Modeling the complexity of sickle cell disease pathophysiology and severity is difficult. Using data from 3380 patients accounting for all common genotypes of sickle cell disease, Bayesian network modeling of 25 clinical events and laboratory tests was used to estimate sickle cell disease severity, which was represented as a score predicting the risk of death within 5 years. The reliability of the model was supported by analysis of 2 independent patient groups. In 1 group, the severity score was related to disease severity based on the opinion of expert clinicians. In the other group, the severity score was related to the presence and severity of pulmonary hypertension and the risk of death. Along with previously known risk factors for mortality, like renal insufficiency and leukocytosis, the network identified laboratory markers of the severity of hemolytic anemia and its associated clinical events as contributing risk factors. This model can be used to compute a personalized disease severity score allowing therapeutic decisions to be made according to the prognosis. The severity score could serve as an estimate of overall disease severity in genotype-phenotype association studies, and the model provides an additional method to study the complex pathophysiology of sickle cell disease.
Introduction
Sickle cell disease is caused by mutations in the β-hemoglobin gene (HBB). Individuals homozygous for the HBB glu6val mutation (HbS) have sickle cell anemia; compound heterozygotes for HBB glu6val and glu6lys (HbC) mutations have sickle cell–HbC (HbSC) disease. Both of these types of sickle cell disease, and the sickle-β thalassemias, have extremely variable phenotypes. Some individuals have mild disease that can be clinically unapparent; others can have most of the known severe complications such as pulmonary hypertension, priapism, stroke, leg ulceration, acute painful episodes, acute chest syndrome, and avascular necrosis of bone.1,2 While the median age of death in the United States was estimated to be in the fifth decade for patients with sickle cell anemia,3 some individuals die young and others live into their eighth or ninth decade. Therefore, to forecast the severity of sickle cell disease and the risk of near-term death, it would be clinically useful to understand the relationships among clinical and laboratory measures of disease expression and to identify genetic variants that impact the disease severity.
An impediment to this objective has been the inability to integrate the many clinical and laboratory dimensions of the disease into a single measure of disease severity using traditional statistical methods.4,,,–8 In this study, we developed a predictive model of disease severity using a Bayesian network modeling approach that considered 13 laboratory tests, 7 clinical events, and demographic and treatment information in 3380 patients with sickle cell disease. Bayesian network modeling has been used to develop diagnostic9 and prognostic tools,10,,–13 and, compared with a traditional regression model, it offers the advantage of describing the mutual relationships among the variables and identifying those variables that are directly associated with a disease or a disease subphenotype.
Our analysis shows the complex network of associations between laboratory tests and clinical events that modulate the risk of death in sickle cell disease. As suggested by previous studies, renal insufficiency, leukocytosis, and the intensity of the hemolytic anemia are related to the severity of sickle cell disease and near-term death.7,14,–16 Using this model, we computed the risk of death within 5 years and consider this risk as a disease severity score, which ranges from 0 (least severe) to 1 (most severe). The predictive value (ie, accuracy of forecasting death based on a clinical and laboratory profile) of the model was validated in 2 unrelated sets of patients and shows that the model accurately forecasts the risk of death for a subject given their clinical and laboratory profile. Our model could help clinicians formulate a prognosis that could be used for planning treatment.
Patients, materials, and methods
Patient databases
Patients followed in the Cooperative Study of Sickle Cell Disease (CSSCD) were the primary data source and included 3380 adult and pediatric patients with sickle cell anemia with or without coincident α thalassemia and also patients with HbSC disease.17,18 This was an observational study, designed to describe the clinical and laboratory features of sickle cell disease. Recruitment started in 1978 and continued for some patient groups into 1988; both community-based, clinic patients and a newborn cohort were enrolled. Patients were followed on average for 5 years. Treatment of complications was not specified and hydroxyurea was not available for treating sickle cell anemia in that era. In this database, 283 patients had died and were used to compute the risk of death. Sepsis was among the most frequent case of death (14%), followed by cerebrovascular accident (10%). In the data set, 1753 patients were younger than 19 years of age, and 67 patients who died were younger than 19 years of age.
Complications were defined by standardized definitions, and a schedule for laboratory testing was prescribed by the protocol. From the database, we extracted information concerning acute chest syndrome, leg ulceration, avascular necrosis of bone, acute painful episodes, priapism, sepsis, stroke, age at enrollment, sex, and selected laboratory variables. Laboratory testing was performed in the “steady state” at least 3 weeks after an acute sickle cell event such as a painful episode. Sepsis was defined as a positive blood culture result not associated with a known source of infection (such as osteomyelitis, septic arthritis, pneumonia, or meningitis). We used the median values of the routine measures and the most recent value of serum creatinine that is related to age.19 Laboratory values were categorized into low, normal, and high by using a mix of normal reference values in the general population and expected ranges in sickle cell disease derived from this same population (Tables 1,2; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).19
We used 2 unrelated data sets to confirm and validate our results. The first validation set comprised 140 patients, aged 20 years or more, followed at Boston Medical Center for their medical care. This was a clinic-based population of all genotypes of sickle cell disease treated by Boston Medical Center investigators (A.H.A., L.C.M.), with a median length of follow-up of 10.5 years. Five of these patients (aged > 26 years) died during the follow-up. Clinic visits and laboratory testing (steady-state values are reported) were dictated by each patients' clinical course, and 27% of the patients were treated with hydroxyurea. The clinical severity of these patients, estimated as mild, intermediate, or severe, was assessed by expert clinicians using the frequency of hospital visits and clinical events.
The second validation set comprised 210 subjects, older than 18 years of age, from the National Institutes of Health (NIH) Pulmonary Hypertension Screening Study with all genotypes of sickle cell disease who were followed for an average of 27 months.20 The purpose of the study was to define the prevalence of pulmonary hypertension and its associated clinical and laboratory findings in adult sickle cell disease, and recruitment was from the community. Treatment was not specified, and 48% of these adult patients were prescribed hydroxyurea. Nineteen patients died during the study (aged between 18 and 74 years). Severity was assessed by clinical criteria and measurement of the tricuspid regurgitant jet velocity, an estimation of pulmonary artery systolic pressure that has been associated with increased risk of death.20 The tricuspid regurgitant jet velocity was used as follows: no pulmonary hypertension, tricuspid regurgitant jet velocity less than 2.5 m/s; mild pulmonary hypertension, tricuspid regurgitant jet velocity greater than 2.5 m/s but less than 3.0 m/s; severe pulmonary hypertension, tricuspid regurgitant jet velocity 3.0 m/s or greater. This classification was shown to have sensitivity between 79% and 100% and specificity between 60% and 98%.21
These studies were approved by the Institutional Review Boards of the participating institutions in accordance with the Declaration of Helsinki.
Statistical analysis
We used Bayesian networks to model the relationships between clinical complications of sickle cell disease, laboratory tests, and the risk of death. In contrast to a regression model, which can only represent the dependency of 1 outcome variable on 1 or more predictor variables, a Bayesian network can represent the mutual and hierarchal relationships among many variables using probabilistic rules and thus, in many instances, is more appropriate for prognostic and diagnostic applications.
Description of the network
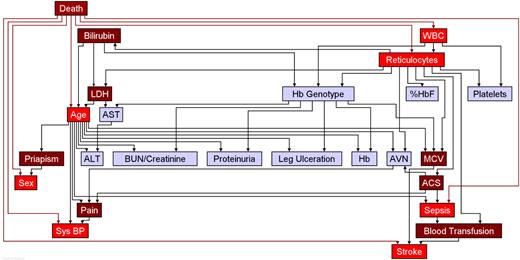
Figure 1 displays the network that was generated using the program Discoverer (http://www.bayesware.com). The nodes (boxes) in the graph represent the variables in the data set, and the directed arcs (arrows) represent probabilistic dependencies from parents (the nodes with outgoing arcs) to children (nodes with incoming arcs). The parent node has a directed arc pointing to its child nodes, and a child node has incoming arcs that are quantified by the conditional probability distributions of the child nodes, given the possible configurations of the parent nodes. For example, the node “Platelets,” which represents the variable platelet counts, is a child of the nodes “Reticulocytes” and “WBC” (white blood cell), and this dependency is an indication that the 3 variables are associated. We represent this association in the network by Table 3, the table of conditional probability distributions of “Platelets” (the child node), given the possible combinations of values of the parent nodes. Table 3 shows 4 of these 9 conditional distributions. These tables of conditional probability distributions, 1 for each node in the network, represent the uncertainty in making prediction about individual subjects. For example, for a subject with low WBC and reticulocyte counts, we can predict low platelets counts with probability 0.83, and for a subject with low WBC counts but normal reticulocyte counts we can predict low platelets counts with probability 0.60 (Table 3).
The parent-child connections in the network can represent either prognostic (a cause leading to an effect) or diagnostic (an effect leading to a cause) relations. For example, the node “WBC” in the network is a child of the node “Death” (yes or no), meaning that the 2 variables are associated and the dependency is quantified by the conditional probability table of the node “WBC,” given the possible combinations of values of the parent node (Table 4). The dependency of “WBC” (the possible cause) on the node “Death” (the possible outcome) represents the diagnostic rather than prognostic relation. The node “Death” is the parent of the 7 variables highlighted in bright red in Figure 1: Age, Reticulocytes, “Sex,” “Sepsis,” “Stroke,” “Systolic Blood Pressure,” and “WBC.” The dependencies show that these 7 variables are directly associated with death; however, these are not all of the variables that modulate the risk for death. In fact, the distribution of these 7 variables is also a function of other parent nodes, for example the node “Age” is a child node of “Bilirubin” and “LDH” (lactate dehydrogenase): these parent nodes become related to the node “Death” because of the common child node and affect “Death” only conditionally on the common child nodes. Based on this consideration, the variables “ACS,” “Bilirubin,” “Blood Transfusion,” “LDH,” mean corpuscular volume (“MCV”), “Pain,” and “Priapism” are associated with the node “Death” through the common child nodes and are colored in dark red in the network. This set of 14 nodes separates the node “Death” from all of the remaining variables and is sufficient to compute the risk for death.
Prognostic and diagnostic use of the network
The conditional probability distributions of each child node can be multiplied together to obtain the joint probability distribution of the variables in the network, which can be used to show how 1 or more events effect each other in the production of 1 or more outcomes. The algorithm to make this calculation is based on Bayes theorem—hence the name Bayesian networks—that updates the prior (“marginal”) probability of each event or node into the posterior (“conditional”) probability of the event, which is influenced by other events. We can employ this algorithm to use the network as a prognostic tool and compute the risk for death within 5 years given a particular clinical profile. This risk ranges between 0 (least severe) and 1 (most severe) and we consider this measure as a severity score. In the absence of any information, the risk for death is fixed to the referent value, 0.5, and changes according to the clinical profile. For example, the risk of death within 5 years of a male younger than 18 years of age with all “normal-range” laboratory results and no history of complications is 0.04, indicating a mild clinical profile. This increases to 0.12 if the patient has a history of stroke and painful episodes. Some examples that can lead to a severe clinical profile, with a score greater than 0.5, are shown in Table 5. We provide the complete set of conditional probability tables that define the network in Document S2.22 The network can also be used as a diagnostic tool to try to identify the most likely symptoms of disease severity.
We provide a “disease severity score calculator” on the web22 for the simple computation of this score in any patient, given the availability of clinical and laboratory data.
Network building from data
We estimated how nodes were interdependent and the conditional probability tables using the database of 3380 sickle cell disease patients and the Bayesian algorithm introduced by Cooper and Herskovitz.23 This was implemented with the program Bayesware Discoverer (http://www.bayesware.com/). The algorithm of Cooper et al23 explores multiple networks, scores each individual network by its probability based on the data, and returns the most probable network. Variables that define candidate parent nodes are first ordered as described previously. We assumed that the node “Death” could be a parent of every other node. We then ordered the remaining variables by their variance, so that a parent node must have larger variance than its child nodes.
The initial ordering process focuses attention on diagnostic rather than prognostic models and has the advantage of including more variables compared with a prognostic model. For example, the best logistic regression model we could fit using stepwise regression contained only 7 predictors, including key laboratory variables such as the WBC or LDH. However, clinical events such as priapism were not included. Unlike the network, the regression model does not suggest a comprehensive chain of events associated with the likelihood of a near-term death (Table S3).
The final network was selected after exploring more than 1000 networks and was more than 150 times more probable than the second-best network. The probability of each network and the associated conditional probability tables were calculated as described by Sebastiani et al.24 (See Cowell et al,25 Sebastiani et al,26 and Document S2 for additional technical details and the list of 25 conditional probability tables.) We report here only the marginal effect estimates and approximate confidence intervals for the variables that are predictive of the risk for death. However, the network includes a multitude of synergistic and antagonistic interactions among the variables. As an example, Table 5 shows the calculation of risk for different combinations of clinical events and laboratory results.
Network validation and reliability
We validated the predictive value and reliability of the network by comparing the severity score assigned by the network with the clinically assessed level of severity in patients followed at the Boston Medical Center and in the NIH–pulmonary hypertension patients and by the sensitivity in identifying patients at high risk in both patient groups. In the NIH–pulmonary hypertension cases, we also compared the disease severity score with the measurement of tricuspid regurgitant jet velocity.
To compute the error rate, we repeatedly divided the original data set into 2 nonoverlapping sets of 2880 and 500 patients that were randomly chosen. In each iteration, we found the most likely network using the set of 2880 patients. We then computed the score of the remaining 500 patients and estimated the error as the overall number of false negatives (subjects with score ≤ 0.5 who died) and false positives (subjects with score > 0.5 who survived). The error rate was the average frequency of errors. However, while we expected patients who died from a complication of the disease during the study to have a high score, we could have patients with a severe medical condition who did not die. Therefore, comparison of the severity score against survival can only be used to estimate the sensitivity but not the specificity of the network.
To assess the accuracy of the network against more specific measures of severity, we compared the distribution of the disease severity score in our 2 other patient groups clinically classified as having severe, intermediate, or mild disease. In these groups, we had information on 5 and 19 subjects who died during the follow-up. We used these data to assess the sensitivity of our network, which was defined by the relative frequency of subjects with a score greater than 0.5 who died. We used the number of subjects with a score less than 0.5 who were classified as mild as an indication of the specificity. We computed the correlation between the score and the tricuspid regurgitant jet velocity and compared the score distribution in the groups of patients with absent, mild, or severe pulmonary hypertension.
Results
Disease severity score
Patients from the Boston Medical Center have a milder clinical profile compared with subjects in the primary data source (CSSCD in Tables 1,2). This is likely to be due to the higher prevalence of HbSC disease in this group (41% compared with 26% in the Cooperative Study patients) and the approval of hydroxyurea as treatment for sickle cell anemia, which was not available for use in patients of the Cooperative Study. Patients in the NIH–pulmonary hypertension sample appear to have more severe disease with a higher prevalence of complications and an increased mortality rate. These 2 contemporary patient data sets have complementary features and represent an urban-American sickle cell disease population.
Laboratory characteristics of patient groups
| Variable . | CSSCD, n = 3380 . | BMC, n = 140 . | NIH-PHT, n = 210 . | |||
|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | Mean . | SD . | |
| Age, y | 20.00 | 14.04 | 21.40 | 14.91 | 35.99* | 11.45 |
| ALT level, (Sgpt), U/L | 35.03 | 52.53 | 25.72* | 24.37 | 27.09* | 14.49 |
| AST level, (Sgot), U/L | 44.51 | 23.74 | 45.14 | 30.88 | 41.71 | 21.74 |
| Bilirubin level, mg/dL | 2.71 | 1.89 | 3.10 | 3.19 | 2.75 | 1.74 |
| BUN level, mg/dL | 9.36 | 5.36 | 9.68 | 4.73 | 10.76 | 10.62 |
| Creatinine level, mg/dL* | 0.70 | 0.64 | 0.48* | 0.35 | 0.93† | 1.30 |
| Hb level, g/dL | 9.13 | 1.81 | 11.71 | 24.37 | 9.44 | 1.88 |
| % HbF | 6.12 | 5.78 | 6.15 | 7.57 | 7.41 | 6.96 |
| LDH level, U/L† | 459.51 | 206.34 | 406.7† | 289.89 | 361.9* | 164.04 |
| MCV, fL | 87.42 | 8.75 | 88.03 | 11.36 | 91.94* | 11.30 |
| Platelet count, 109/L | 399.30 | 131.17 | 372.70 | 156.30 | 365.30 | 140.13 |
| Reticulocyte count, % RRC | 9.56 | 5.85 | 9.05 | 9.11 | 250.1† | 130.13 |
| Sys BP, mmHg* | 105.70 | 12.35 | 115.9* | 14.91 | 120.5* | 17.82 |
| WBC count, 109/L | 11.17 | 3.31 | 10.94 | 4.73 | 10.39† | 3.62 |
| Variable . | CSSCD, n = 3380 . | BMC, n = 140 . | NIH-PHT, n = 210 . | |||
|---|---|---|---|---|---|---|
| Mean . | SD . | Mean . | SD . | Mean . | SD . | |
| Age, y | 20.00 | 14.04 | 21.40 | 14.91 | 35.99* | 11.45 |
| ALT level, (Sgpt), U/L | 35.03 | 52.53 | 25.72* | 24.37 | 27.09* | 14.49 |
| AST level, (Sgot), U/L | 44.51 | 23.74 | 45.14 | 30.88 | 41.71 | 21.74 |
| Bilirubin level, mg/dL | 2.71 | 1.89 | 3.10 | 3.19 | 2.75 | 1.74 |
| BUN level, mg/dL | 9.36 | 5.36 | 9.68 | 4.73 | 10.76 | 10.62 |
| Creatinine level, mg/dL* | 0.70 | 0.64 | 0.48* | 0.35 | 0.93† | 1.30 |
| Hb level, g/dL | 9.13 | 1.81 | 11.71 | 24.37 | 9.44 | 1.88 |
| % HbF | 6.12 | 5.78 | 6.15 | 7.57 | 7.41 | 6.96 |
| LDH level, U/L† | 459.51 | 206.34 | 406.7† | 289.89 | 361.9* | 164.04 |
| MCV, fL | 87.42 | 8.75 | 88.03 | 11.36 | 91.94* | 11.30 |
| Platelet count, 109/L | 399.30 | 131.17 | 372.70 | 156.30 | 365.30 | 140.13 |
| Reticulocyte count, % RRC | 9.56 | 5.85 | 9.05 | 9.11 | 250.1† | 130.13 |
| Sys BP, mmHg* | 105.70 | 12.35 | 115.9* | 14.91 | 120.5* | 17.82 |
| WBC count, 109/L | 11.17 | 3.31 | 10.94 | 4.73 | 10.39† | 3.62 |
Patients enrolled in the CSSCD included children aged younger than 1 year and adults with sickle cell anemia with or without coincident α thalassemia (deletion of 1 or 2 α-globin genes [HBA2, HBA1]) and also patients with HbSC disease. Two additional independent, longitudinally followed contemporaneous patient groups were examined. One-hundred forty patients were from BMC and 210 patients participated in the NIH-PHT. These patients also had either sickle cell anemia or HbSC disease. The NIH-PHT patients had echocardiographic assessment of pulmonary hypertension. Reticulocyte counts in the NIH-PHT patients are presented as absolute numbers. This data was not available in the CSSCD. To convert to SI units: bilirubin, ×17.1 (μmol/L); BUN, ×0.357 (mmol/L); creatinine, ×88.4 (μmol/L); hemoglobin, ×10 (g/L). The reticulocytes are expressed as a percent of all red blood cells.
CSSCD indicates Cooperative Study of Sickle Cell Disease; BMC, Boston Medical Center; NIH-PHT, NIH-Pulmonary Hypertension Screening Study; ALT, alanine aminotransferase; Sgpt, serum glutamic pyruvic transaminase; AST, aspartate aminotransferase; Sgop, serum glutamic oxoloacetic transaminase; BUN, blood urea nitrogen; Hb, hemoglobin; and Sys BP, systolic blood pressure.
P < .001 relative to the CSSCD data.
01 < P < .05 relative to the CSSCD data.
Clinical characteristics of patient groups
| Variables . | Proportion, % . | ||
|---|---|---|---|
| CSSCD, n = 3380 . | BMC, n = 140 . | NIH-PHT, n = 210 . | |
| ACS | 62 | 54 | 82 |
| AVN | 32 | 21 | 25 |
| Blood transfusion | 13 | 3 | 58 |
| Death | 8 | 3 | 9 |
| Hb genotype, HbSC | 26 | 41 | 20 |
| Leg ulceration | 12 | 4 | 23 |
| Pain | 83 | 93 | 82 |
| Priapism | 15 | 12 | 45 |
| Sepsis | 13 | 10 | Unknown |
| Sex, female | 52 | 46 | 58 |
| Stroke | 6 | 5 | 14 |
| PHT | 28 | ||
| Variables . | Proportion, % . | ||
|---|---|---|---|
| CSSCD, n = 3380 . | BMC, n = 140 . | NIH-PHT, n = 210 . | |
| ACS | 62 | 54 | 82 |
| AVN | 32 | 21 | 25 |
| Blood transfusion | 13 | 3 | 58 |
| Death | 8 | 3 | 9 |
| Hb genotype, HbSC | 26 | 41 | 20 |
| Leg ulceration | 12 | 4 | 23 |
| Pain | 83 | 93 | 82 |
| Priapism | 15 | 12 | 45 |
| Sepsis | 13 | 10 | Unknown |
| Sex, female | 52 | 46 | 58 |
| Stroke | 6 | 5 | 14 |
| PHT | 28 | ||
ACS indicates acute chest syndrome; and AVN, avascular necrosis of bone; PHT, pulmonary hypertension.
Figure 1 shows the network of associations among the 25 variables included in the analysis (see Tables 3 and 4). The nodes in bright and dark red highlight the variables that are sufficient to compute the disease severity score. They include clinical features like age, sex, chronic blood transfusion, acute chest syndrome, priapism, painful episodes, stroke, and sepsis and laboratory tests like WBC, MCV, reticulocyte count, bilirubin level, LDH level, and systolic blood pressure. The marginal and interactive effects of these variables are summarized in Tables 5 and 6.
The network of associations between death, clinical complications, and laboratory findings in sickle cell disease. The arc (arrow) direction specifies the conditional probability tables that are sufficient to compute the overall distribution. Colored in red are the nodes that alone are sufficient to predict the risk for death (severity score). Nodes in blue are associated with predictive nodes in red. For example, the Hb genotype is associated with several laboratory variables including WBC and LDH and thus modulates disease severity indirectly through these nodes. See Tables 3,4. ACS indicates acute chest syndrome; AVN, avascular necrosis; BUN/creatinine, ratio of BUN to creatinine; Sys BP, systolic blood pressure; Hb, total hemoglobin concentration; %HbF, percentage of fetal hemoglobin; WBC, leukocyte count; Hb genotype, sickle cell anemia, sickle cell anemia-α thalassemia, HbSC disease.
The network of associations between death, clinical complications, and laboratory findings in sickle cell disease. The arc (arrow) direction specifies the conditional probability tables that are sufficient to compute the overall distribution. Colored in red are the nodes that alone are sufficient to predict the risk for death (severity score). Nodes in blue are associated with predictive nodes in red. For example, the Hb genotype is associated with several laboratory variables including WBC and LDH and thus modulates disease severity indirectly through these nodes. See Tables 3,4. ACS indicates acute chest syndrome; AVN, avascular necrosis; BUN/creatinine, ratio of BUN to creatinine; Sys BP, systolic blood pressure; Hb, total hemoglobin concentration; %HbF, percentage of fetal hemoglobin; WBC, leukocyte count; Hb genotype, sickle cell anemia, sickle cell anemia-α thalassemia, HbSC disease.
Distribution of the node “Platelets”
| Reticulocyte count . | WBC count . | Platelets counts . | ||
|---|---|---|---|---|
| Less than 400 . | 400-490 . | Greater than 490 . | ||
| <4.8 | <10.8 | 0.83 | 0.11 | 0.06 |
| <4.8 | 10.8-13.5 | 0.60 | 0.23 | 0.17 |
| <4.8 | >13.5 | 0.41 | 0.35 | 0.24 |
| 4.8-13 | <10.8 | 0.60 | 0.22 | 0.18 |
| Reticulocyte count . | WBC count . | Platelets counts . | ||
|---|---|---|---|---|
| Less than 400 . | 400-490 . | Greater than 490 . | ||
| <4.8 | <10.8 | 0.83 | 0.11 | 0.06 |
| <4.8 | 10.8-13.5 | 0.60 | 0.23 | 0.17 |
| <4.8 | >13.5 | 0.41 | 0.35 | 0.24 |
| 4.8-13 | <10.8 | 0.60 | 0.22 | 0.18 |
Distribution of the node “WBC”
| Death . | WBC Counts . | ||
|---|---|---|---|
| Less than 10.8 . | 10.8-13.5 . | Greater than 13.5 . | |
| Yes | 0.37 | 0.31 | 0.32 |
| No | 0.49 | 0.29 | 0.22 |
| Death . | WBC Counts . | ||
|---|---|---|---|
| Less than 10.8 . | 10.8-13.5 . | Greater than 13.5 . | |
| Yes | 0.37 | 0.31 | 0.32 |
| No | 0.49 | 0.29 | 0.22 |
Disease severity scores for different clinical profiles
| Variable . | Profile 1 . | Profile 2 . | Profile 3 . | Profile 4 . | Profile 5 . | Profile 6 . |
|---|---|---|---|---|---|---|
| ACS | No | No | Yes | No | No | No |
| Age, y | 2–18 | 2–18 | 2–18 | 18–40 | 18–40 | 18–40 |
| Bilirubin level | Normal | Normal | High | Normal | Normal | Normal |
| Blood transfusion | No | No | No | No | No | No |
| LDH level | Normal | Normal | High | Normal | Normal | Normal |
| MCV | Normal | Normal | Normal | Normal | High | High |
| Pain | No | Yes | No | Yes | Yes | Yes |
| Priapism | No | No | NA | No | No | No |
| Reticulocyte count | Normal | Low | Normal | Low | High | Normal |
| Sepsis | No | No | Yes | No | No | Yes |
| Sex | Male | Male | Female | Male | Male | Male |
| Stroke | No | Yes | No | Yes | Yes | Yes |
| Sys BP | Normal | Normal | Normal | Normal | Normal | Normal |
| WBC count | Normal | Normal | Normal | Normal | Very high | Very high |
| Disease severity score | 0.04 | 0.116 | 0.581 | 0.64 | 0.76 | 1.00 |
| Variable . | Profile 1 . | Profile 2 . | Profile 3 . | Profile 4 . | Profile 5 . | Profile 6 . |
|---|---|---|---|---|---|---|
| ACS | No | No | Yes | No | No | No |
| Age, y | 2–18 | 2–18 | 2–18 | 18–40 | 18–40 | 18–40 |
| Bilirubin level | Normal | Normal | High | Normal | Normal | Normal |
| Blood transfusion | No | No | No | No | No | No |
| LDH level | Normal | Normal | High | Normal | Normal | Normal |
| MCV | Normal | Normal | Normal | Normal | High | High |
| Pain | No | Yes | No | Yes | Yes | Yes |
| Priapism | No | No | NA | No | No | No |
| Reticulocyte count | Normal | Low | Normal | Low | High | Normal |
| Sepsis | No | No | Yes | No | No | Yes |
| Sex | Male | Male | Female | Male | Male | Male |
| Stroke | No | Yes | No | Yes | Yes | Yes |
| Sys BP | Normal | Normal | Normal | Normal | Normal | Normal |
| WBC count | Normal | Normal | Normal | Normal | Very high | Very high |
| Disease severity score | 0.04 | 0.116 | 0.581 | 0.64 | 0.76 | 1.00 |
As some examples of how the severity score calculated by the network model (Figure 1) is dependent on the variables in the network, the table shows that the occurrence of stroke is associated with a wide severity spectrum, according to changes in laboratory variables, age, and other complications. Note that while sepsis is a very strong indicator of disease severity, its contribution to severity changes according to the other variables in the network (see the severity scores of profiles 3 and 6).
NA indicates not applicable.
Summary of the strength of associations in the network
| Variable* . | Effect, OR . | Effect, OR . | Referent group . |
|---|---|---|---|
| ACS | 1.18 (1.14;1.24) | † | No ACS |
| Age, y | 2.67 (2.44;2.91) [18-40] | 7.61 (5.11;1.34) [>40] | 2-18 years |
| Bilirubin level | 1.43 (1.28;1.60) [1.3-3.4] | 1.59 (1.02;2.49) [>3.4] | Normal |
| Blood transfusion | 2.01 (1.78;2.27) | † | No chronic BT |
| LDH level | 0.85 (0.76;0.94) [<300] | 1.1 (0.99;1.24) [>600] | Normal |
| MCV | 0.54 (0.49;0.60) [<80] | 1.98 (1.71; 2.29) [>98] | Normal |
| Pain | 1.61 (1.45;1.77) | † | No pain |
| Priapism | 1.35 (1.09;1.67) | † | No priapism |
| Reticulocyte count | 0.5 (0.44;0.55) [<4.8] | 1.51 (1.39;1.65) [>13] | Normal |
| Sepsis | 67.19 (57.66;78.29) | † | No sepsis |
| Sex | 1.16 (1.08;1.25) | † | Females |
| Stroke | 3.81 (3.20;4.54) | † | No stroke |
| Sys BP | 2.84 (1.67;4.82) [low] | 3.41 (2.71;4.27) [high] | Normal |
| WBC count | 1.37 (1.24;1.51) [10.8-13.5] | 1.92 (1.44;2.55) [>13.5] | Normal |
| Variable* . | Effect, OR . | Effect, OR . | Referent group . |
|---|---|---|---|
| ACS | 1.18 (1.14;1.24) | † | No ACS |
| Age, y | 2.67 (2.44;2.91) [18-40] | 7.61 (5.11;1.34) [>40] | 2-18 years |
| Bilirubin level | 1.43 (1.28;1.60) [1.3-3.4] | 1.59 (1.02;2.49) [>3.4] | Normal |
| Blood transfusion | 2.01 (1.78;2.27) | † | No chronic BT |
| LDH level | 0.85 (0.76;0.94) [<300] | 1.1 (0.99;1.24) [>600] | Normal |
| MCV | 0.54 (0.49;0.60) [<80] | 1.98 (1.71; 2.29) [>98] | Normal |
| Pain | 1.61 (1.45;1.77) | † | No pain |
| Priapism | 1.35 (1.09;1.67) | † | No priapism |
| Reticulocyte count | 0.5 (0.44;0.55) [<4.8] | 1.51 (1.39;1.65) [>13] | Normal |
| Sepsis | 67.19 (57.66;78.29) | † | No sepsis |
| Sex | 1.16 (1.08;1.25) | † | Females |
| Stroke | 3.81 (3.20;4.54) | † | No stroke |
| Sys BP | 2.84 (1.67;4.82) [low] | 3.41 (2.71;4.27) [high] | Normal |
| WBC count | 1.37 (1.24;1.51) [10.8-13.5] | 1.92 (1.44;2.55) [>13.5] | Normal |
The second and third columns report, for each variable in column 1, the marginal effect on the disease severity score measured by the odds ratio for death within 5 years, and 95% confidence intervals in parentheses. Names in square brackets describe the category compared with the referent group when the variable has more than 2 categories: for example 2.67 in column 2, row 2, are the odds for death in 5 years of a subject aged between 18 and 40 years, compared with a subject less than age 18 years. Note that the effect of each variable changes when the other variables change (see Table 5).
Variables sufficient to compute the disease severity score.
When the variable has only 2 categories, column 2 reports the odds for death in 5 years compared with the referent group in column 4.
In Figure 1, nodes in blue are associated with predictive nodes in red. For example, while the genotype of sickle cell disease (node “Hb Genotype”) obviously must contribute to disease severity, this contribution is through its effects on clinical events and laboratory test results. For this reason, we included all genotypes of sickle cell disease in our analysis. With the exception of sepsis, the other variables associated with disease severity have, individually, a moderate effect on severity but their co-occurrence becomes strongly predictive (Table 5).
Predictive value
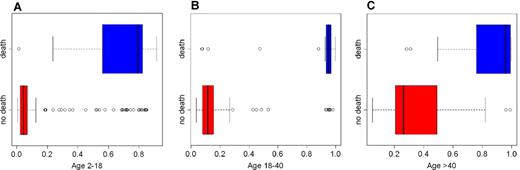
The error rate was 7.5%, suggesting that the network accurately detects subjects at risk. Box plots in Figure 2 show the distribution of the severity score by age group and vital status and highlight the predictive value of the network in 1 test set that was generated during the computation of the error rate. Subjects who died during observation had a high severity score, suggesting an accurate positive predictive value. The differences in severity score in the 3 age groups were all statistically significant (t tests: −6.6, −11.1, −5.9; P < .001). Eleven subjects aged between 2 and 11 years (Figure 2A) and 7 subjects aged between 18 and 30 years (Figure 2B) were assigned a score greater than 0.75 but survived until the end of the study. Because the information is censored to an average follow-up of 5 years, the fact that these subjects survived cannot be taken as an indication of low specificity. In fact, consistent with a high severity score, all subjects had sepsis, and all but 1 of the subjects had a serious clinical profile, with more than 3 serious complications.
Distribution of disease severity score in 1 validation test set. Each box plot displays the observations between the first and the third quartile in the rectangle, with a line for the median. The whiskers extend to 1.5 times the interquartile range from each end of the rectangle. Circles represent outliers beyond the end of the whiskers. Box plots in blue display the score distribution for patients who died, and box plots in red display the score distribution for survivors. There is a separation between the scores of subjects in these groups that is especially clear in patients aged 18 to 40 years (B). In the group aged 2 to 18 years (A), 2 subjects with a score below 0.5 died. Both subjects (1 HbSC, score 0.01; 1 with sickle cell anemia, score 0.24) died for unknown causes. In the group aged 18 to 40 years (B), 2 of the 4 subjects with low severity score died for unknown causes as did 2 of the 3 subjects with low severity score in the group aged older than 40 years (C). Many young subjects with a high severity score survived until the end of the follow-up, consistent with the high survival rate of children even with a severe clinical profile.
Distribution of disease severity score in 1 validation test set. Each box plot displays the observations between the first and the third quartile in the rectangle, with a line for the median. The whiskers extend to 1.5 times the interquartile range from each end of the rectangle. Circles represent outliers beyond the end of the whiskers. Box plots in blue display the score distribution for patients who died, and box plots in red display the score distribution for survivors. There is a separation between the scores of subjects in these groups that is especially clear in patients aged 18 to 40 years (B). In the group aged 2 to 18 years (A), 2 subjects with a score below 0.5 died. Both subjects (1 HbSC, score 0.01; 1 with sickle cell anemia, score 0.24) died for unknown causes. In the group aged 18 to 40 years (B), 2 of the 4 subjects with low severity score died for unknown causes as did 2 of the 3 subjects with low severity score in the group aged older than 40 years (C). Many young subjects with a high severity score survived until the end of the follow-up, consistent with the high survival rate of children even with a severe clinical profile.
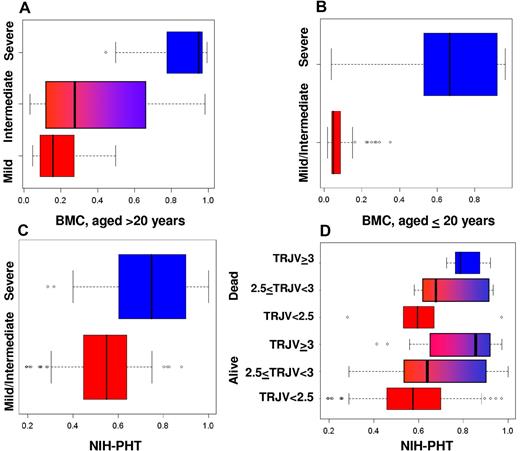
Figure 3 summarizes the distribution of the disease severity in patients from Boston Medical Center and the NIH–Pulmonary Hypertension Screening Study. Figure 3A-B displays the score of the 135 surviving Boston Medical Center patients. Figure 3A displays the distribution of the score according to the clinical assessment (mild, intermediate, severe) of 45 patients, 21 years or older. The median score of 0.95 for the 12 severe cases was substantially higher than the median score of 0.28 for the 22 intermediate cases and 0.16 for the 11 mild cases and significantly different from both (t tests −8.5 and −5.8, P < .001). Although the score distribution of the mild and intermediate subjects shows some overlapping, the scores were significantly different (t test −21.4, P = .04). Furthermore, none of the mild cases were assigned a score greater than 0.5, suggesting 100% specificity. Figure 3B displays the score distribution in 90 patients, 20 years or younger. These subjects were dichotomized as mild/intermediate or severe cases because of their shorter clinical history. The median score in the 80 mild/intermediate cases was 0.05 versus a median score of 0.67 in the 10 severe cases, and the mean scores were significantly different (t test −4.9, P < .001). In the severe cases, our network detected 2 patients at low risk with scores of 0.03 and 0.04. The clinical presentation of 1 of these patients might have been influenced by treatment with hydroxyurea. In the other patient, we lacked information on LDH level, a strong predictor of disease severity. Our model assigned a score greater than 0.5 to 4 of 5 patients who died since 2000, thus reaching a sensitivity of 80%. One patient with HbSC disease died suddenly with multiorgan failure due to pulmonary fat embolus. His score (0.36) was higher than the median score of intermediate cases but less than 0.5, so this patient would not have been considered high risk. The fact that no mild or intermediate patients were assigned a score greater than 0.5 suggests high specificity, even in the younger patients.
Validations in independent patient groups. (A,B) Validation in patients from the Boston Medical Center. (A) Distribution of severity score (x-axis) for groups of adults (aged ≥ 18 years) whose clinical status was assessed as mild, intermediate, or severe. (B) Distribution of severity score (x-axis) for the pediatric groups (aged < 18 years) whose clinical status was assessed as mild/intermediate (bottom) and severe. (C,D) Validation in patients from the NIH. (C) Distribution of severity score (x-axis) for patients whose clinical status was determined as mild/intermediate (bottom box plot) and severe (top box plot). (D) Distribution of severity score of the 19 subjects who died during the follow-up (top 3 box plots) and the 191 subjects who survived the follow-up (bottom 3 box plots), grouped as follows: no pulmonary hypertension (tricuspid regurgitant jet velocity ≤ 2.5 m/s); mild pulmonary hypertension (2.5 < tricuspid regurgitant jet velocity < 3 m/s); severe pulmonary hypertension (tricuspid regurgitant jet velocity ≥ 3.0 m/s).
Validations in independent patient groups. (A,B) Validation in patients from the Boston Medical Center. (A) Distribution of severity score (x-axis) for groups of adults (aged ≥ 18 years) whose clinical status was assessed as mild, intermediate, or severe. (B) Distribution of severity score (x-axis) for the pediatric groups (aged < 18 years) whose clinical status was assessed as mild/intermediate (bottom) and severe. (C,D) Validation in patients from the NIH. (C) Distribution of severity score (x-axis) for patients whose clinical status was determined as mild/intermediate (bottom box plot) and severe (top box plot). (D) Distribution of severity score of the 19 subjects who died during the follow-up (top 3 box plots) and the 191 subjects who survived the follow-up (bottom 3 box plots), grouped as follows: no pulmonary hypertension (tricuspid regurgitant jet velocity ≤ 2.5 m/s); mild pulmonary hypertension (2.5 < tricuspid regurgitant jet velocity < 3 m/s); severe pulmonary hypertension (tricuspid regurgitant jet velocity ≥ 3.0 m/s).
In the NIH group (Figure 3C), patients classified as mild/intermediate on the basis of their clinical assessment had a significantly lower median score (0.054) than severe patients (0.71; t test = 8.1; P < .001). None of the mild/intermediate patients had a score greater than 0.5 or a tricuspid regurgitant jet velocity greater than 2.5, and these results suggest that these 2 methods have the same 100% specificity.
Figure 3D shows the distribution of the score in the same patients as in Figure 3C grouped by survival and severity of pulmonary hypertension. The plots show an agreement between our score and severity of pulmonary hypertension in the group of living subjects; the median score is 0.57 in 135 patients without pulmonary hypertension, 0.64 in 40 patients with mild pulmonary hypertension, and 0.86 in 15 patients with severe pulmonary hypertension. The difference in average score between living patients with mild pulmonary hypertension and without pulmonary hypertension is significant (P = .02). However, the difference in average score between subjects with mild or severe pulmonary hypertension is not significant (t test 1.6, P = .12). The same increasing trend is noticeable in the subjects who died during follow-up: 0.6 in 6 subjects without pulmonary hypertension, 0.68 in 8 subjects with mild pulmonary hypertension, and 0.79 in 5 subjects with severe pulmonary hypertension. We did not assess the significance of the score difference because of the small sample sizes. A risk measure based only on the tricuspid regurgitant jet velocity would have missed 6 of the 19 patients, reaching a sensitivity of 0.68. The network assigned a score greater than 0.5 to 18 of them, with a sensitivity of 95%.
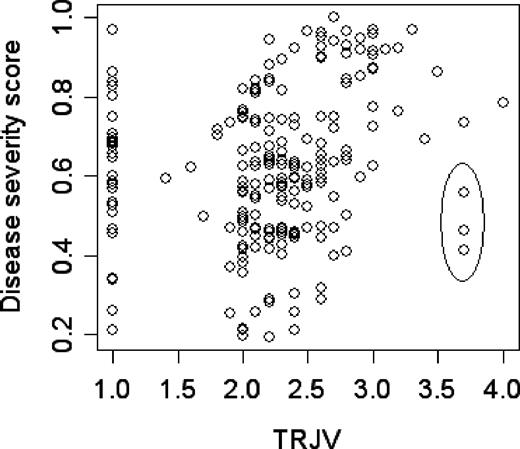
The greater sensitivity of our network compared with the tricuspid regurgitant jet velocity alone is also supported by the ability of the network to assign a score to subjects for whom the tricuspid regurgitant jet velocity could not be measured. Figure 4 displays the relationship between the severity score and tricuspid regurgitant jet velocity, including 29 patients with severity scores between 0.2 and 1 who did not have a detectable tricuspid regurgitant jet. In 3 subjects with high tricuspid regurgitant jet velocity, their low severity score was concordant with their clinical profile, which was not typical of sickle cell disease–associated pulmonary hypertension (Figure 4 legend). Excluding these 32 patients, the correlation between the severity score and the tricuspid regurgitant jet velocity is 0.43, suggesting a relationship between these 2 estimates of disease severity; however, the network has a higher sensitivity than tricuspid regurgitant jet velocity at detecting overall disease severity.
Scatter plot of the disease severity score (y-axis) versus the tricuspid regurgitant jet velocity (m/s; x-axis) in the 210 subjects of the NIH–Pulmonary Hypertension Screening Study. For 29 subjects, the tricuspid regurgitant jet velocity could not be measured and was set equal to 1. The score of these 29 subjects ranges from 0.2 to 0.97; more than 75% of these subjects have a score above 0.5 and would be judged as severe. The 3 points highlighted by an ellipse represent a discordance in assessing the severity between our score and the tricuspid regurgitant jet velocity. While these patients have a tricuspid regurgitant jet velocity greater than 3 m/s (high risk of death), our model assigns them scores of 0.41, 0.46 (not at risk), and 0.60 (mild risk). One subject (score 0.41) had mitral valve insufficiency, subsequently treated surgically, so that the high tricuspid regurgitant jet velocity was due to cardiac disease. The second subject (score 0.46) had very severe pulmonary hypertension associated with very severe obstructive sleep apnea requiring tracheostomy. The third subject (score 0.6) had undergone apparently successful nonmyeloablative bone marrow transplantation since enrollment. She appeared to have typical sickle cell disease–associated pulmonary hypertension but was on chronic transfusion at the time of enrollment.
Scatter plot of the disease severity score (y-axis) versus the tricuspid regurgitant jet velocity (m/s; x-axis) in the 210 subjects of the NIH–Pulmonary Hypertension Screening Study. For 29 subjects, the tricuspid regurgitant jet velocity could not be measured and was set equal to 1. The score of these 29 subjects ranges from 0.2 to 0.97; more than 75% of these subjects have a score above 0.5 and would be judged as severe. The 3 points highlighted by an ellipse represent a discordance in assessing the severity between our score and the tricuspid regurgitant jet velocity. While these patients have a tricuspid regurgitant jet velocity greater than 3 m/s (high risk of death), our model assigns them scores of 0.41, 0.46 (not at risk), and 0.60 (mild risk). One subject (score 0.41) had mitral valve insufficiency, subsequently treated surgically, so that the high tricuspid regurgitant jet velocity was due to cardiac disease. The second subject (score 0.46) had very severe pulmonary hypertension associated with very severe obstructive sleep apnea requiring tracheostomy. The third subject (score 0.6) had undergone apparently successful nonmyeloablative bone marrow transplantation since enrollment. She appeared to have typical sickle cell disease–associated pulmonary hypertension but was on chronic transfusion at the time of enrollment.
Discussion
Stroke, acute chest syndrome, sepsis, painful episodes, pulmonary hypertension, and priapism are characteristic of severe sickle cell disease.1,3,7,14,20,27,–29 Many typically abnormal laboratory test results are also found.30,31 Nevertheless, each individual clinical complication of sickle cell disease, except perhaps pulmonary hypertension, or any isolated laboratory measurement, has limited predictive value when defining disease severity and the likelihood of near-term death. By integrating individual disease complications and the results of selected laboratory tests, the severity score computed by our network allows computation of a personalized measure of sickle cell disease severity and an estimate of the likelihood of death within 5 years. To validate and further test the reliability of the network score, we used it to estimate the severity of 2 independent patient groups. The Bayesian network is more specific than expert clinician assessment alone and has the virtue of integrating many clinical and laboratory findings and providing a quantifiable estimate of disease severity. When compared with an objective marker of severe disease, like the tricuspid regurgitant jet velocity (a measure of the severity of pulmonary hypertension and a predictor of mortality), the network-derived score was a more useful measure of disease severity, especially when it was not possible to measure tricuspid regurgitant jet velocity or tricuspid regurgitant jet velocity was high for reasons other than sickle cell disease (Figure 4).
Prior work suggested that vasoocclusion/blood viscosity–related events like acute chest syndrome and the frequency of acute painful episodes are associated with a poor prognosis.3,8 Other studies have suggested that the hemolysis of sickle cell disease, perhaps via its role in pulmonary hypertension and sickle vasculopathy, is an important contributor to mortality.20,32 Our network identifies complications that can be associated with both hemolysis and sickle vasoocclusion/blood viscosity as related to the risk of death. It also identified the association of sepsis with mortality. Perhaps sepsis is a manifestation of tissue necrosis, endothelial damage, and the presence of venous access devices that could signify more severe disease.
The pathophysiologic implications of hemolysis in sickle cell disease were underappreciated until recent work suggested the importance of dysregulated nitric oxide homeostasis.33,–35 Pulmonary hypertension, priapism, leg ulceration, and stroke, all subphenotypes of sickle cell disease and identified in our network, have been associated with the intensity of hemolysis.20,36,–38 Elevated systolic blood pressure increased the odds of death within 5 years by 3.4, consistent with hypertension as a marker of early death in sickle cell anemia39 and its association with pulmonary hypertension and reduced nitric oxide bioavailability.20 LDH level, aspartate aminotransferase (AST) level, and bilirubin level reflect the severity of hemolytic anemia; nevertheless, they can also be indicative of liver disease.
Reticulocyte count is another estimate of the severity of hemolysis. The presence of both hemolysis- and viscosity-related events in our network supports the likelihood that they are interconnected. Perhaps the nexus of this relationship is provided by the sickle reticulocyte, whose numbers rise as hemolysis increases. Reticulocytes bear adhesive ligands promoting intracellular interactions. Also, fetal hemoglobin inhibits sickle hemoglobin polymerization, thereby reducing erythrocyte injury, increasing red cell lifespan, and reducing reticulocytosis.2,8,40
The hemoglobin genotype (node “Hb genotype,” Figure 1) occupies a central place in the network and modulates severity through its effects on hemolysis, leukocyte count, and fetal hemoglobin level. Consistent with previous findings,8 our network shows that subjects with sickle cell anemia are at greatest risk for death compared with subjects with sickle cell anemia and concurrent α thalassemia and with subjects with HbSC disease. Assuming similar clinical profiles, our network assigns twice the odds of death to sickle cell anemia cases compared with HbSC disease, and 1.12 times the odds of death to sickle cell anemia subjects compared with sickle cell anemia–α thalassemia. Our network predicts that the presence of proteinuria in the urine increases the odds of death by 63% in subjects with sickle cell anemia and confirms the adverse prognosis associated with this measure of renal disease.15,41,42
Beyond its potential clinical utility, our analysis begins to dissect the complex pathophysiology underlying the extreme variability of the multiple phenotypes of sickle cell disease. In our analysis, severity is determined by a network of interactions among 14 variables (Figure 1; Tables 5,6). The prognostic role of some of these variables, for example the effect of WBC and bilirubin as risk factors for early death in pediatric patients, has been reported,3 but a measure of an all-encompassing severity of sickle cell disease has been difficult to capture.8 While confirming some earlier results, our analysis is generalizable to a wide spectrum of sickle cell disease patients.
Several limitations of this model should be considered. Our network does not integrate the genetic polymorphisms that are likely to modulate the laboratory variables and clinical events included in the model. When these are incorporated, the utility of applying such a model to the youngest subjects in whom certain disease complications have yet to appear might be greatly enhanced. The 2 validation groups are small compared with the primary patient group and 1 consisted only of adults with a mean age of 40 years. They also had the benefit of treatments not available to participants in the Cooperative Study of Sickle Cell Disease. Patient data were derived from residents of the United States. Although we believe our patients are representative of this population, the model might not be applicable to individuals in other locales. Nevertheless, this could be tested. Patients with certain disease complications like stroke or acute chest syndrome were more likely to be chronically transfused. This is likely to have reduced their risk of most complications.
Automated scoring systems of disease severity like APACHE (Acute Physiology and Chronic Health Evaluation)43 or the Glasgow coma score44 have been useful tools in critical care but our method of network modeling is distinct from the derivation of these scores. The high specificity and sensitivity of our model suggest that it could become a useful decision support system to help clinicians design individual treatments, although more evaluation is needed in other patient populations.
The network can be used as an unbiased assessment of the clinical severity of patients with sickle cell disease, given any clinical and laboratory profile, employing a simple tool that is freely available.22 Users need only to insert the values of the variables to compute the severity score and update when there are changes in the patient profile. A potential application is as a quantitative assessment of the severity of disease that could be used in candidate gene or genome-wide genotype-phenotype association studies. Another use of the network score is for prognostic purposes. By knowing that an individual is at high risk for near-term death, a practitioner might choose to more vigorously explore therapeutic options, like chronic blood transfusion, hydroxyurea, or stem cell transplantation, each of which has its own special risks.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Heart, Lung and Blood Institute (NHLBI) grants R21 HL080463 (P.S.), R01 HL68970 (M.H.S.), U54 HL70819 (M.H.S.), and T32 HL007501 (V.G.N.).
We thank Maureen McKenna, Dimitri Bacos, Traci Sutherland, Alice Bisbee, and John Farrell for assistance with data collection and management and the investigators of the Cooperative Study of Sickle Cell Disease who obtained clinical and laboratory data and blood samples for DNA-based studies during the first 10 years of that program.
Authorship
Contribution: P.S. designed the study, developed the Bayesian network and the methods of statistical analysis, drafted the paper, and participated in its revisions. V.G.N. assisted in statistical analysis and worked with our databases. C.T.B. was involved with study design. M.M.A.-G. was involved with programming. L.W. assisted with Bayesian analysis. A.H.A. and L.C.M. evaluated patient disease severity. L.A.F. helped revise the paper. J.G.T., G.J.K., and M.T.G. collected NIH patients, did echocardiography, participated in the design of some analysis, and made critical revisions. M.H.S. helped design the study, interpret the analysis, and drafted and revised the manuscript.
Conflict-of-interest disclosure: P.S. is a shareholder and cofounder of Bayesware Discoverer. The other authors declare no competing financial interests.
Correspondence: Martin H. Steinberg, Department of Medicine, Boston University School of Medicine, Rm E248, 88 East Newton Street, Boston, MA 02118; e-mail:mhsteinb@bu.edu.