platelet binding. They show similar levels of binding of hD1D2, which is approximately one third that of CRP. This is consistent with binding of a single hD1D2 across the 3 triplet motif, and CRP binding 3 hD1D2 molecules across its full length. III-04 and III-49 each have an OGPOGP motif and bind similar levels of hD1D2 to III-01 and III-09. However, III-21, III-22, and III-41 also contain OGPOGP motifs and do not bind hD1D2. III-30 and III-40 show hD1D2 binding that is approximately double that of III-01 and III-09 and two-thirds of CRP, perhaps indicating adhesion of 2 hD1D2 molecules, consistent with the presence of 2 OGP/GPO loci present in these peptides. However, other peptides containing comparable groupings (eg, III-22, III-26, III-41) do not support hD1D2 binding. Hence, any hypothesis on the relationship between amino acid sequence and GpVI reactivity must account for more than the level and location of OGP/GPO sequences. Adhesion of hD1D2 to III-16 dramatically reinforces this view.
The role of HYP in GpVI-dependent binding was investigated by modifying III-30 with HYP→PRO and p8 with HYP→ALA substitutions. We modified III-01 since it contains the greatest level of HYP while not binding murine platelets at all. For human platelets, changing GPO content in III-01 and III-30 did not dramatically change binding, although there was an increase in binding to III-01a with more OGP/GPO and a reduction in III-30a and III-30c each with one less HYP. Platelet binding to III-30b was largely unaffected, making it an excellent GpVI substrate with only 2 HYP residues within 2 GPOGP motifs spaced 5 triplets apart, a feature it shares with III-26, which did not bind platelets. This spacing is too wide to accommodate a single GpVI,16 indicating 2 distinct GpVI loci on III-30. Peptides III-01b and III-01c were designed to reproduce the spacing of GPO and GPOGPO present in III-30; however, III-01b bound murine platelets and III-01c did not, further supporting our contention that GpVI binding is not determined solely by location and quantity of OGP/GPO motifs.
All modified p8 peptides showed reduced platelet binding. p8-A534 (cf III-30c) lost all binding, while p8-A546 (cf III-30b) retained best binding. Although these results reflect those with the III-30 peptides, precise comparisons cannot be drawn, since the substituting amino acid is ALA, allowing greater flexibility of the peptide backbone, compared with PRO.
D1D2 binding to modified peptides was comparable for human and mouse. Among the III-30 analogues, binding was lowest to III-30a and highest to III-30c. This suggests that the GPOGPOGP motif in III-30c can bind D1D2. The loss of the single GPO motif abolishes murine but not human platelet binding, highlighting potential differences between murine and human GpVI function.
A K59E substitution in human GpVI reduces its affinity for collagen.15 This suggests that negative charges in collagen may coordinate with K59 in hD1D2. Conspicuously, III-30 contains a GLU residue between its 2 OGP/GPO clusters, although there is a LYS in the same region. Peptide III-16, which bound hD1D2 effectively, also has 3 GLU residues close to its OGP motif. Any role for charged residues in the GpVI-binding site can be adequately addressed only by the synthesis of new peptides in conjunction with structural studies.
III-30 also has a glycine in the X position between the 2 OGP/GPO clusters. GLY-GLY motifs increase instability of the collagen triple helix,33 resulting in extra flexibility. This could allow GpVI receptors to align more freely, enabling an interaction between them, thereby increasing the overall affinity.
Aggregation studies enabled a comparison of the activation of human and murine platelets by the modified cross-linked peptides. CRP-XL was a more potent agonist than III-30-XL, particularly in human platelets. III-30b-XL was slightly less potent than III-30-XL in human platelets, but equipotent in murine platelets. It is striking that III-30b-XL, which contains 2 HYP residues in OGP/GPO motifs, activated murine platelets, whereas III-01a-XL, which contains 6 such HYP residues, did not. The lack of activity of III-01a-XL is unlikely to be due to differences in the extent of cross-linking since it was effective in human platelets, albeit weakly. Furthermore, 2 different preparations of III-01a-XL with visibly different levels of cross-linking generated indistinguishable results. The disparity in activity between III-30a-XL, III-30b-XL, and III-30c-XL is also noteworthy. These data suggest that the particular spatial arrangement generated when collagen clusters GpVI is a significant determinant of the subsequent activation of the platelet and further support the view that the interactions of human and murine GpVI with collagen differ significantly.
CRP also interacts with nonplatelet receptors. Leukocyte-associated Ig-like receptor (LAIR-1, also a member of the Ig-like superfamily34 ) binds collagen and CRP, but not GPP. Although CRP may provide a useful template for the interaction of collagen with Ig-like receptors, the specificity of these interactions is probably not dependent solely on OGP/GPO content.35
Peptide III-30 may prove a valuable tool for modeling and understanding the GpVI-collagen interaction, and provide insights into the differences between human and murine GpVI binding to collagen. As a more physiologically agonist, it may be useful for the investigation of GpVI function in ways that CRP is not.
In conclusion, we have identified the major GpVI-binding locus in human collagen type III. III-30 is an effective substrate for both human and murine GpVI. The GpVI-binding site cannot be explained simply by the location and number of OGP/GPO motifs in collagen, but other factors must also contribute. Further work will identify those features of III-30 that contribute to GpVI-binding activity.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Barry Coller for generously supplying the antibody 6F1.
This work was supported by the British Heart Foundation (G.E.J., D.J.O., P.A.S.), the Wellcome Trust (peptide synthesis), and the Medical Research Council.
Wellcome Trust
Authorship
Contribution: G.E.J. performed research, designed research and reagents, analyzed data, and wrote the paper; N.R. synthesized peptides; J.P.L., D.J.O., and A.A. performed research; P.A.S. developed bioanalytical reagents; R.W.F. was the group head, attracted funding, designed research and reagents, assisted in writing, and reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gavin E. Jarvis, Department of Biochemistry, University of Cambridge, Downing Site, Cambridge CB2 1QW, United Kingdom; e-mail: gej1000@cam.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal