Abstract
The ability to rearrange the germ-line DNA to generate antibody diversity is an essential prerequisite for the production of a functional repertoire. While this is essential to prevent infections, it also represents the “Achilles heal” of the B-cell lineage, occasionally leading to malignant transformation of these cells by translocation of protooncogenes into the immunoglobulin (Ig) loci. However, in evolutionary terms this is a small price to pay for a functional immune system. The study of the configuration and rearrangements of the Ig gene loci has contributed extensively to our understanding of the natural history of development of myeloma. In addition to this, the analysis of Ig gene rearrangements in B-cell neoplasms provides information about the clonal origin of the disease, prognosis, as well as providing a clinical useful tool for clonality detection and minimal residual disease monitoring. Herein, we review the data currently available on both Ig gene rearrangements and protein patterns seen in myeloma with the aim of illustrating how this knowledge has contributed to our understanding of the pathobiology of myeloma.
Introduction
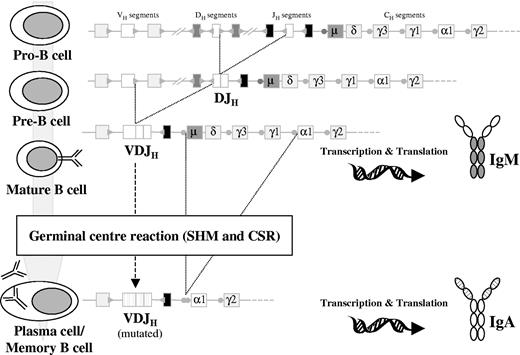
During early B-cell differentiation in the bone marrow (BM) the variable (V), diversity (D), and joining (J) gene segments of the immunoglobulin (Ig) genes are rearranged in an ordered fashion to generate the primary Ig repertoire. Ig heavy chain gene (IGH) rearrangement precedes Ig light chain gene rearrangement, and DH to JH joining precedes VH to DJH joining.1,2 These rearrangements are mediated by a tightly regulated enzymatic machinery involving several different proteins operating at the DNA level, which are controlled by the recombination signal sequences (RSSs) flanking each gene segment (Figure 1).3,4 The process of allelic exclusion ensures that once a functional VDJH rearrangement has been achieved, the other IGH allele is generally excluded from further recombination attempts.5 Following successful IGH recombination, the Ig light chain loci proceed to rearrange; initial attempts occur at the Ig kappa locus (IGK), and if a functional IGK rearrangement is not achieved the Ig lambda locus (IGL) undergoes recombination.6,7 Usually, rearrangements of the IGL locus are accompanied by deletion of the nonfunctional IGK rearrangements (Figure 2).7,8
Schematic diagram ofIGHgene rearrangement, CSR, and SHM during B-cell development. DH to JH recombination occurs at the pro-B stage, producing an incomplete DJH rearrangement. This is followed by VH to DJH recombination at the pre-B stage that will give rise to a complete VDJH rearrangement. If this VDJH is functional, the cell will develop into a mature B-cell–expressing surface IgM (IgM+). These cells can enter the GCs where SHM and CSR occur, thus becoming VH-mutated, class-switched (eg, IgA) memory B or plasma cells. Circles preceding the CH gene segments (except δ) represent the switch regions. Dotted lines on the VH, DH, and JH segments and the variable region of the Ig protein represent hypermutated genes after the SHM process.
Schematic diagram ofIGHgene rearrangement, CSR, and SHM during B-cell development. DH to JH recombination occurs at the pro-B stage, producing an incomplete DJH rearrangement. This is followed by VH to DJH recombination at the pre-B stage that will give rise to a complete VDJH rearrangement. If this VDJH is functional, the cell will develop into a mature B-cell–expressing surface IgM (IgM+). These cells can enter the GCs where SHM and CSR occur, thus becoming VH-mutated, class-switched (eg, IgA) memory B or plasma cells. Circles preceding the CH gene segments (except δ) represent the switch regions. Dotted lines on the VH, DH, and JH segments and the variable region of the Ig protein represent hypermutated genes after the SHM process.
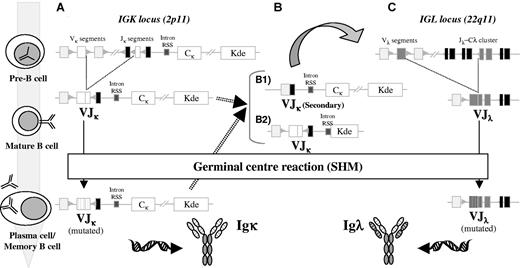
Schematic diagram of the orderedIGKandIGLgene rearrangement and SHM during B-cell development. At the pre-B cell stage, after IGH rearrangements have been completed and a μ heavy chain is produced, recombination at the IGK locus occurs between a VK and a JK segment. If such rearrangement is functional, the cell can enter the GCs and undergo SHM, with the end result a mutated IgK+ memory B or plasma cell (A). However, if the IGK rearrangements at the pre-B stage produce nonfunctional Igκ chains, additional rearrangements can occur in both IGK alleles, either before entering the GC or during the GC reaction (B). We can distinguish between 2 different types of rearrangements. First, if 5′ VK and 3′ JK segments remain still available, secondary VJK rearrangements can occur, rendering a new (functional) VJK rearrangement (B1). Alternatively, deletional rearrangements via Kde can occur, with excision of the CK region as shown or with deletion of the entire JK-CK region through direct VK to Kde recombination (B2). Such deletions are normally accompanied by rearrangement of the IGL locus in 22q11 to rescue B cells from apoptosis, which can then enter the GCs undergoing affinity maturation via the SHM process (C).
Schematic diagram of the orderedIGKandIGLgene rearrangement and SHM during B-cell development. At the pre-B cell stage, after IGH rearrangements have been completed and a μ heavy chain is produced, recombination at the IGK locus occurs between a VK and a JK segment. If such rearrangement is functional, the cell can enter the GCs and undergo SHM, with the end result a mutated IgK+ memory B or plasma cell (A). However, if the IGK rearrangements at the pre-B stage produce nonfunctional Igκ chains, additional rearrangements can occur in both IGK alleles, either before entering the GC or during the GC reaction (B). We can distinguish between 2 different types of rearrangements. First, if 5′ VK and 3′ JK segments remain still available, secondary VJK rearrangements can occur, rendering a new (functional) VJK rearrangement (B1). Alternatively, deletional rearrangements via Kde can occur, with excision of the CK region as shown or with deletion of the entire JK-CK region through direct VK to Kde recombination (B2). Such deletions are normally accompanied by rearrangement of the IGL locus in 22q11 to rescue B cells from apoptosis, which can then enter the GCs undergoing affinity maturation via the SHM process (C).
Assembly of a functional IgH-IgL protein complex on the cell surface, the so-called B-cell receptor (BCR), allows B cells to escape apoptosis and proceed with maturation.9 Immature yet immunocompetent B cells expressing surface Ig exit the BM environment and move to the secondary lymphoid organs. In the lymph node B cells pass through the mantle zone to reach the germinal center (GC), where affinity maturation occurs, following encounter with their cognate antigen.10 This process involves close interaction between B cells, antigen-processing dendritic cells, and T cells, allowing B cells carrying antigen-specific receptors on their surface to survive and proliferate. At this stage, 2 major molecular events occur: somatic hypermutation (SHM) and class switch recombination (CSR).11-14 SHM introduces point mutations in the rearranged Ig genes, producing high-affinity antibodies that recognize and optimally bind to foreign antigens. After initial contact with antigen, low-affinity IgM is produced, and CSR is the mechanism that changes that isotype to IgG-, IgA-, or IgE-generating specific antibodies with different functional characteristics (Figure 1).
As a result of having undergone the processes of SHM and CSR, the Ig genes in plasma cells (PCs) from myeloma patients are characterized by heavily mutated VH regions with no intraclonal variation and carry isotype-switched IGH genes (IgG or IgA).15 A significant number of myelomas (50% to 70%) also carry translocations targeting the switch regions of the IGH genes located at chromosome 14q32.16,17 These aberrant rearrangements juxtapose oncogenes into the proximity of the powerful IGH enhancers, driving abnormal expression of the translocated oncogenes. These characteristics together with the known biased usage for certain V and D gene segments establish a molecular archaeology for myeloma in which positive and negative selection processes shape the Ig repertoire prior to the acquisition of the malignant phenotype.18-20
Paraprotein patterns in myeloma
Heavy chain expression patterns
Recently, the International Myeloma Working Group has collected more than 10 000 cases as part of the International Staging System (ISS) study.21 In this series 60% of cases had an IgG paraprotein, 24% had IgA, 11% produced only a light chain, 3% had IgD, and 2% had biclonal or another isotype.21 It is also possible to distinguish 4 different subclasses of IgG and 2 IgA subclasses. Pooling the published data the following distribution can be seen: 68% IgG1, 17% IgG2, 11% IgG3, and 4% IgG4 for the IgG group and 93% IgA1 and 7% IgA2 for the IgA cases.22,23 These percentages largely reflect the proportions of Ig produced per day in normal individuals and, overall, they correlate with previous findings by different groups.24,25
Myeloma has been considered to be a disease of postswitch cells, but it is now well established that preswitched IgM and IgD myelomas exist, though they are uncommon (0.5% and 2%, respectively, in the Mayo Clinic series).24 Similarly, only 39 IgE myelomas have been described to date. Despite their low frequency, IgE, IgD, IgM, and nonsecretory myelomas are of special interest due to their associations and biologic implications. A collection of 33 IgE, IgD, IgM, and nonsecretory myelomas showed an overrepresentation of t(11;14)(q13;q32) compared with IgG/IgA myelomas, with 83% of cases carrying the translocation.26
Nonsecretory and light chain–only myelomas are seen in 1% to 3% and 10% to 20% of the total, respectively.21,24,27 These data have to be interpreted with caution because new techniques, such as the serum-free light chain assay, can actually detect portions of the Ig molecule in nonsecretory myelomas. Furthermore, to conclusively define a case of true nonsecretory myeloma, the absence of a paraprotein must be maintained, with M protein developing in up to 24% of patients during follow-up.24 Consequently, true nonsecretory myeloma may account for less than 1% of all myelomas.
Light chain expression patterns
Neoplastic PCs produce either κ or λ Ig light chain at a similar ratio to that normally produced in healthy individuals, 63% IgL-κ and 37% IgL-λ.24 Although such distribution is thought to be similar for the different myeloma subtypes, a bias toward Igλ expression has been noted in IgD-only myeloma. This could theoretically result as a consequence of receptor revision.28 Three IgDλ myelomas have been analyzed in detail, and no indication of receptor revision was found.29 The Vκ-Jκ rearrangements identified were all out-of-frame and without somatic mutations and, in support of this, a study on normal B cells reported similar observations.30 Therefore, it remains to be established why IgD myeloma has a bias for Igλ expression or why Cμ deletion, which seems to be the basis of the IgD-only phenotype, occurs preferentially in Igλ+ B cells.
Biclonal/dual IgH expression
Based on protein electrophoresis data, up to 2% of myelomas can secrete 2 different IgH isotypes or subclasses, with most being IgG+/IgA+ (53%) or IgG+/IgM+ (26%).31 The existence of true biclonal myeloma based on the presence of more that 1 IgH subtype must be carefully reviewed because both serum M components may share the same clonal origin with an identical variable region rather than being 2 independent clones. Thus, at the RNA level, clonotypic Cμ transcripts can be found in the BM of 68% of patients with IgG+ myeloma, consistent with the idea that these cells could be the clonogenic origin of the tumor clone.32,33 Similarly, in IgM+ myelomas Cγ transcripts with a clonogenic CDR3 region can be identified, consistent with the cell of origin being able to undergo the CSR process.34 Unfortunately, myelomas secreting 2 M components as detected by protein electrophoresis have not been studied in enough detail to allow us to comment on their clonal relationship. In this sense, they could also represent truly independent myeloma clones, with ongoing CSR detectable in subclones derived from the same myeloma progenitor or, alternatively, lack of allelic exclusion within the same myeloma cell.35
Biclonal Igκ/Igλ expression
In contrast to cases showing 2 different IgH subtypes, cases characterized by 2 different IgL peaks can be considered as truly biclonal, because no molecular mechanism for changing light chain expression within the same myeloma clone has been described so far. Biclonal myeloma cases expressing both kappa and lambda are not exceptional, although its frequency has been estimated to be only around 1% to 2%.21,36 This low frequency has led to difficulty in interpreting the clinical characteristics of these patients; some authors having found no difference in the clinical characteristics and prognosis,31 while others suggest a poor response to therapy.37 Two cases expressing cytoplasmic Igκ and Igλ but secreting only IgGλ have been described by ourselves (M.v.d.B., unpublished results, June 2002), suggesting that it is possible to produce both light chain isotypes. This observation can be explained by the fact that the Igκ light chain was unable to assemble with the Ig heavy chain and illustrates how a functional IgH-IgL assembly may be one of the mechanisms leading to allelic exclusion and monospecificity.
IGH gene rearrangements in myeloma
The IGH locus contains 27 DH segments, 6 JH segments, and up to 44 functional VH gene segments.38 Rearranged VDJH genes represent the variable domain of the IgH molecules (Figure 1), with the V-D-J joining area—or complementarity-determining region (CDR)—constituting the hypervariable domain that determines the Ig specificity for a particular antigen.2
Patterns of IGH rearrangements
Most malignant PCs secrete mature antibodies (class switched and hypermutated) coded by a functional VDJH rearrangement.1,39 Southern blot analysis of IGH rearrangements in mature B-cell malignancies shows the presence of JH rearrangements in virtually all samples, with up to 80% containing biallelic rearrangements.40 It has recently been shown by polymerase chain reaction (PCR) that up to 60% of myeloma samples contain a single DJH rearrangement on the nonproductively rearranged allele. The remaining biallelic samples usually contain 2 VDJH rearrangements, 1 of which is nonfunctional.20,41 These data are consistent with the theoretical and observed frequencies of VDJH and DJH rearrangements in normal B cells, which again supports the regulated model of allelic exclusion during B-cell development (reviewed by Jung et al42 ).
VH, DH, and JH gene segment usage
Preferential usage of Ig gene segments has been shown during the different stages of normal B-cell maturation.43,44 The VH3 family contains more members than any other and, consequently, it comprises most VH gene segments found in myeloma, followed by the VH4 and VH1 families (Table 1).18-20,45,46 VH3-30 has been consistently found to be preferentially rearranged in myeloma, followed by VH5-51, VH1-69, and VH3-23 (Table 2).18,20,46 VH3-30 and VH3-23 are the most frequently used gene segments both in normal and malignant B-cell populations,47-50 suggesting that their preferential usage is not a myeloma-specific feature but reflects the frequency seen in normal VDJH recombination. Probably the most interesting finding in this area of research is the complete absence of VH4-34 in the functional VDJH repertoire of myeloma cells,18,51 although it can be found in nonfunctional or extensively hypermutated IGH rearrangements from myeloma samples.20 The fact that VH4-34–producing cells are prevented from differentiating into antibody-producing plasma cells in healthy individuals (although not in patients with systemic lupus erythematosus)52 means that the lack of VH4-34 in the functional repertoire of myeloma cells is not a disease-specific feature but a normal B-cell differentiation mechanism of negative selection. It is well known that VH4-34 encodes antibodies that are intrinsically autoreactive, and this finding has also been proposed as an explanation for the paucity of autoimmune phenomena in myeloma. Moreover, VH4-34 is extensively represented in the normal B-cell repertoire49,52-55 and in B-cell malignancies derived from all preplasma cell stages of differentiation.56,57 Taken together, these data indicate that myeloma cells arise from a B cell in a late stage of differentiation in which negative selection of autoreactive clones has already occurred, as opposed to other B-cell malignancies, where VH4-34 is overrepresented.
VH family usage in IGH rearrangements in multiple myeloma
| . | VH1, no. (%) . | VH2, no. (%) . | VH3, no. (%) . | VH4, no. (%) . | VH5, no. (%) . | VH6, no. (%) . | No. of patients . |
|---|---|---|---|---|---|---|---|
| Baker et al, 199445 | 5 (17.9) | 1 (3.6) | 15 (53.6) | 6 (21.4) | 0 (0) | 1 (3.6) | 28 |
| Rettig et al, 199618 | 13 (18.1) | 4 (5.6) | 34 (47.2) | 14 (19.4) | 6 (8.3) | 1 (1.4) | 72 |
| Kiyoi et al, 199619 | 4 (17.4) | 1 (4.3) | 10 (43.5) | 4 (17.4) | 4 (17.4) | 0 (0.0) | 23 |
| Gonzalez et al, 200520 | 6 (8.2) | 5 (6.8) | 42 (57.5) | 15 (20.6) | 5 (6.8) | 0 (0.0) | 73 |
| Hadzidimitriou et al, 200646 | 14 (18.9) | 6 (8.1) | 31 (41.9) | 16 (21.6) | 6 (8.1) | 1 (1.4) | 74 |
| Total | 42 (15.6) | 17 (6.3) | 132 (48.9) | 55 (20.4) | 21 (7.8) | 3 (1.1) | 270 |
| . | VH1, no. (%) . | VH2, no. (%) . | VH3, no. (%) . | VH4, no. (%) . | VH5, no. (%) . | VH6, no. (%) . | No. of patients . |
|---|---|---|---|---|---|---|---|
| Baker et al, 199445 | 5 (17.9) | 1 (3.6) | 15 (53.6) | 6 (21.4) | 0 (0) | 1 (3.6) | 28 |
| Rettig et al, 199618 | 13 (18.1) | 4 (5.6) | 34 (47.2) | 14 (19.4) | 6 (8.3) | 1 (1.4) | 72 |
| Kiyoi et al, 199619 | 4 (17.4) | 1 (4.3) | 10 (43.5) | 4 (17.4) | 4 (17.4) | 0 (0.0) | 23 |
| Gonzalez et al, 200520 | 6 (8.2) | 5 (6.8) | 42 (57.5) | 15 (20.6) | 5 (6.8) | 0 (0.0) | 73 |
| Hadzidimitriou et al, 200646 | 14 (18.9) | 6 (8.1) | 31 (41.9) | 16 (21.6) | 6 (8.1) | 1 (1.4) | 74 |
| Total | 42 (15.6) | 17 (6.3) | 132 (48.9) | 55 (20.4) | 21 (7.8) | 3 (1.1) | 270 |
Preferential VH gene segment usage in IGH rearrangements in MM
| . | Hadzidimitriou et al, 200646 . | Gonzalez et al, 200520 . | Rettig et al, 199618 . | No.* (%) . |
|---|---|---|---|---|
| 3-30† | 8 | 8 | 6 | 22 (10) |
| 5-51 | 6 | 4 | 2 | 12 (5.5) |
| 3-23 | 2 | 4 | 5 | 11 (5) |
| 1-69 | 6 | 1 | 4 | 11 (5) |
| 3-15 | 3 | 5 | 2 | 10 (4.6) |
| 3-09 | 3 | 2 | 4 | 9 (4.1) |
| 4-59 | 3 | 4 | 2 | 9 (4.1) |
| 3-11 | 2 | 4 | 1 | 7 (3.2) |
| Total per series | 33 | 32 | 26 | 91 (41.6) |
| . | Hadzidimitriou et al, 200646 . | Gonzalez et al, 200520 . | Rettig et al, 199618 . | No.* (%) . |
|---|---|---|---|---|
| 3-30† | 8 | 8 | 6 | 22 (10) |
| 5-51 | 6 | 4 | 2 | 12 (5.5) |
| 3-23 | 2 | 4 | 5 | 11 (5) |
| 1-69 | 6 | 1 | 4 | 11 (5) |
| 3-15 | 3 | 5 | 2 | 10 (4.6) |
| 3-09 | 3 | 2 | 4 | 9 (4.1) |
| 4-59 | 3 | 4 | 2 | 9 (4.1) |
| 3-11 | 2 | 4 | 1 | 7 (3.2) |
| Total per series | 33 | 32 | 26 | 91 (41.6) |
Total = 219.
Includes the polymorphic VH segment 3-30.3.
DH3 and DH2 families are overrepresented in both the VDJH and DJH rearrangements in myeloma,19,20,46 a finding most likely reflecting normal frequencies of IGH rearrangements at the pro-B cell stage. However, some particular segments like DH1-07 are found in DJH rearrangements at normal frequencies whereas they are completely excluded from the VDJH repertoire.20,46 These data again suggest that particular gene segments can be negatively selected after VDJH recombination in precursor cells. Taking into account all of the above findings, the obvious conclusion is that environmentally encountered antigen plays at least some part in the etiology of myeloma and/or normal PC differentiation.
Somatic hypermutation
All VDJH rearrangements in myeloma are hypermutated, containing higher mutation rates than any other B-cell malignancy, with an average rate of 9%, with up to 23% mutations being seen in some samples.41,51,58,59 Several studies have indicated that the pattern of SHM in IGH rearrangements can be accounted for by antigen selection according to a binomial distribution model.60 However, about 40% of myeloma patients do not display such pattern in the IGH genes.
As opposed to VDJH rearrangements, most incomplete DJH rearrangements in myeloma are characterized by the absence of SHM.41 This finding has been applied in clinical strategies to improve detection rates aimed at demonstrating clonality and for PCR-based minimal residual disease (MRD) follow-up studies.61,62 Nonetheless, about 10% of DJH rearrangements show some evidence of mutation, containing less than 98% homology to the closest germ-line gene.41 This finding may have biologic significance or simply reflect the presence of previously unknown polymorphisms. However, the nature of this phenomenon is currently uncertain.
Molecular explanation of IgH-negative myeloma
Lack of IgH protein production in nonsecretory myeloma can theoretically be caused by abnormalities at various levels (ie, at the genomic level, at the transcriptional level, at the translational level, or through rapid degradation of newly synthesized IgH protein). In a series of 9 IgH-negative myeloma cases and cell lines investigated to address this issue, lack of IGH transcription was suggested as the major cause.63 However, in a study of 12 primary IgH-negative myeloma cases in which the IGH locus was analyzed in detail, gene rearrangement defects at the DNA level were confirmed as the principal cause. In 1 case a biallelic deletion of the JH region was found, whereas in 5 others only incomplete DJH rearrangements were observed, mostly associated with deletion of the other allele. This finding of exclusive DJH rearrangements in combination with mutated IGK genes can only be explained in terms of normal B-cell development through deletion of the functional IGH allele. Also, the use of CH probes showed that rearrangements of the JH and CH regions were not concordant in 5 cases, probably due to illegitimate CSR resulting in chromosomal translocation into the IGH locus affecting the other allele.64 Consistent with this, using fluorescence in situ hybridization (FISH) most myeloma samples and cell lines lacking IgH production have been found to contain chromosome translocations.65,66 Therefore, the available data suggest that aberrations at the DNA level are the major cause of nonsecretory myeloma.
IGK and IGL gene rearrangements
Both the IGK and IGL loci contain a large set of V and several J gene segments but, unlike the IGH locus, there are no D gene segments (Figure 2). The human IGK light chain locus comprises many Vκ gene segments, grouped into 7 families, and 5 Jκ gene segments.67 In addition, the kappa-deleting element (Kde), located near an RSS several kilobases downstream of the Jκ-Cκ region, can rearrange to Vκ gene segments (Vκ-Kde) or to an isolated RSS in the Jκ-Cκ intron (intron-Kde).68-70 Both types of Kde gene rearrangements lead to inactivation of the IGK allele, through deletion of the entire Jκ-Cκ area or the Cκ region only, respectively (Figure 2B). The IGL locus contains many functional and pseudo Vλ gene segments, grouped into 10 families and organized in 3 large separate clusters, as well as 7 Jλ-Cλ regions, of which only the J-Cλ1, J-Cλ2, J-Cλ3, and J-Cλ7 regions are functional.71,72
It can be anticipated that the IGK and IGL gene rearrangement patterns in Igκ+ and Igλ+ myeloma are similar to those found in other mature B-cell malignancies.46,59 Thus, Igκ+ myeloma cells harbor functional Vκ-Jκ gene rearrangements and hardly any IGL gene rearrangements, whereas Igλ+ myeloma cases show functional Vλ-Jλ gene rearrangements. Deletional rearrangements in the IGK locus are found on the nonexpressed second allele in Igκ+ myeloma cases but are even more frequent in Igλ+ cases, reflecting the end phase of nonsuccessful recombination of the IGK locus prior to IGL rearrangement (Figure 2B). In a series of 84 myeloma patients analyzed by Southern blotting, 57% of cases showed monoclonal Kde rearrangements to the Vκ/intron-RSS segments (21% monoallelic and 36% biallelic rearrangements, respectively). All patients lacking this IGK deletion and those with monoallelic deletion showed an Igκ monoclonal component in serum and/or urine, while all cases with biallelic Kde rearrangements showed an Igλ monoclonal component (36%).73
Gene segment usage and somatic hypermutation
Several reports have produced detailed analysis of Vκ and Vλ gene segment usage in myeloma. Most of the studies show a clear preference for Vκ1 family usage (53% of the total), followed by Vκ3 (25%) (Table 3).46,59,74-78 These percentages are in line with Vκ family usage in normal B cells and other B-cell lymphoproliferative disorders, largely reflecting the distribution of Vκ gene segments in the germ-line pool. In addition, a few Vκ1 gene segments seem to be used very often; these include the Vκ1-33/1D-33, Vκ1-39/1D-39, and Vκ1-6 gene segments.59,76,78 Also, the Vκ3-20 segment, believed to be indicative of autoimmunity, was seen relatively often in 3 studies.46,59,78 This finding would seem to contrast to that seen at the IGH locus, where no evidence of VDJH rearrangements associated with autoantibodies is seen. However, the number of cases in the collective studies is too small to provide evidence for the involvement of particular (self-) antigens or epitopes in the etiology of myeloma. The studies dealing with Vλ usage in Igλ myeloma cases show largely equal Vλ1 (27%), Vλ2 (29%), and Vλ3 (41%) family usage (Table 4).46,59,77,78 Within these families, no clear preference for individual gene segments was observed.
IGK gene segment usage in Igκ+ myeloma
| Study . | Vκ family usage . | Jκ gene segment usage . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vκ1 . | Vκ2 . | Vκ3 . | Vκ4 . | Vκ5-7 . | Jκ1 . | Jκ2 . | Jκ3 . | Jκ4 . | Jκ5 . | |
| Cannell et al, 199474 | 2/3 | — | — | 1/3 | — | ND | ND | ND | ND | ND |
| Wagner et al, 199475 | 3/4 | — | 1/4 | — | — | 1/4 | 1/4 | — | 2/4 | — |
| Kosmas et al, 199676 | 9/11 | — | — | 2/11 | — | ND | ND | ND | ND | ND |
| Sahota et al, 199759 | 5/9 | — | 4/9 | — | — | 2/9 | 2/9 | 1/9 | 3/9 | 1/9 |
| Kiyoi et al, 199878 | 8/17 | 3/17 | 4/17 | 1/17 | 1/17* | 5/17 | 2/17 | 3/17 | 3/17 | 4/17 |
| Hadzidimitriou et al, 200646 | 19/42 | 6/42 | 9/42 | 8/42 | — | ND | ND | ND | ND | ND |
| Total | 46/86 | 9/59 | 18/72 | 12/73 | 1/17 | 8/30 | 5/30 | 4/30 | 8/30 | 5/30 |
| Study . | Vκ family usage . | Jκ gene segment usage . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vκ1 . | Vκ2 . | Vκ3 . | Vκ4 . | Vκ5-7 . | Jκ1 . | Jκ2 . | Jκ3 . | Jκ4 . | Jκ5 . | |
| Cannell et al, 199474 | 2/3 | — | — | 1/3 | — | ND | ND | ND | ND | ND |
| Wagner et al, 199475 | 3/4 | — | 1/4 | — | — | 1/4 | 1/4 | — | 2/4 | — |
| Kosmas et al, 199676 | 9/11 | — | — | 2/11 | — | ND | ND | ND | ND | ND |
| Sahota et al, 199759 | 5/9 | — | 4/9 | — | — | 2/9 | 2/9 | 1/9 | 3/9 | 1/9 |
| Kiyoi et al, 199878 | 8/17 | 3/17 | 4/17 | 1/17 | 1/17* | 5/17 | 2/17 | 3/17 | 3/17 | 4/17 |
| Hadzidimitriou et al, 200646 | 19/42 | 6/42 | 9/42 | 8/42 | — | ND | ND | ND | ND | ND |
| Total | 46/86 | 9/59 | 18/72 | 12/73 | 1/17 | 8/30 | 5/30 | 4/30 | 8/30 | 5/30 |
Values are number/total number of cases.
— indicates no single case found; ND, not determined.
Vκ5 family usage.
IGL gene segment usage in Igλ+ myeloma
| Study . | Vλ family usage . | Jλ gene segment usage . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Vλ1 . | Vλ2 . | Vλ3 . | Vλ4-10 . | Jλ1 . | Jλ2 . | Jλ3 . | Jλ4-6 . | Jλ7 . | |
| Sahota, 199759 | 2/6 | 2/6 | 2/6 | — | 1/6 | 3/6 | — | — | 2/6 |
| Kosmas, 199877 | 2/6 | 1/6 | 2/6 | 1/6* | ND | ND | ND | ND | ND |
| Kiyoi, 199878 | 4/16 | 5/16 | 6/16 | 1/16† | 5/16 | 11/16‡ | 11/16‡ | — | — |
| Hadzidimitriou, 200646 | 11/43 | 12/43 | 19/43 | 1/43§ | 6/43 | 37/43‡ | 37/43‡ | — | — |
| Total | 19/71 | 20/71 | 29/71 | 3/65 | 6/22 | 14/22 | 14/22 | 0/22 | 2/22 |
| Study . | Vλ family usage . | Jλ gene segment usage . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Vλ1 . | Vλ2 . | Vλ3 . | Vλ4-10 . | Jλ1 . | Jλ2 . | Jλ3 . | Jλ4-6 . | Jλ7 . | |
| Sahota, 199759 | 2/6 | 2/6 | 2/6 | — | 1/6 | 3/6 | — | — | 2/6 |
| Kosmas, 199877 | 2/6 | 1/6 | 2/6 | 1/6* | ND | ND | ND | ND | ND |
| Kiyoi, 199878 | 4/16 | 5/16 | 6/16 | 1/16† | 5/16 | 11/16‡ | 11/16‡ | — | — |
| Hadzidimitriou, 200646 | 11/43 | 12/43 | 19/43 | 1/43§ | 6/43 | 37/43‡ | 37/43‡ | — | — |
| Total | 19/71 | 20/71 | 29/71 | 3/65 | 6/22 | 14/22 | 14/22 | 0/22 | 2/22 |
Values are number/total number of cases.
— indicates no single case found; ND, not determined.
Vλ6 family usage.
Vλ7 family usage.
No discrimination was made between Jλ2 and Jλ3.
Vλ5 family usage.
Ample evidence exists for SHM in Vκ and Vλ gene segments of myeloma cases.59,74-78 However, the degree of somatic mutation can be variable between individual cases, with IgD/λ myeloma cases being renowned for extensive mutation in their VH and Vλ regions.28,29 Similar to VDJH rearrangements, there are no indications of intraclonal heterogeneity of the Vκ or Vλ gene segments, implying that there is no ongoing SHM in myeloma cells. Clustering of replacement mutations has been found in the CDR regions of both Vκ and Vλ gene segments. Probability calculations according to Chang and Casali60 using the Replacement/Silent mutation distribution revealed evidence for antigen selection in 8 of 17 (47%) myeloma cases in the studies of Kosmas et al76,77 and 4 of 15 (27%) in the study of Sahota et al.59 In the latter, it was demonstrated that if significant clustering of mutation was present, it was in either VH or VL but not both. Notably, sequence changes in the IGH and IGL CDR3, which are not considered in the binomial model, can also drastically alter antigen binding, leading to an underestimation of those Ig gene rearrangements that have been selected for by antigen.50,59,79,80 Taking together these data and the data from the IGH genes, it can be concluded that most myeloma clones have been selected for by antigen.
Jκ and Jλ gene segment usage and IGL isotype
Theoretically, the differential usage of Jκ segments has no implications for the Igκ isotype, because all Vκ-Jκ gene rearrangements will eventually use the same Cκ segment. Accordingly, published data have shown the 5 different Jκ gene segments used at similar frequencies in Igκ+ myeloma, with a slight predominance of Jκ1 and Jκ4 gene segments (Table 3).59,75,78 In the case of Igλ, however, Jλ usage has direct implications for the Cλ isotype used, as a result of the Jλ segments being located directly upstream of the respective Cλ segments. Several studies of Igλ+ myelomas by Southern blotting and PCR have shown that J-Cλ2 and, to a lesser extent, J-Cλ3 are found more frequently (Table 4).59,78,81 This Jλ2 and Jλ3 usage in myeloma cases is similar to other B-cell malignancies. Remarkably, however, myeloma cases show predominance of Jλ2 over Jλ3 usage, whereas an inverse pattern is seen in other types of B-cell malignancies.81
Molecular explanations for Ig light chain negativity and aberrations in myeloma
Compared with IgH, the lack of Ig light chains has been studied much less extensively. Of the 3 true nonproducer cases studied in a series of 12 IgH-negative myelomas, no functional Ig light chain rearrangements could be amplified in 1 case.64 In the other 2 cases an in-frame IGK and biallelic IGL gene rearrangements could be found, suggesting that genomic aberrations may not be the major factor in Ig light chain negativity. A clue toward the unusual posttranslational modification of Ig light chains has come from a report describing a light chain disease case with a truncated Igκ chain due to unusual cleavage by signal peptidase.82 Although deletions in the V gene segments of the light chain loci have been shown to be a cause for truncated Ig light chain proteins in some cases of heavy chain disease (reviewed by Cogne et al83 ), this has not been shown to happen in myeloma, probably due to the low total number of cases so far studied.
IGH translocations and class switch recombination
Translocations involving the IGH locus have been reported to be the most common genetic lesion in myeloma, including t(11;14), 15% to 21%; t(4;14), 10% to 14%; t(16;14), less than 5%; and t(6;14), less than 5% of all cases.84-86 Each IGH translocation is associated with the deregulation of a specific gene (or genes) mediating ectopic expression via the influence of the powerful IGH enhancers. The t(11;14)(q13;q32) leads to the overexpression of cyclin D1,87 although it has been proposed that another gene at 11q13, MYEOV, can also be deregulated via this translocation.88 The karyotypically cryptic t(4;14)(p16;q32) involves FGFR3 and WHSC1, and it is well characterized at the molecular level.89 WHSC1 contains a SET domain and is overexpressed in t(4;14) myelomas in the form of an IGH-WHSC1 hybrid fusion transcript.89 Interestingly, the presence of IGH-WHSC1 transcripts without expression of FGFR3 is observed in a significant number (25% to 33%) of t(4:14) cases.90,91 This has been shown to occur through deletions of the whole der14 or the translocated FGFR3 gene, although different mechanisms have also been seen in some cases.90,92 The incidence of such cases (one third of all t(4;14) myelomas) and the fact that FGFR3 gene deletion is present in more than 80% of the myeloma cells in these cases suggests that it is a very early event soon after the translocation, making FGFR3 overexpression dispensable for myeloma progression/evolution. Finally, t(6;14)(p21;q32) deregulates cyclin D3 and has been reported in about 4% of the patients, whereas t(14;16)(q32;q23) deregulates c-maf and is present in about 5% of the patients. Deregulation of MYC by t(8;14)(8q24;q32) is more of a secondary event associated with tumor progression—being more commonly seen in cell lines—and is characterized by a different structure at the underlying breakpoints.93-95
In myeloma, the breakpoints on 14q32 are localized downstream of the VDJ region, in the more centromeric switch (S) regions situated upstream of all the constant genes except δ. Physiological CSR occurs between 2 S regions producing a hybrid S region; by convention, there is breakage of DNA in Sμ and a downstream Sα or Sγ region (Figure 1). Errors in the process of breakage and reassembling the double-stranded DNA during CSR may cause DNA from another chromosome to become involved, resulting in a translocation event, so-called aberrant or illegitimate CSR. Given that most myeloma cells secrete a functional IgG or IgA molecule, any illegitimate CSR must therefore occur on the other (nonfunctional) allele. Furthermore, FISH analyses have shown that 14q32 translocations are normally present in most of the myeloma clone, indicating that they are one of the initiating events in myeloma pathogenesis.
Altogether, these data point toward defects during the physiological CSR process being responsible for the translocation events seen in most myeloma cases and that these translocations into the IGH locus provide one of the initial immortalizing events in the pathogenesis of myeloma. However, late acquisition of translocations may occur during progression of the disease (eg, t(8;14)), although in these cases the structure of the breakpoints seems to be different than in primary translocations (reviewed by Gabrea et al96 ) and begs the question about the underlying mechanisms involved in these translocations.
Ontogeny of multiple myeloma
Any hypothesis about the etiology of myeloma should be able to explain the molecular characteristics found in myeloma cells that we have described so far: (a) evidence of antigen-driven selection with restricted VH segment usage and highly hypermutated Ig heavy and light chain genes with no intraclonal variation; (b) class-switched functional Ig molecules (mainly IgG or IgA) in most cases, with detectable preswitch (Cμ) clonotypic transcripts in some cases; and (c) aberrant Ig translocations in 40% to 70% of samples.
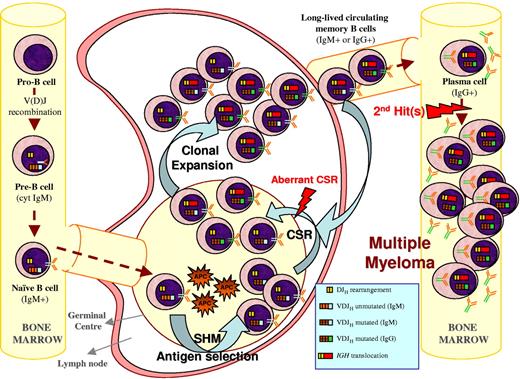
One such hypothesis is illustrated in Figure 3 and is further described here: during the GC reaction, mature B cells are selected by foreign antigens and undergo the SHM and CSR processes to provide a functional IgG or IgA plasma cell that subsequently migrates to the BM. At this stage, cells carrying Ig molecules specific for certain antigens (ie, VH4-34–positive cells) are excluded from becoming PCs. If illegitimate CSR occurs during the GC reaction while the cell can still undergo maturation to a memory B or plasma cell, it may exit the lymph node with an acquired ability to survive and proliferate as a consequence of oncogene deregulation. Alternatively, translocations might occur in the Ig light chain loci in 2p11 or 22q11 during the process of receptor editing.93 The acquired survival/proliferative ability would allow this premalignant clone of PCs to accumulate secondary hits, which will eventually occur in and deregulate critical genes, leading to the emergence of a malignant myeloma clone in the BM. The lack of intraclonal variation in the Ig genes of myeloma cells is consistent with this idea. A feature of this hypothesis is that the final malignant transformation events occur within the BM after the cell has abandoned the GC microenvironment. This hypothesis is similar to ideas put forward to explain the findings in follicular and mantle cell lymphoma, where Ig translocations are suggested to happen at the pro-B cell stage without altering the differentiation process to mature B cells.97 In fact, the t(14;18) is frequently detected in healthy individuals without evidence of developing follicular lymphoma.98
Multiple myeloma ontogeny's hypothesis exemplified by a 14q translocation–positive case. A naive B cell harboring an incomplete DJH and a functional VDJH rearrangement enters the GC where it undergoes SHM and antigen selection (eg, selection against VH4-34 IGH rearrangements), followed by CSR. During the process of physiological CSR, double-strand breaks on the switch regions of the nonfunctional allele (DJH in the example) can be resolved by joining with a different chromosome resulting in a 14q32 translocation. If this illegitimate recombination occurs prior to legitimate CSR on the functional IGH allele, a mixture of different subpopulations (eg, IgM+ and IgG+ or IgA+) will have a survival or proliferative advantage due to the translocation, which will make them long-lived memory B or plasma cells without becoming fully malignant. After one of these subpopulations leaves the GC to become a plasma cell homing to the BM (normally IgG+ or IgA+), secondary genetic hits may occur that render such cells malignant plasma cells.
Multiple myeloma ontogeny's hypothesis exemplified by a 14q translocation–positive case. A naive B cell harboring an incomplete DJH and a functional VDJH rearrangement enters the GC where it undergoes SHM and antigen selection (eg, selection against VH4-34 IGH rearrangements), followed by CSR. During the process of physiological CSR, double-strand breaks on the switch regions of the nonfunctional allele (DJH in the example) can be resolved by joining with a different chromosome resulting in a 14q32 translocation. If this illegitimate recombination occurs prior to legitimate CSR on the functional IGH allele, a mixture of different subpopulations (eg, IgM+ and IgG+ or IgA+) will have a survival or proliferative advantage due to the translocation, which will make them long-lived memory B or plasma cells without becoming fully malignant. After one of these subpopulations leaves the GC to become a plasma cell homing to the BM (normally IgG+ or IgA+), secondary genetic hits may occur that render such cells malignant plasma cells.
Switch translocations in myeloma occur preferentially in the nonfunctional allele, because only a small proportion of myeloma cases lack paraprotein expression. This suggests that at the time of the translocation, the cells are still dependent on BCR signaling, which, in turn, implies the need for a functional Ig molecule to be present at the cell surface. The presence and the molecular characteristics of the translocations in the switch regions of the IGH genes in myeloma suggest that they occur during the process of physiological CSR within the GC. On the basis of these and other observations, it can be concluded that translocation events in myeloma constitute early alterations being responsible for tumor initiation without being responsible for complete tumorigenic transformation. It is obvious, however, from gene expression and genomic studies that these translocations define subgroups of the disease with highly distinct gene signatures and outcomes.85,86,99
The idea that myeloma-specific translocations are insufficient to produce a fully malignant phenotype is also supported by the observation that the frequency of translocations in monoclonal gammopathy of undetermined significance (MGUS) and myeloma is similar, with only a small number of MGUS patients progressing to myeloma. A possible exception is the low frequency of MGUS cases with a t(4;14), which seem to progress more rapidly to myeloma. In this context, the t(4;14) could be associated with a higher rate of genomic instability that might facilitate the acquisition of new genomic alterations leading more rapidly to myeloma.
The hypothesis put forward herein is also consistent with the experimental observations of clonotypic IgM+ VDJH rearrangements in IgG or IgA myeloma patients. The explanation is that there are residual clonotypic “preswitch” cells—which may or may not contain the Ig translocation—that have differentiated into long-lived memory B cells or PCs without becoming fully malignant (Figure 3). These cells are predicted to be dependent on BCR signaling, and the consistent and ongoing interaction of these cells with the foreign antigen they are specific for may lead to clonal expansion and recirculation of these cells through the peripheral blood and BM, allowing their detection by PCR methods. Further experimental observation that would strengthen and support these ideas would be the detection of translocations involving the switch region of the IGH genes in a small percentage of memory B and nonmalignant PCs in myeloma patients as well as in PCs from healthy individuals. However, to the best of our knowledge, such studies are yet to be published.
Conclusions and future directions
Studying the “molecular archaeology” of myeloma using Ig gene rearrangement and mutation status has been particularly useful because of the lack of other tools with which to examine this process. In particular, analysis of the MGUS/myeloma transition has been particularly informative. The presence of ongoing mutation within the PCs in some MGUS cases strongly suggests that the precursor cell in MGUS is continually exposed to the process of SHM and, by inference, must exist at a level still related to the GC.100 Nonetheless, it does not exclude the possibility that cells acquire mutations in their Ig regions via other routes, as yet not fully defined. What is certain, however, is that in myeloma this apparent intraclonal diversity is absent and, on relapse, most myeloma samples secrete the same paraprotein and carry the same primary chromosomal aberrations as at diagnosis, consistent with the outgrowth of a single clone of PCs located within the BM. In this sense, this PC would seem to be the logical target for treatment, and eradicating this cell should be the focus of therapeutic approaches. However, this does not fully exclude the possibility that late relapses could be caused by a myeloma “stem cell” with a different phenotype than the myeloma PC, for which new treatment strategies may prove to be more effective.
The study of Ig gene rearrangements can also help us to understand the impact of treatment on the malignant clone. The clonal Ig gene rearrangements provide specific targets to monitor the tumor cells. In this context, PCR is a sensitive tool to detect clonal rearrangements at these loci, and the lack of SHM in most DJH rearrangements increases the rate of clonality detection by PCR and offers excellent targets for monitoring MRD by consensus real-time quantitative-PCR, thus overcoming the limitations of designing specific probes for each individual patient, which is significantly more expensive and time consuming.41,61,62
Both patients and clinicians alike are keen to understand what causes myeloma, and while classical epidemiologic approaches have established some candidate factors, there is still a lot to learn about the etiology of this disease. Studies of Ig gene rearrangements can provide insights into this process and perhaps allow us to better focus classical and molecular epidemiologic studies. In this respect, Ig gene segment usage in myeloma appears to reflect some degree of both positive and negative selection by environmentally encountered antigen. These data, together with the presence of a high degree of SHM in IGH and IGK/IGL functional rearrangements and characteristic translocations targeting the switch regions of the IGH locus in most of the myeloma cases, indicate that processes involving the Ig genes, such as BCR signaling, CSR, and SHM, play a fundamental role in early myeloma pathogenesis.
Authorship
Contribution: M.v.d.B., R.G.-S., J.A.F., A.W.L., M.G., J.J.M.v.D., and J.F.S.M. wrote and reviewed the paper; and D.G. and G.J.M. designed, wrote, edited, and reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gareth J. Morgan, Section of Haemato-Oncology, Institute of Cancer Research, 15 Cotswold Rd, Sutton, Surrey, SM2 5NG, United Kingdom; e-mail: gareth.morgan@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal