Translocations involving the immunoglobulin heavy chain locus (IGH@) at chromosome band 14q32 are common in mature B-cell neoplasms, but are rare in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Here, we report the translocation, t(6;14)(p22;q32), involving IGH@ as a novel recurrent translocation in 13 BCP-ALL patients. Fluorescence in situ hybridization and long-distance inverse polymerase chain reaction (PCR) identified ID4 as the partner gene. Breakpoints were scattered over a 19kb region centromeric of ID4. Quantitative real-time PCR showed up-regulation of ID4 mRNA. All patients had deletions of CDKN2A and PAX5 located on the short arm of chromosome 9, frequently as a result of an isochromosome, i(9)(q10) (9/13, 69%). This study defines a new subgroup of BCP-ALL characterized by ID4 over-expression and CDKN2A and PAX5 deletions. Preliminary survival data suggest that this subgroup may be associated with a good response to therapy.
Introduction
Chromosomal translocations lead to oncogene activation in hematologic malignancies, where they play an important role both at diagnosis and as an indicator of prognosis. Rearrangements involving the immunoglobulin heavy chain locus (IGH@) at chromosome band 14q32 are frequently observed in mature B-cell neoplasms,1,2 although a number are now emerging in B-cell precursor acute lymphoblastic leukemia (BCP-ALL).3,,,,,,–10
The translocation, t(6;14)(p22;q32), has been reported in 2 independent cases of BCP-ALL.7,8 It was shown that as a consequence of juxtaposing to the IGH@ enhancer, the partner gene, ID4, was overexpressed.7 We report here a further 13 cases, indicating the recurrent nature of this translocation in BCP-ALL. Moreover, we note an association with other consistent genetic features, including deletions of CDKN2A and PAX5.
Methods
Patient samples
Samples were received from patients with the translocation, t(6;14)(p22;q32), entered in the UK childhood (UKALLXI, ALL97, or ALL2003) or adult (UKALLXII) ALL treatment trials or the German ALL-BFM 2000 trial (Table 1), after informed consent was obtained in accordance with the Declaration of Helsinki. Institutional Review Board approval was provided by each participating institution (University of Southampton, University of Leicester, and University-Hospital Schleswig-Holstein).
Cytogenetic and clinical features of BCP-ALL with t(6;14)(p22;q32)
| Patient no. . | Trial . | Age, y/sex . | WBC ×109/L . | Karyotype* . | FISH . | EFS/OS, mo . | |||
|---|---|---|---|---|---|---|---|---|---|
| IGH@† . | IGH@-ID4 . | CDKN2A‡ . | PAX5§ . | ||||||
| 2125 | UKALLXI | 11/F | 3 | 47,XX,t(2;13)(p11;q1?),+5,t(6;14)(p22;q32),i(9)(q10)[cp5] | 1R1G1F (69%) | 1R1G2F (57%) | 0R2G0F (86%) | ND | 27/68¶ |
| 2817 | ALL97 | 14/M | 4 | 46,XY,i(9)(q10),t(6;14)(p22;q32)[8] | 1R1G1F (20%) | 1R1G2F (32%) | 1R2G0F (17%) | ND | 84+ |
| 3297 | ALL97 | 6/M | 6 | 46,X,-Y,der(4)t(4;9)(q2?;?),del(5)(q31), der(6)t(6;9)(p2?5;?),t(6;14)(p22;q32),add(9)(p2?), der(9)t(4;9)(q2?;p?)t(5;9)(q31;q22),-17,+22,add(22)(q?), der(22)t(10;22)(q11;q1?3),+mar,inc[cp3] | 1R1G1F (40%) | 2R1G1F (34%)‖ | 0R2G0F (32%) | 0R0G1F (32%) | 93+ |
| 3666 | UKALLXII | 45/M | 4 | 45,X,-Y,del(3)(q1?q2?9), der(6)del(6)(q1?3q?21)ins(6;3)(q1?3;q?q?) t(6;14)(p22;q32),i(9)(q10),add(12)(p13),1der(14)ins(14;12)(q32;?) t(6;14)(p22;q32),i(18)(q10)[cp7] | 1R1G1F (77%) | 1R1G2F (91%) | 1R2G0F (93%) | ND | 81+ |
| 3739 | ALL97 | 13/M | 5 | 46,XY,i(9)(q10),t(6;14)(p22;q32), del(17)(p11.1)[10]/47,idem,+5[2] | NA | NA | NA | NA | 66+ |
| 4341 | UKALLXII | 16/M | 1 | 48,XY,t(6;14)(p22;q32),+8,i(9)(q10),del(13)(q22q32),+22[7] | NA | NA | NA | NA | 66+ |
| 4767 | UKALLXII | 16/M | 11 | 46,XY,t(6;14)(p22;q32),i(9)(q10)[4] | NA | NA | NA | NA | 54+ |
| 6120 | MRD PILOT | 15/F | 2 | 47,XY,add(3)(q12),add(4)(q12),+5,+6,dic(6;9)(q1?5;p13), t(6;14)(p22;q32),i(9)(q10),add(10)(q22), der(10)t(4;10)(q21;q22),add(11)(p14)[13] | 1R1G1F (83%) | 1R1G2F (63%) | 0R2G0F (86%) | ND | 31+ |
| 7091 | UKALLXII | 34/F | 3 | 46,XX,-X,inv(1)(p1?q4?),add(4)(p?),+5,+del(5)(q2?), t(6;14)(p22;q32),dic(7;12)(p1?;p1?),add(8)(p?), der(9)t(X;9)(q1?3;p1?3),-13,+mar[cp6] | 1R1G1F (83%) | 1R1G2F (58%) | 1R2G0F (81%) | 0R0G1F (90%) | 2¶ |
| 7294 | ALL2003 | 15/M | 3 | 46–47,XY,add(3)(p2?6),del(3)(q2?5), +5,del(6)(q2?1),t(6;14)(p22;q32),inc[cp5] | 1R1G1F (28%) | 1R1G2F (58%) | 1R2G0F (59%) | 0R0G1F (31%) | NA |
| 12284 | ALL2003 | 19/F | 3 | 47,XX,t(6;14)(p22;q32),add(9)(p11),+mar[4] | 1R1G1F (65%) | 1R1G2F (65%) | 0R2G0F (40%) | 0R0G1F (64%) | NA |
| 11746 | UKALLXII | 48/M | 1 | 46,XY,t(6;14)(p22;q32),i(9)(q10),del(12)(p13),del(13)(q12q14)[16] | 1R1G1F (94%) | 1R1G2F (88%) | 1R2G0F (89%) | ND | NA |
| 16503 | ALL-BFM 2000 | 19/F | 3 | 45,XX,t(6;14)(p22;q32),i(9)(q10),del(13)(q12q33),-20[15] | 1R1G1F (82%) | 1R1G2F (88%) | 1R2G0F (74%) | ND | 30+ |
| Patient no. . | Trial . | Age, y/sex . | WBC ×109/L . | Karyotype* . | FISH . | EFS/OS, mo . | |||
|---|---|---|---|---|---|---|---|---|---|
| IGH@† . | IGH@-ID4 . | CDKN2A‡ . | PAX5§ . | ||||||
| 2125 | UKALLXI | 11/F | 3 | 47,XX,t(2;13)(p11;q1?),+5,t(6;14)(p22;q32),i(9)(q10)[cp5] | 1R1G1F (69%) | 1R1G2F (57%) | 0R2G0F (86%) | ND | 27/68¶ |
| 2817 | ALL97 | 14/M | 4 | 46,XY,i(9)(q10),t(6;14)(p22;q32)[8] | 1R1G1F (20%) | 1R1G2F (32%) | 1R2G0F (17%) | ND | 84+ |
| 3297 | ALL97 | 6/M | 6 | 46,X,-Y,der(4)t(4;9)(q2?;?),del(5)(q31), der(6)t(6;9)(p2?5;?),t(6;14)(p22;q32),add(9)(p2?), der(9)t(4;9)(q2?;p?)t(5;9)(q31;q22),-17,+22,add(22)(q?), der(22)t(10;22)(q11;q1?3),+mar,inc[cp3] | 1R1G1F (40%) | 2R1G1F (34%)‖ | 0R2G0F (32%) | 0R0G1F (32%) | 93+ |
| 3666 | UKALLXII | 45/M | 4 | 45,X,-Y,del(3)(q1?q2?9), der(6)del(6)(q1?3q?21)ins(6;3)(q1?3;q?q?) t(6;14)(p22;q32),i(9)(q10),add(12)(p13),1der(14)ins(14;12)(q32;?) t(6;14)(p22;q32),i(18)(q10)[cp7] | 1R1G1F (77%) | 1R1G2F (91%) | 1R2G0F (93%) | ND | 81+ |
| 3739 | ALL97 | 13/M | 5 | 46,XY,i(9)(q10),t(6;14)(p22;q32), del(17)(p11.1)[10]/47,idem,+5[2] | NA | NA | NA | NA | 66+ |
| 4341 | UKALLXII | 16/M | 1 | 48,XY,t(6;14)(p22;q32),+8,i(9)(q10),del(13)(q22q32),+22[7] | NA | NA | NA | NA | 66+ |
| 4767 | UKALLXII | 16/M | 11 | 46,XY,t(6;14)(p22;q32),i(9)(q10)[4] | NA | NA | NA | NA | 54+ |
| 6120 | MRD PILOT | 15/F | 2 | 47,XY,add(3)(q12),add(4)(q12),+5,+6,dic(6;9)(q1?5;p13), t(6;14)(p22;q32),i(9)(q10),add(10)(q22), der(10)t(4;10)(q21;q22),add(11)(p14)[13] | 1R1G1F (83%) | 1R1G2F (63%) | 0R2G0F (86%) | ND | 31+ |
| 7091 | UKALLXII | 34/F | 3 | 46,XX,-X,inv(1)(p1?q4?),add(4)(p?),+5,+del(5)(q2?), t(6;14)(p22;q32),dic(7;12)(p1?;p1?),add(8)(p?), der(9)t(X;9)(q1?3;p1?3),-13,+mar[cp6] | 1R1G1F (83%) | 1R1G2F (58%) | 1R2G0F (81%) | 0R0G1F (90%) | 2¶ |
| 7294 | ALL2003 | 15/M | 3 | 46–47,XY,add(3)(p2?6),del(3)(q2?5), +5,del(6)(q2?1),t(6;14)(p22;q32),inc[cp5] | 1R1G1F (28%) | 1R1G2F (58%) | 1R2G0F (59%) | 0R0G1F (31%) | NA |
| 12284 | ALL2003 | 19/F | 3 | 47,XX,t(6;14)(p22;q32),add(9)(p11),+mar[4] | 1R1G1F (65%) | 1R1G2F (65%) | 0R2G0F (40%) | 0R0G1F (64%) | NA |
| 11746 | UKALLXII | 48/M | 1 | 46,XY,t(6;14)(p22;q32),i(9)(q10),del(12)(p13),del(13)(q12q14)[16] | 1R1G1F (94%) | 1R1G2F (88%) | 1R2G0F (89%) | ND | NA |
| 16503 | ALL-BFM 2000 | 19/F | 3 | 45,XX,t(6;14)(p22;q32),i(9)(q10),del(13)(q12q33),-20[15] | 1R1G1F (82%) | 1R1G2F (88%) | 1R2G0F (74%) | ND | 30+ |
FISH data are genetic changes detected in the 4 genes shown, with percentage occurrence in parentheses.
R indicates red signal; G, green signal; F, fusion signal; EFS, event free survival; OS, overall survival; NA, not available; and ND, not done.
The normal clone has been omitted from the abnormal karyotypes.
1R1G1F indicates the presence of a translocation involving IGH@
0R2G0F and 1R2G0F indicate bi- and mono-allelic deletions, respectively, of CDKN2A.
0R0G1F indicates a monoallelic deletion of PAX5.
Patient has died.
This variant signal pattern was seen in interphase, the expected pattern (1R1G2F) was seen in metaphase with a faint IGH@ (G) signal present on the derived chromosome 14.
Cytogenetics and fluorescence in situ hybridization
Cytogenetics and fluorescence in situ hybridization (FISH) were performed on the same diagnostic samples. The involvement of IGH@ was determined by interphase FISH, using the commercially available IGH@ break-apart probe (Abbott Diagnostics, Abbott Park, IL). A home-grown dual-color, break-apart probe (consisting of BACs: RP11-377N20 and RP3-498I24; all clones from Sanger Institute, Hinxton, United Kingdom) and a dual-color, dual-fusion probe (IGH@-ID4; Figure 1A) were designed to detect the presence of the translocation in metaphase and interphase, respectively. A commercially available probe to CDKN2A (Abbott Diagnostics) and a home-grown probe to PAX5 (BACs: RP11-344B23 and RP11-297B17) were used to evaluate deletions of 9p. FISH mapping was carried out as reported previously.9
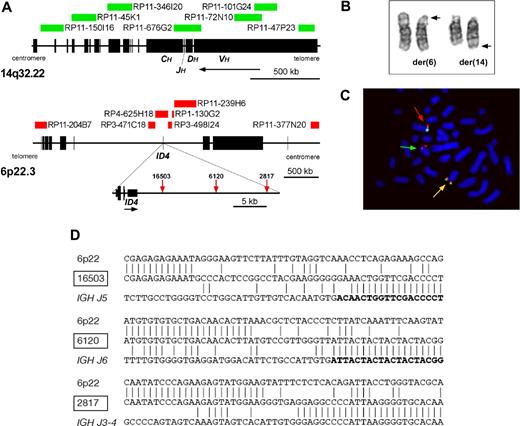
Involvement of ID4 in the translocation, t(6;14)(p22;q32). (A) Diagram showing the location of clones used in the dual-color, dual-fusion FISH probe and breakpoints cloned by LDI-PCR from IGHJ6 segments. (B) Partial G-banded karyotype showing normal and derived copies of chromosomes 6 and 14. The normal copies of each chromosome are on the left and the rearranged copies are denoted by arrows showing the breakpoints. (C) Metaphase chromosomes hybridized with clones RP11-377N20 (Spectrum green) and RP3-498I24 (Spectrum red). Two red/green fusion signals are shown, one located on the normal chromosome 6 (red arrow) and the other on the derived chromosome 6 (yellow arrow). A red signal is seen on the derived chromosome 14 (green arrow) indicating that the breakpoint is located within RP3-498I24 close to ID4. Image was acquired by staining with DAPI (Vector Laboratories, Burlingame, CA), with a Zeiss Axioskop microscope (Zeiss, Welwyn Garden City, United Kingdom) fitted with a 100× oil objective, a CCD camera (Applied Imaging, Newcastle, United Kingdom), and MacProbe image acquisition version 4.3 software (Applied Imaging). (D) Nucleotide sequences of the ID4-IGHJ junctions. Vertical lines show nucleotide identity. IGHJ segments are shown in boldface letters. Patient 2817 had an inverted fragment froms IGHJ2-3 prior to IGHJ4 segment.
Involvement of ID4 in the translocation, t(6;14)(p22;q32). (A) Diagram showing the location of clones used in the dual-color, dual-fusion FISH probe and breakpoints cloned by LDI-PCR from IGHJ6 segments. (B) Partial G-banded karyotype showing normal and derived copies of chromosomes 6 and 14. The normal copies of each chromosome are on the left and the rearranged copies are denoted by arrows showing the breakpoints. (C) Metaphase chromosomes hybridized with clones RP11-377N20 (Spectrum green) and RP3-498I24 (Spectrum red). Two red/green fusion signals are shown, one located on the normal chromosome 6 (red arrow) and the other on the derived chromosome 6 (yellow arrow). A red signal is seen on the derived chromosome 14 (green arrow) indicating that the breakpoint is located within RP3-498I24 close to ID4. Image was acquired by staining with DAPI (Vector Laboratories, Burlingame, CA), with a Zeiss Axioskop microscope (Zeiss, Welwyn Garden City, United Kingdom) fitted with a 100× oil objective, a CCD camera (Applied Imaging, Newcastle, United Kingdom), and MacProbe image acquisition version 4.3 software (Applied Imaging). (D) Nucleotide sequences of the ID4-IGHJ junctions. Vertical lines show nucleotide identity. IGHJ segments are shown in boldface letters. Patient 2817 had an inverted fragment froms IGHJ2-3 prior to IGHJ4 segment.
Long-distance inverse polymerase chain reaction
Long-distance inverse polymerase chain reaction (LDI-PCR) from IGHJ was carried out as previously described.9
Quantitative real-time PCR
Total cellular RNA, extracted from the same diagnostic samples, was available from 2 patients: 6120 and 16503. Quantitative analysis was performed using Applied Biosytems (Foster City, CA) MGB probes, as previously described.9
Results and discussion
Patient details
Thirteen BCP-ALL patients with t(6;14)(p22;q32) were identified (Figure 1B), indicating the recurrent nature of this translocation. Clinical and demographic data are shown in Table 1. All patients had a common/pre-B ALL immunophenotype with positive expression of CD10, CD19, HLA-DR and TdT where data were available. No patient showed positivity for surface IgM, T-cell, or myeloid markers. White blood cell counts were low (median, 3 × 109/L; range, 1-11 × 109/L) and patients were older than expected for BCP-ALL (median 16 years, range 6-48). All patients for whom outcome data were available (n = 10) achieved a complete remission (CR). Two patients died in CR, while 8 remain in CR; 5 for more than 5 years. Although the number of patients is small, their outcome appears to be good, especially among this age group.
Molecular characterization of 6p22 breakpoints using FISH, LDI-PCR, and quantitative real-time PCR
The involvement of IGH@ was confirmed by a positive interphase FISH result in 10 patients with available material (Table 1). The 5′ IGH@ signal relocated to 6p in metaphases.
Because a previous report had indicated involvement of ID4,7 the dual-color, break-apart FISH probe to ID4 was applied to metaphases (Figure 1C). This confirmed that the breakpoint was located close to ID4. The novel dual-color, dual-fusion probe (Figure 1A) showed a positive signal pattern in all 10 IGH@ positive cases (Table 1), indicating the reliability of this probe for the identification of this subtle translocation in interphase.
Breakpoint cloning by LDI-PCR was successful in the 3 cases with available high-molecular weight DNA. The breakpoints of IGH@ were within the IGHJ segments, while those in 6p22 were located centromeric (3′) of ID4 (Figure 1A,D).
Quantitative real-time PCR confirmed over-expression of ID4 (Assay ID, Hs_00155465, Applied Biosystems) mRNA in 2 patients with t(6;14)(p22;q32) (Table 2).
Quantitative real-time PCR analysis of ID4 and PAX5 mRNA levels
| Sample* . | Fold change of ID4 expression† . | Fold change of PAX5 expression‡ . |
|---|---|---|
| 6120 | 20.97 | 61.39 |
| 16503 | 142.68 | 33.13 |
| Normal thyroid | 111.69 | ND |
| BCP-ALL patient without t(6;14) | 0.00 | ND |
| BCP-ALL patient without t(6;14) | 0.01 | ND |
| BCP-ALL patient with no PAX5 deletion | ND | 478.82 |
| BCP-ALL patient with no PAX5 deletion | ND | 869.07 |
| Normal bone marrow§ | 1.00 | 1.00 |
| Sample* . | Fold change of ID4 expression† . | Fold change of PAX5 expression‡ . |
|---|---|---|
| 6120 | 20.97 | 61.39 |
| 16503 | 142.68 | 33.13 |
| Normal thyroid | 111.69 | ND |
| BCP-ALL patient without t(6;14) | 0.00 | ND |
| BCP-ALL patient without t(6;14) | 0.01 | ND |
| BCP-ALL patient with no PAX5 deletion | ND | 478.82 |
| BCP-ALL patient with no PAX5 deletion | ND | 869.07 |
| Normal bone marrow§ | 1.00 | 1.00 |
ND indicates not done.
The endogenous control gene, B2M, was used to normalize the results of both patients and controls (Applied Biosystems).
Controls were 2 patients without t(6;14) and normal thyroid tissue (seen to highly express ID4 according to the GeneHub-GEPIS, http://www.rbvi.ucsf.edu/Research/genentech/genehub-gepis/index.html11 ).
Controls were 2 patients with no PAX5 deletion.
ID4 and PAX5 mRNA levels were compared with expression in normal bone marrow (BD Biosciences, Oxford, United Kingdom).
Associated cytogenetic changes
An abnormality of the short arm of chromosome 9 (9p) was visible by conventional cytogenetics in 12 patients. Interestingly, this manifested as an isochromosome [i(9)(q10)], a rare mechanism of 9p loss, in 9 patients, also reported in one previously published case.8 FISH showed either a monoallelic (n = 6) or biallelic (n = 4) deletion of CDKN2A in all 10 patients tested (Table 1), including the single case (7294) without a cytogenetically visible deletion of 9p, and an additional submicroscopic 9p deletion in cases 2125 and 12284. A monoallelic deletion of PAX5 was indicated in all cases, either from the presence of an i(9)(q10) (n = 9) or by FISH (n = 4) (Table 1). 2 of the 4 patients tested by FISH for PAX5 deletions showed biallelic deletions of CDKN2A.
PAX5 mRNA (Assay ID, Hs_01045950, Applied Biosystems) expression was found to be decreased in patients 6120 and 16503 (Table 2).
ID4 is one of 4 members of the basic helix-loop-helix (bHLH) family of transcription factors, which act as transcription inhibitory proteins.12 They play a role in regulation of numerous cellular processes, including growth, differentiation, senescence and apoptosis.13 ID proteins have recently been shown to play a unique role in hematopoeitic cell development14 and to induce apoptosis in a variety of cell types,15,16 including B-lymphocytes.17 Consistent with a tumor-suppressor role, down-regulation of ID4 has been reported in both mouse and human leukemias at a high frequency, arising from aberrant methylation within the promoter region of the gene.18 In contrast, ID gene family members have been reported to show overexpression in a range of cancer types,19,,,–23 as well as one case of BCP-ALL with t(6;14)(p22;q32).7
Collectively, these observations indicate that the effects of ID4 appear to be context dependent; leading to either increased proliferation or increased cell death according to which cell lineage is involved. It could be that ID4 expression differentially affects lymphoid cells compared with myeloid cells. In this regard, ID4 appears to act in a similar manner to the CEBP proteins, whose expression in most instances results in growth suppression but, as we have previously postulated, aberrant expression of CEBP proteins may lead to transformation in some B-cell precursors.9,10
Although CDKN2A and PAX5 deletions have been well documented in all subgroups of ALL,24,–26 their consistent loss in association with t(6;14)(p22;q32) indicates an interaction between these pathways and a pivotal role for one or both genes. A recent extensive study showed a decrease in PAX5 mRNA expression in 31.7% of childhood BCP-ALL.26 They reported diminished PAX5 expression or loss of PAX5 functions occurring via a number of different mechanisms including: deletion (with retention of an apparently normal second allele, suggesting haploinsufficiency); translocation, resulting in PAX5 fusion genes; epigenetic silencing; or, more rarely, mutation. The mutation status of the second PAX5 allele could not be determined in this study due to lack of material.
This is the first report of a recurrent, novel subgroup of IGH@ positive BCP-ALL with t(6;14)(p22;q32), resulting in deregulated expression of ID4 in cooperation with loss of CDKN2A and PAX5. These findings implicate ID4 as a dominant oncogene in these patients.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank all members of the UK Cancer Cytogenetics Group for providing cytogenetic data and the Clinical Trial Service Unit (CTSU, University of Oxford, United Kingdom) for providing clinical data. This study could not have been performed without the dedication of the United Kingdom Children's Cancer and Leukemia Group and Adult Leukemia Working Party and their members, who designed and coordinated the clinical trials through which these patients were identified and in which they were treated.
This work was supported by Leukaemia Research (London, United Kingdom), Medical Research Council (London, United Kingdom), Deutsche Krebshilfe (Bonn, Germany), and Kider-Krebs-Initiative Buchholx (Holm-Seppensen, Germany).
Authorship
Contribution: L.J.R., C.J.H., M.J.S.D., and R.S. designed the research and wrote the paper. L.J.R., L.H., I.N., S.G., T.A., A.M., K.S., and E.L.K. carried out the experiments and analyzed the data. F.R., H.M., and A.C. provided clinical material. A.V.M provided clinical data. J.C.S played a technical supervisory role. All authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor Christine J. Harrison, PhD, FRCPath, Leukaemia Research Cytogenetics Group, Cancer Sciences Division, University of Southampton, MP822, Duthie Building, Southampton General Hospital, Tremona Road, Southampton, SO16 6YD, United Kingdom; e-mail: Harrison@soton.ac.uk.