Shwachman-Diamond syndrome (SDS) is a rare multisystem disorder characterized by exocrine pancreatic insufficiency, multilineage hematopoietic dysfunction, and metaphyseal chondrodysplasia. Bone marrow dysfunction is present in nearly all patients with SDS, with neutropenia being the most common abnormality. The majority of patients with SDS have mutations in the Shwachman Bodian Diamond syndrome (SBDS) gene. We have developed a strategy to examine the consequences of lentiviral-mediated RNA interference (RNAi) of Sbds on hematopoiesis. Here, we report that both Sbds RNA and protein expression can be efficiently inhibited in primary murine hematopoietic cells using lentiviral-mediated RNAi. Inhibition of Sbds results in a defect in granulocytic differentiation in vitro and impairs myeloid progenitor generation in vivo. In addition, short-term hematopoietic engraftment was impaired, which is due in part to reduced homing of hematopoietic progenitors to the bone marrow. Finally, we show that inhibition of Sbds is associated with a decrease in circulating B lymphocytes, despite evidence of normal B lymphopoiesis. These data provide the first evidence that loss of Sbds is sufficient to induce abnormalities in hematopoiesis.
Introduction
Shwachman Diamond syndrome (SDS, also known as Shwachman-Bodian-Diamond syndrome) is a rare multiorgan syndrome characterized by exocrine pancreatic insufficiency, skeletal abnormalities, and multilineage hematopoietic dysfunction.1,,–4 The most common hematopoietic abnormality is intermittent neutropenia, and it occurs in approximately 95% of patients with SDS.3 Although neutropenia is thought to be the major cause of increased susceptibility to recurrent infections in patients with SDS, defects in B- and T-lymphocyte function and survival have been reported.5 Patients with SDS occasionally display a variety of additional hematopoietic phenotypes, including anemia, thrombocytopenia, and trilineage cytopenia. As with other bone marrow failure syndromes, SDS markedly increases the risk of the development of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) (reviewed in Dror6 )
SDS is inherited in an autosomal recessive fashion. In a landmark study, Boocock et al7 reported mutations of the Shwachman-Bodian-Diamond Syndrome (SBDS) gene in approximately 90% of patients with SDS, a finding that has been confirmed in subsequent genetic analyses.8,,–11 The most common mutations (183-184TA > CT and 258 + 2T > C) introduce early stop codons that truncate the majority of the SBDS protein. The remaining mutations are missense and are distributed throughout the SBDS gene. The genotype of approximately 60% of patients with SDS is compound heterozygosity for the 183-184TA > CT and 258 + 2T > C mutations, and most patients carry at least one of these truncation alleles.7,9,10 These observations form the basis of the hypothesis that loss of SBDS function is the primary molecular cause of SDS. However, it remains a possibility that the common 183-184TA > CT and 258 + 2T > C alleles produce truncated SBDS protein with residual or neomorphic activity. Because these N-terminal truncation products have not been studied in vivo, their pathogenic potential is unknown.
SBDS encodes a highly conserved, novel protein of unknown cellular function with homologs from archaea to mammals. Human SBDS protein localizes to the cytoplasm and nucleus with nucleolar accumulation.12 Global gene expression studies of the yeast SBDS homolog indicate that it is coordinately regulated with RNA-processing genes in Saccharomyces cerevisiae.13 Finally, the archaeal crystal structure reveals that SBDS has a nucleic acid binding groove.14,15 Collectively, these data support an existing hypothesis that SBDS functions in the processing or transport of RNA. Consistent with this hypothesis, it recently was reported that SBDS associates with 28S ribosomal RNA in mammalian cells.16 Moreover, a recent study showed that the yeast SBDS ortholog Sdo1 is required for the recycling of the nucleolar shuttling factor Tif6 from pre-60S ribosomes.17 There also is evidence suggesting that SBDS may play an important role in cell motility or chemotaxis or both.18,19 In accordance with this observation, defects in mobility and chemotaxis are well documented in circulating neutrophils isolated from patients with SDS.18,20,,,–24
Although it is clear that SBDS mutations are associated with SDS, experimental models have not yet been generated to study their necessity or sufficiency in the development of hematopoietic phenotypes. A recent study has shown a critical role for Sbds in development, because complete loss of Sbds in mice results in early lethality at embryonic day 6.5.25 However, because of this early lethality, the role of SBDS in hematopoiesis could not be evaluated in that study. Herein, we used lentiviral-mediated RNA interference (RNAi) to determine whether inhibition of Sbds function is sufficient to generate hematopoietic defects. We show that Sbds can be efficiently inhibited by RNAi in primary murine hematopoietic cells, resulting in delayed granulocytic differentiation in vitro. Moreover, reduction of Sbds is associated with reduced homing to the bone marrow and impaired short-term engraftment of hematopoietic progenitor cells (HPCs) in vivo. Finally, in mice reconstituted with Sbds RNAi-transduced bone marrow cells, decreased myeloid progenitors and circulating B lymphocytes were observed. Together, these data provide the first experimental evidence that loss of Sbds is sufficient to produce a subset of the hematopoietic phenotypes commonly associated with SDS.
Materials and methods
Mice
Wild-type C57BL/6 mice and a congenic strain of C57BL/6 that have the Ly5.1 gene (B6.SJL-PtPrc*Pep3bBoyJ) were obtained from Jackson Laboratories (Bar Harbor, ME). All mice were reared in a specific pathogen-free facility and monitored daily by the veterinary staff. All animal work was approved by the Washington University Animal Studies Committee.
Lentiviral constructs
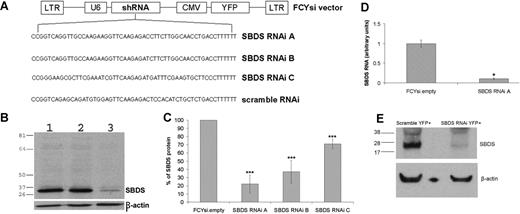
Three pairs of antisense oligonucleotides were designed to generate short hairpin RNA (shRNA) complementary to the Mus musculus Sbds mRNA transcript (GenBank NCBI accession NM_023248, here ATG start codon assigned 1; Sbds RNAi A, 187-206; Sbds RNAi B, 187-206, with G::U wobble engineered at 187; Sbds RNAi C, 71-90 with G::U wobble at positions 71 and 83. Sequences reported in Figure 1A). A control shRNA with a nonspecific nucleotide sequence was also designed (scramble RNAi; Figure 1A). Basic local alignment search tool analysis26 verified that these oligonucleotides were specific for Sbds and that the scramble RNAi primers were not homologous to any region of the mouse genome. Annealed oligonucleotides were directionally cloned into the genomic lentiviral vector pFCYsi (generous gift from J. Millbrandt). pFCYsi was generated by replacing the internal ribosomal entry sequence–yellow fluorescent protein (YFP) cassette in the lentiviral pFSPsi vector27 with cytomegalovirus (CMV)–YFP.
Generation of high-titer lentivirus
293T cells were transfected with the packaging plasmid pΔ8.9, viral envelope plasmid pMD.G,28 and genomic plasmid pFCYsi by calcium phosphate DNA precipitation.29,30 Viral supernatant was collected 48 and 72 hours after transfection and concentrated by centrifugation at 76 000g for 2 hours at 4°C (SW32 rotor in Optima LE-80K ultracentrifuge; Beckman Coulter, Palo Alto, CA).
Peripheral blood, spleen, and bone marrow sampling
Peripheral blood was obtained by retro-orbital venous plexus sampling. Complete blood counts were measured using a Hemavet automated cell counter (CDC Technologies, Oxford, CT). Whole spleens were extracted and homogenized in phosphate-buffered saline (PBS). Red blood cells were lysed in tris [(hydroxymethyl) aminomethane-buffered ammonium chloride, pH 7.2]. Bone marrow was flushed from femurs with PBS containing 0.2% bovine serum albumin (BSA).
Flow cytometry and cell sorting
Flow cytometry.
Cells were incubated with the indicated primary antibody for 1 hour at 4°C in PBS containing 0.2% BSA and 0.1% sodium azide. The following primary antibodies were used: c-Kit, Gr-1, B220, CD3, CD45.1, CD45.2 (BD Bioscience, San Diego, CA). Cells were analyzed on a FACScan flow cytometer (BD Bioscience).
Transduction efficiency.
Bone marrow cells were harvested 72 hours after lentiviral transduction and stained with allophycocyanin (APC)–conjugated c-Kit and a panel of phycoerythrin (PE)–conjugated lineage markers (PE-Lin): B220 (B lymphocytes), CD3 (T lymphocytes), Gr-1 (granulocytes), and Ter119 (erythrocytes). The percentage of c-Kit+, lineage− cells expressing YFP was measured by flow cytometry.
Cell sorting.
Approximately 24 hours after lentiviral transduction, cells were stained with APC–c-Kit and PE-Lin antibodies as described in “Transduction efficiency.” c-Kit+, lineage− cells were sorted based on YFP expression using a MoFlo high-speed cell sorter (DAKO, Fort Collins, CO).
Real-time RT-PCR
RNA was extracted from cells using the High Pure RNA Isolation Kit, according to manufacturer's protocol (Roche, Mannheim Germany), and Sbds RNA levels were determined by real-time reverse transcription–polymerase chain reaction (RT-PCR) (Taqman kit, 7300 real-time PCR system; Applied Biosystems, Foster City, CA). The following RT-PCR primers and probes were used: Sbds forward, 5′-CCCAACAAGAGCACAAAGCAA-3′; Sbds reverse, 5′-TCGTTCACTGGCAGGATGAAG-3′; Sbds PCR product was detected with SYBR green (Molecular Probes, Eugene, OR); β-actin forward, 5′-ACCAACTGGGACGATATGGAGAAGA-3′; β-actin reverse, 5′-TACGACCAGAGGCATACAGGGACAA-3′; β-actin probe, 5′-AGCCATGTACGTAGCCATCCAGGCTG-3′.
Immunoblotting
Murine hematopoietic cell transduction
Bone marrow cells were incubated with the PE-lineage antibody panel. After washing, cells were incubated with anti-PE colloid, and lineage+ cells were depleted using the AutoMacs system (Miltenyi Biotec, Auburn, CA). The lineage− fraction was collected and cultured overnight in α-MEM containing stem-cell factor (SCF; 100 ng/mL), interleukin-3 (10 ng/mL), thrombopoietin (10 ng/mL), and Flt3 ligand (50 ng/mL) (PeproTech, Rocky Hill, NJ). Cells were transduced with virus 12 and 36 hours later at the indicated multiplicity of infection (MOI) by spinoculation31 (centrifugation of cells plus virus at 1000g for 90 minutes at 30°C).
Bone marrow transplantation
Syngeneic C57BL/6 recipients were exposed to a single dose of 1000 cGy from a 137Cesium source at a rate of 95 cGy/minute 12 to 24 hours before transplantation. Unsorted, transduced bone marrow cells (5 × 105) were injected into the tail vein of each irradiated recipient. At least 4 mice per experimental group were transplanted; results from 2 to 4 independent transplantations were analyzed separately and pooled, unless otherwise noted. Antibiotics (trimethoprim-sulfamethoxazole; Alpharma, Baltimore, MD) were administered in the drinking water for 2 weeks after transplantation.
Secondary transplantations were performed as described, except 2 × 106 unfractionated bone marrow cells from primary donors 6 months after transplantation were infused into the tail vein of each irradiated secondary recipient.
Progenitor colony-forming assays
CFU-Cs (complete colony-forming units).
Bone marrow cells (25 000) or c-Kit+, lineage− cells (1000) were plated in MethoCult M3434 (Stem Cell Technologies, Vancouver, BC). To identify BFU-E (erythroid blast-forming unit) colonies, CFU-C cultures were stained with a benzidine solution composed of 0.02 M benzidine (Sigma) and 13% (vol/vol) acetic acid. Colonies staining dark blue/black 3 minutes after addition of stain were scored as BFU-Es.
CFU-Gs (granulocyte colony-forming units).
c-Kit+, lineage− cells (2000) were plated in MethoCult M3231 (Stem Cell Technologies) supplemented with 100 ng/mL granulocyte colony-stimulating factor (G-CSF) (Amgen, Thousand Oaks, CA). For pre-B CFUs, bone marrow cells (150 000) were plated in MethoCult M3630.
CFU-GMs (granulocyte/macrophage colony-forming units).
c-Kit+, lineage− cells (1000) were plated in MethoCult M3231 supplemented with 20 ng/mL GM-CSF (PeproTech).
In each case, colonies containing at least 50 cells were scored on day 10 of culture.
Homing assays
Donor bone marrow cells were infected with lentivirus as described in “Murine hematopoietic cell transduction.” A portion (2000) of these cells was plated directly in MethoCult M3434, and the percentage of YFP+ CFU-Cs was determined on day 10 using a fluorescence microscope (Nikon Eclipse TE 300; Nikon, Tokyo, Japan). The remainder of the transduced bone marrow cells was transplanted into lethally irradiated recipients (106 per mouse). Bone marrow was harvested from recipient mice 24 hours after bone marrow transplantation, 5 × 105 cells were plated in methylcellulose, and the percentage of YFP+ CFU-Cs was determined. Homing efficiency is reported as the ratio of the percentage of YFP+ CFU-Cs after transplantation to before transplantation.
Progenitor transmigration assay
Transwell migration of HPCs in response to CXCL12α (or stromal-derived factor-1) was performed, essentially as described previously.32 In brief, transduced unsorted lineage− bone marrow cells (15 000) were loaded into the upper chamber of 5-μm-pore transwell inserts (Corning Life Sciences, Acton, MA). Recombinant human recombinant CXCL12α was added to the lower chamber at a concentration of 500 ng/mL, and the chamber was incubated for 4 hours at 37°C. The number of YFP− and YFP+ CFU-Cs in the original cell preparation and in the migrated cell population was determined.
Granulocytic differentiation in vitro
Growth of primary murine hematopoietic cells in vitro was analyzed as previously described.33 In brief, c-Kit+, lineage− cells were cultured in α-MEM containing SCF and G-CSF (100 ng/mL each). After 7 days in culture, cells were sorted based on YFP expression, and manual differentials were performed on Wright-Giemsa–stained cytospin preparations.
Data analysis
Unless otherwise noted, all data are presented as the mean (± standard deviation [SD]). A 2-tailed Student t test was applied to individual datasets to determine statistical significance. Survival curves were analyzed using the log-rank test.
Results
Lentiviral-mediated RNAi efficiently inhibits Sbds expression in primary hematopoietic cells
A bicistronic lentiviral vector, in which the U6 pol3 promoter drives ubiquitous expression of an Sbds-specific antisense shRNA and the CMV promoter drives expression of YFP, was used to determine whether Sbds expression could be inhibited. Lentiviruses containing the lentiviral vector alone (FCYsi.empty), a nonspecific shRNA control (scramble RNAi), and 3 independent shRNAs directed against the Sbds mRNA were designed (Sbds RNAi A-C; Figure 1A). High-titer lentivirus was generated, and the ability to reduce Sbds expression using RNAi was first assessed in BaF3 cells, a pro-B murine cell line. Compared with both parental BaF3 cells and cells transduced with FCYsi.empty, Sbds RNAi A reduced Sbds protein levels by 78% (± 11%; Figure 1B,C); Sbds RNAi B and Sbds RNAi C reduced Sbds protein levels by 63% (± 14%) and 29% (± 6%), respectively (Figure 1C). A similar decrease in Sbds mRNA was observed; in BaF3 cells transduced with Sbds RNAi A, RNA was reduced 89.5% (± 3.5%; Sbds RNAi A; Figure 1D) compared with FCYsi.empty control. On the basis of these results, Sbds RNAi A (hereafter simply termed Sbds RNAi) was used in all subsequent experiments.
RNAi inhibition of Sbds expression. (A) Schematic of FCYsi lentiviral vector showing long terminal repeats (LTRs), U6 promoter-short hairpin RNA (shRNA) cassette, and cytomegalovirus (CMV)–YFP cassette. The sequences of the 3 Sbds-specific and the scramble shRNAs are shown in the lower panel. (B) Representative immunoblot of Sbds in parental BaF3 cells (1) and BaF3 cells transduced with FCYsi.empty (2) or Sbds RNAi A (3) vectors. β-Actin loading control is shown in the lower panel. (C) Quantification of Sbds protein levels; Sbds protein abundance in BaF3 cells transduced with FCYsi.empty was assigned a value of 100% (n = 4-6). (D) Sbds mRNA expression relative to β-actin mRNA in BaF3 cells was measured by real-time RT-PCR (n = 2). (E) Murine HPCs were transduced with scramble RNAi or Sbds RNAi A; YFP+ cells were sorted from cultures 72 hours after transduction and were cultured for 7 days. Shown is a representative immunoblot for Sbds and β-actin. *P ≤ .05, ***P ≤ .005, compared with control cells. All data represent the mean (± SD).
RNAi inhibition of Sbds expression. (A) Schematic of FCYsi lentiviral vector showing long terminal repeats (LTRs), U6 promoter-short hairpin RNA (shRNA) cassette, and cytomegalovirus (CMV)–YFP cassette. The sequences of the 3 Sbds-specific and the scramble shRNAs are shown in the lower panel. (B) Representative immunoblot of Sbds in parental BaF3 cells (1) and BaF3 cells transduced with FCYsi.empty (2) or Sbds RNAi A (3) vectors. β-Actin loading control is shown in the lower panel. (C) Quantification of Sbds protein levels; Sbds protein abundance in BaF3 cells transduced with FCYsi.empty was assigned a value of 100% (n = 4-6). (D) Sbds mRNA expression relative to β-actin mRNA in BaF3 cells was measured by real-time RT-PCR (n = 2). (E) Murine HPCs were transduced with scramble RNAi or Sbds RNAi A; YFP+ cells were sorted from cultures 72 hours after transduction and were cultured for 7 days. Shown is a representative immunoblot for Sbds and β-actin. *P ≤ .05, ***P ≤ .005, compared with control cells. All data represent the mean (± SD).
The efficacy of this lentiviral construct to inhibit Sbds expression in primary murine myeloid cells was next examined. Murine bone marrow cells were transduced with the Sbds RNAi or scramble RNAi lentivirus. Transduced (YFP+) cells were sorted and expanded in media containing G-CSF, SCF, interleukin-3, and TPO, and Sbds protein expression was assessed on day 7 of culture. Compared with scramble RNAi-transduced cells, Sbds protein expression was reduced 86% (± 13.7%; average ± SEM; n = 3) in Sbds RNAi cells (Figure 1E).
Sbds RNAi impairs the granulocytic differentiation of primary myeloid cells in vitro
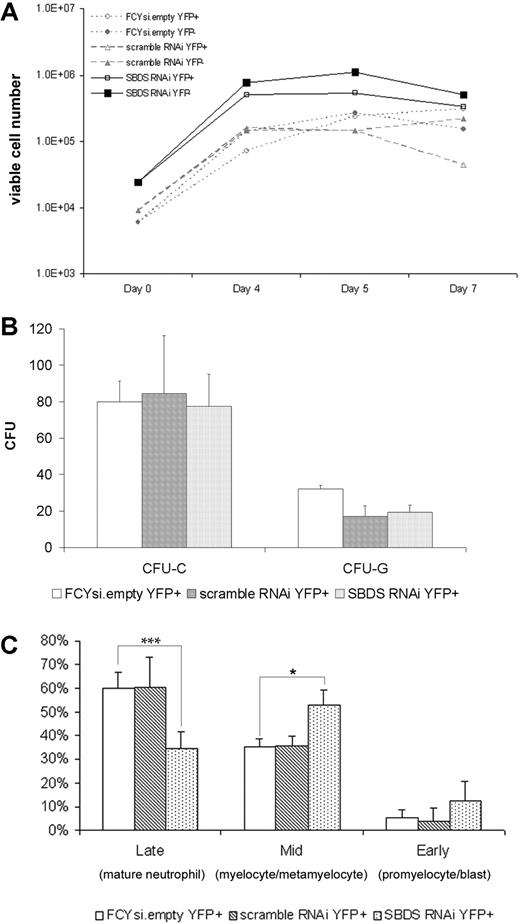
Neutropenia is the most common hematopoietic defect in patients with SDS.3 To determine the effect of reduced murine Sbds expression on granulopoiesis, transduced hematopoietic progenitors were grown in liquid culture containing G-CSF and SCF, conditions that support granulocytic differentiation. As expected based on previous studies,33 FCYsi.empty and scramble RNAi-transduced cells expanded 42-fold (± 3.2) and 38-fold (± 19), respectively (Figure 2A). A similar fold expansion was noted in cultures of Sbds RNAi-transduced cells (38- ± 13-fold). Of note, the number of apoptotic cells, as measured by surface Annexin V expression, was small and comparable in all cultures (data not shown). To further assess the effect of Sbds inhibition on the cytokine responsiveness and growth of myeloid progenitors, c-Kit+ lineage− cells were plated into methylcellulose cultures immediately after transduction (Figure 2B). Inhibition of Sbds had no effect on the number of colonies formed in response to a cocktail of myeloid cytokines (CFU-C) or G-CSF alone (CFU-G) (Figure 2B).
Sbds RNAi impairs in vitro granulocytic cell differentiation. Murine bone marrow cells were transduced with the indicated lentivirus, sorted for YFP expression, and either plated in suspension cultures containing SCF and G-CSF (A, C) or methylcellulose (B). (A) The number of viable cells was measured on initiation of the culture (day 0) and at the indicated times (representative of 3 independent experiments). (B) The number of CFU-Cs and CFU-Gs per 1000 c-Kit+, lineage− cells is shown (n = 3-5). (C) After 7 days in vitro granulocytic differentiation, cells were resorted based on YFP expression, and leukocyte differentials were performed (n = 2-4). *P ≤ .05, ***P ≤.005. All data represent the mean (± SD).
Sbds RNAi impairs in vitro granulocytic cell differentiation. Murine bone marrow cells were transduced with the indicated lentivirus, sorted for YFP expression, and either plated in suspension cultures containing SCF and G-CSF (A, C) or methylcellulose (B). (A) The number of viable cells was measured on initiation of the culture (day 0) and at the indicated times (representative of 3 independent experiments). (B) The number of CFU-Cs and CFU-Gs per 1000 c-Kit+, lineage− cells is shown (n = 3-5). (C) After 7 days in vitro granulocytic differentiation, cells were resorted based on YFP expression, and leukocyte differentials were performed (n = 2-4). *P ≤ .05, ***P ≤.005. All data represent the mean (± SD).
To determine the effect of Sbds RNAi on granulocytic differentiation, YFP+ cells were resorted after 7 days in culture, and leukocyte differentials were performed in a blinded fashion. As expected, the majority of cells in both the FCYsi.empty and scramble RNAi cultures were mature (band or segmented) neutrophils (60% ± 6.9% and 60% ± 13%, respectively), the remaining cells were granulocytic precursors (Figure 2C). The percentage of mature neutrophils in cultures of Sbds RNAi-transduced cells was significantly reduced (35% ± 7.3%). The remaining cells in this culture were granulocytic precursors, with a significant accumulation of myelocytes and metamyelocytes and a trend toward an increase in promyelocytes and blasts (Figure 2C). These data show that inhibition of Sbds results in normal granulocytic cell proliferation but impaired differentiation in vitro.
Sbds RNAi reduces short-term hematopoietic engraftment
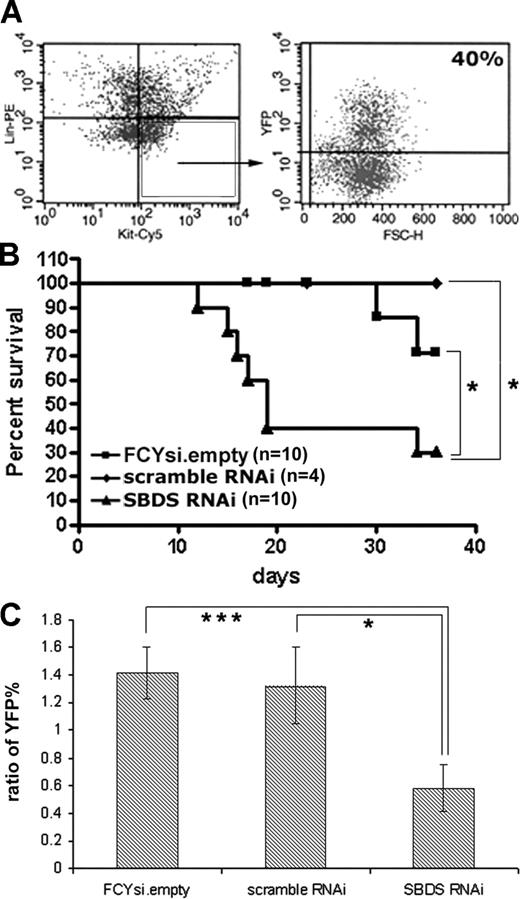
The effect of Sbds RNAi on hematopoiesis in vivo was determined through bone marrow transplantation experiments. Donor bone marrow was first infected at a relatively high concentration of virus (MOI of 20:1). Transduction efficiency was estimated by determining the percentage of c-Kit+, lineage− cells that expressed YFP (Figure 3A). Importantly, in each cohort of animals that received a transplant the transduction efficiency was similar between all 3 experimental groups (average of 43% ± 26%). Unsorted, transduced bone marrow cells (containing both transduced and wild-type cells) were transplanted into syngeneic recipients, and survival was assessed (Figure 3B). Although the majority of mice that received a transplant with cells transduced with the FCYsi.empty and scramble RNAi controls survived the engraftment period, short-term survival was significantly reduced in mice that received a transplant with Sbds RNAi-transduced cells (Figure 3B). Whereas advanced postmortem changes precluded analysis of most mice, we were able to characterize hematopoiesis in 2 morbidly ill mice that received a transplant of Sbds RNAi-transduced cells at 17 and 19 days after transplantation. Compared with control mice, mice that received a transplant of Sbds RNAi-transduced cells displayed trilineage aplasia. A trend toward a reduction in the red blood cell count [3.2 ± 0.8 × 106 red cells/mL (FCYsi.empty) versus 1.6 ± 0.6 × 106 red cells/mL (Sbds RNAi)], white blood cell count [0.46 ± 0.1 × 106 cells/mL (FCYsi.empty) versus 0.25 ± 0.1 × 106 cells/mL (Sbds RNAi)] and platelets [33 ± 25 × 106/mL (FCYsi.empty) versus 21 ± 6 × 106/mL (Sbds RNAi)] was observed, although differences were not significant because of a paucity of animals available for analysis.
Reduction in Sbds impairs short-term hematopoietic engraftment. (A) Lineage-depleted bone marrow cells were transduced with lentivirus, and transduction efficiency was assessed at least 48 hours later by flow cytometry. c-Kit+, lineage− cells (box) were analyzed for YFP expression (representative of 5 independent high MOI transductions). (B) Kaplan-Meier survival curve of transplant recipients receiving donor bone marrow transduced with a high MOI (20:1). (C) Engraftment at 3 weeks in mice that received a transplant with cells transduced at an MOI of 10:1. Shown is the ratio of YFP percentage in the peripheral blood of engrafted recipients to the pretransplant transduction efficiency, as measured in c-Kit+, lineage− donor bone marrow cells (n = 9-11; average ± SEM). *P ≤ .05, ***P ≤ .005.
Reduction in Sbds impairs short-term hematopoietic engraftment. (A) Lineage-depleted bone marrow cells were transduced with lentivirus, and transduction efficiency was assessed at least 48 hours later by flow cytometry. c-Kit+, lineage− cells (box) were analyzed for YFP expression (representative of 5 independent high MOI transductions). (B) Kaplan-Meier survival curve of transplant recipients receiving donor bone marrow transduced with a high MOI (20:1). (C) Engraftment at 3 weeks in mice that received a transplant with cells transduced at an MOI of 10:1. Shown is the ratio of YFP percentage in the peripheral blood of engrafted recipients to the pretransplant transduction efficiency, as measured in c-Kit+, lineage− donor bone marrow cells (n = 9-11; average ± SEM). *P ≤ .05, ***P ≤ .005.
In an attempt to rescue early lethality, the bone marrow transplantation experiments were repeated with a reduced MOI of 10:1. At this MOI, the average transduction efficiency of c-Kit+, lineage− cells was reduced to 13% (± 1.2%) and, again, was similar in all groups. Eighty-nine percent of mice that received a transplant survived at least 6 months. Short-term hematopoietic engraftment was assessed 3 weeks after transplantation. The percentage of circulating peripheral blood cells expressing YFP was measured by flow cytometry and compared with the pretransplant transduction efficiency. Of note, the white blood count and bone marrow cellularity were similar in all groups; thus, the ratio of YFP+ percentage before transplantation to peripheral YFP+ percentage after transplantation accurately reflects YFP maintenance in this model. In mice that received a transplant with cells transduced with FCYsi.empty and scramble RNAi, the percentages of YFP+ cells in the peripheral blood at 3 weeks were 1.4- and 1.3-fold higher than the original transduction efficiency (Figure 3C). In contrast, engraftment of Sbds RNAi-transduced cells was significantly reduced by 3 weeks after transplantation, to only 0.6 of the transduction efficiency before transplantation, a 55% decrease compared with controls (Figure 3C). Collectively, these data show that the inhibition of Sbds significantly reduces short-term hematopoietic engraftment.
Homing to the bone marrow is impaired in Sbds RNAi-transduced HPCs
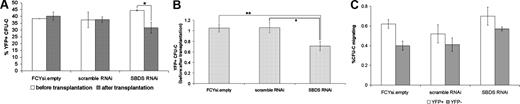
The observed defect in short-term engraftment could be a consequence of Sbds RNAi, reducing the ability of progenitors to either (1) home to the bone marrow or (2) subsequently reconstitute hematopoiesis. To distinguish between these possibilities, homing experiments were conducted in which lethally irradiated recipients received lentiviral-transduced bone marrow and were analyzed 24 hours after transplantation. The percentage of YFP+ CFU-Cs before transplantation and in the bone marrow after transplantation was determined, and the ratio of YFP+ CFU-Cs after transplantation to before transplantation was calculated as a measure of homing efficiency (Figure 4). Importantly, the total number of progenitors that homed to the bone marrow compartment was similar among the 3 experimental groups (data not shown). In progenitors transduced with FCYsi.empty and scramble RNAi controls, the ratio of YFP+ CFU-Cs before transplantation to after transplantation was near 1 (1.1 ± 0.08 and 1.1 ± 0.10, respectively; Figure 4B), indicating that homing was not affected by the lentiviral transduction procedure. In contrast, this ratio was significantly reduced in Sbds RNAi-transduced cells to 0.71 (± 0.09; Figure 4B). These data show that reduction of Sbds inhibits homing of hematopoietic progenitors to the bone marrow.
Reduction in Sbds impairs HPC homing to the bone marrow. Bone marrow cells were transduced with the indicated lentivirus and transplanted into irradiated mice. (A) Bone marrow cells before transplantation and harvested from recipient mice 24 hours after transplantation were plated in methylcellulose, and the percentage of CFU-Cs that expressed YFP was determined. (B) Homing efficiency was measured as the ratio of after transplantation to before transplantation YFP+ CFU-Cs (n = 7-8). (C) A transwell migration assay was performed on transduced lineage− bone marrow cells. Shown is the percentage of YFP+ and YFP− CFU-Cs that migrated in response to CXCL12α (n = 3). *P ≤ .05, **P ≤ .01. All data represent the mean plus or minus SEM.
Reduction in Sbds impairs HPC homing to the bone marrow. Bone marrow cells were transduced with the indicated lentivirus and transplanted into irradiated mice. (A) Bone marrow cells before transplantation and harvested from recipient mice 24 hours after transplantation were plated in methylcellulose, and the percentage of CFU-Cs that expressed YFP was determined. (B) Homing efficiency was measured as the ratio of after transplantation to before transplantation YFP+ CFU-Cs (n = 7-8). (C) A transwell migration assay was performed on transduced lineage− bone marrow cells. Shown is the percentage of YFP+ and YFP− CFU-Cs that migrated in response to CXCL12α (n = 3). *P ≤ .05, **P ≤ .01. All data represent the mean plus or minus SEM.
A chemotaxis defect in SDS hematopoietic cells has been reported.18 BecauseCXCL12α has been implicated in HPC homing to the bone marrow,34,,–37 we assessed the effect of Sbds RNAi on the chemotaxis of HPCs in response to CXCL12α (Figure 4C). A high percentage of CFU-Cs migrated in response to CXCL12α, possibly reflecting the effects of the cytokine stimulation that is required to transduce hematopoietic progenitors. Nonetheless, CFU-C migration was similar for all groups, indicating that CXCL12α-induced chemotaxis is normal in Sbds RNAi-transduced HPCs.
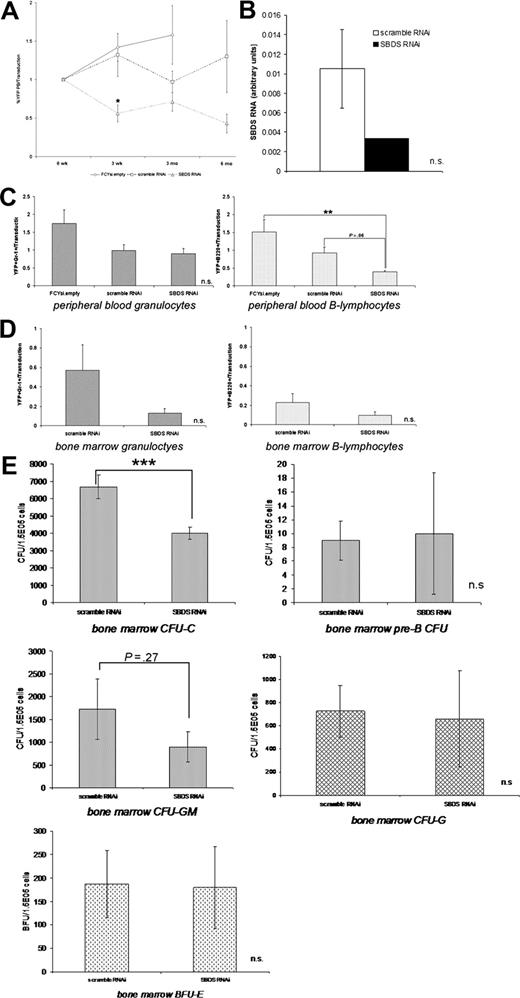
Sbds RNAi reduces the number of myeloid progenitors in vivo
To assess the effect of Sbds inhibition on long-term hematopoietic engraftment, YFP levels in the peripheral blood were measured through 6 months after transplantation. In mice reconstituted with FCYsi.empty and scramble RNAi-transduced cells, the percentage of YFP+ cells in the peripheral blood remained stable throughout the observation period. As shown in Figure 3, the percentage of Sbds RNAi cells in the blood was reduced by 3 weeks after transplantation. However, this reduced level of circulating YFP+ cells remained stable for 6 months (Figure 5A). Importantly, persistent reduction of Sbds in sorted YFP+ cells was documented at both 3 and 6 months after transplantation. Compared with the scramble RNAi control, Sbds RNA expression in sorted YFP+ spleen cells at 3 to 6 months after transplantation was reduced by 68.3% (Figure 5B).
Long-term hematopoietic reconstitution by Sbds RNAi-transduced cells. (A) YFP expression in peripheral blood cells was measured at the indicated time after transplantation (n = 3-11). (B) Sbds mRNA relative to β-actin mRNA expression in mice 3 months after transplantation was measured by real-time RT-PCR (n = 2). The contribution of YFP+ granulocytes and B lymphocytes to the peripheral blood at 3 months (C) and the bone marrow at 6 months (D) was measured. Data were normalized to the pretransplant transduction efficiency. (E) The ability of YFP+ bone marrow cells to generate CFU-Cs and pre-B CFUs (6 months after transplantation), and CFU-GM, CFU-G, and BFU-E colonies (3 months after transplantation) was measured (n = 3-5). *P ≤ .05, **P ≤ .01, ***P ≤ .005. All data represent the mean plus or minus SEM.
Long-term hematopoietic reconstitution by Sbds RNAi-transduced cells. (A) YFP expression in peripheral blood cells was measured at the indicated time after transplantation (n = 3-11). (B) Sbds mRNA relative to β-actin mRNA expression in mice 3 months after transplantation was measured by real-time RT-PCR (n = 2). The contribution of YFP+ granulocytes and B lymphocytes to the peripheral blood at 3 months (C) and the bone marrow at 6 months (D) was measured. Data were normalized to the pretransplant transduction efficiency. (E) The ability of YFP+ bone marrow cells to generate CFU-Cs and pre-B CFUs (6 months after transplantation), and CFU-GM, CFU-G, and BFU-E colonies (3 months after transplantation) was measured (n = 3-5). *P ≤ .05, **P ≤ .01, ***P ≤ .005. All data represent the mean plus or minus SEM.
Granulopoiesis in mice stably reconstituted with transduced bone marrow cells was next examined. Three months after transplantation, Sbds RNAi did not significantly reduce the number of circulating granulocytes (Gr-1+) compared with FCYsi.empty and scramble RNAi controls (Figure 5C). Because the bone marrow is the major site of granulopoiesis, the effect of Sbds RNAi on granulocyte development was examined in the bone marrow of mice at the termination of the experiment, 6 months after transplantation. Note that there were an insufficient number of FCYsi.empty mice to include in this analysis. Compared with the scramble RNAi control, Sbds RNAi decreased the contribution of YFP+ cells to the total granulocyte (Gr-1+) population, although not reaching statistical significance (P = .2; Figure 5D). Furthermore, Sbds RNAi-transduced myeloid bone marrow cells formed significantly fewer CFU-Cs compared with scramble RNAi-transduced control cells (Figure 5E). Additional analysis showed that this reduction in CFU-C number was due to a decrease in CFU-GM colonies; in contrast, no differences were observed in CFU-G or erythroid (BFU-E) colony numbers (Figure 5E). Interestingly, the distribution of YFP+ cells into Gr-1high (mature) and Gr-1low (precursor) populations was similar, suggesting that there was no accumulation of granulocytic precursors (data not shown). Collectively, these data suggest that Sbds RNAi results in a reduction of myeloid progenitors, but only modestly inhibits granulopoiesis in vivo.
Sbds RNAi reduces circulating B lymphocytes
Because lymphocytes are often abnormal in patients with SDS,5 we next measured the contribution of Sbds RNAi-transduced cells to the T- and B-lymphocyte lineages. Whereas T-lymphocyte contribution was comparable to controls (data not shown), a significant reduction in the contribution of Sbds RNAi-transduced cells to the B-lymphocyte lineage was documented. Compared with FCYsi.empty and scramble RNAi controls, the percentage of circulating B220+ cells that expressed YFP was significantly reduced (Figure 5C). In contrast, in the bone marrow, which contains mostly naive and immature B lymphocytes, the contribution of Sbds RNAi-transduced cells to the B-lymphocyte lineage was comparable to control cells (Figure 5D). Moreover, the capacity of Sbds RNAi-transduced cells to form pre-B CFUs was normal (Figure 5E). These data raise the possibility that inhibition of Sbds results in impaired survival or trafficking of mature B cells but does not affect B-cell differentiation in vivo.
Reduction in Sbds inhibits hematopoietic reconstitution after secondary bone marrow transplantation
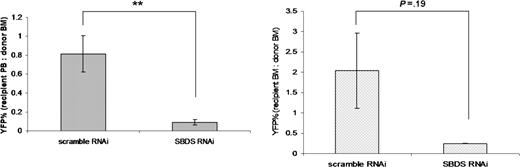
Finally, the ability of hematopoietic cells transduced with Sbds RNAi to reconstitute hematopoiesis in secondary recipients was assessed 6 months after primary bone marrow transplantation. Shown is the percentage of circulating YFP+ cells in the secondary recipient compared with the primary recipient at the time of harvest (Figure 6A). A modest reduction in blood cells expressing YFP was observed in secondary recipients receiving bone marrow cells harvested from scramble RNAi controls (the ratio of YFP+ cells in the recipient peripheral blood to donor bone marrow was 0.70 ± 0.24; Figure 6A). Strikingly, Sbds RNAi significantly impairs secondary engraftment, because this ratio is reduced to 0.10 (± 0.09), which represents an 85% reduction compared with scramble RNAi controls (Figure 6A). Likewise, a reduction in the ratio of YFP+ cells in recipient bone marrow to YFP+ cells in donor bone marrow was observed in Sbds RNAi secondary transplant recipients (0.25 ± 0.04) compared with scramble RNAi controls (2.04 ± 1.4) (Figure 6B).
Sbds RNAi inhibits secondary transplant engraftment. Bone marrow was harvested from primary transplant recipients 6 months after transplantation (n = 3), pooled, and transplanted into secondary recipients. The percentage of YFP+ leukocytes in the (A) blood (n = 8) and (B) bone marrow (n = 2-3) was measured at 6 weeks. Data are presented as the ratio of the percentage of YFP+ cells in the peripheral blood (A) or bone marrow (B) of the recipient to the percentage of YFP+ cells in the bone marrow of the donor (average ± SEM). **P ≤ .01.
Sbds RNAi inhibits secondary transplant engraftment. Bone marrow was harvested from primary transplant recipients 6 months after transplantation (n = 3), pooled, and transplanted into secondary recipients. The percentage of YFP+ leukocytes in the (A) blood (n = 8) and (B) bone marrow (n = 2-3) was measured at 6 weeks. Data are presented as the ratio of the percentage of YFP+ cells in the peripheral blood (A) or bone marrow (B) of the recipient to the percentage of YFP+ cells in the bone marrow of the donor (average ± SEM). **P ≤ .01.
Discussion
Current evidence suggests that mutations of SBDS in patients with SDS are loss-of-function mutations. First, the most common SBDS mutant alleles encode severe C-terminal deletions, removing 75% (183-184TA > CT) and 65% (258 + 2T > C) of the wild-type protein.7 Second, the mutation of S cerevisiae SBDS that is paralogous to the most severe SDS-associated allele (183-184TA > CT) is lethal.15 These observations, as well as the absence of homozygosity for 183 to 184TA > CT in patients with SDS, indicate that complete loss of SBDS function is lethal in eukaryotes. Consistent with this conclusion, Zhang et al25 recently showed that targeted disruption of the Sbds gene in mice results in embryonic lethality. Austin et al12 documented read-through of the most commonly inherited truncation allele (258 + 2T > C), resulting in residual wild-type SBDS protein. Because essentially all (98%) patients with SDS with documented SBDS mutations carry this allele,7,,–10 it is interesting to speculate that this residual activity is essential for survival. Because there are no studies addressing the functional effect of possible neomorphic activity of N-terminal SBDS fragments, the precise molecular pathophysiology of SDS remains unclear.
In the present study, Sbds RNA and protein expression were reduced by lentiviral-mediated RNAi to approximately 15% that of normal controls. Although it is difficult to accurately estimate the percentage of full-length SBDS protein that would be generated from read-through events in vivo, it appears that the level of residual SBDS protein in cell lines derived from patients with SDS may be significantly lower than 15%.12 Even so, some of the hematopoietic abnormalities documented in patients with SDS were recapitulated using our RNAi strategy. Specifically, we observed defects in granulopoiesis, hematopoietic engraftment, and B-lymphocyte homeostasis. Thus, this study provides the first experimental evidence that SBDS function is required for normal mammalian hematopoiesis.
Impaired granulopoiesis is a defining feature of SDS. Chronic or intermittent neutropenia is present in most patients. Bone marrow findings in patients with SDS are highly variable, ranging from normal to granulocyte hypoplasia to granulocyte hyperplasia with a maturation arrest.3 However, the number of myeloid progenitors in the bone marrow is nearly uniformly decreased in patients with SDS.38,,–41 Interestingly, Dror and Freedman41 reported that this decrease in myeloid progenitors is secondary to both a cell intrinsic defect in hematopoietic cells and a defect in bone marrow stromal cells. Consistent with clinical findings in patients with SDS, we show that the number of myeloid progenitors was significantly reduced in mice reconstituted with Sbds RNAi-transduced bone marrow cells. The hypothesis that increased susceptibility to apoptosis is necessary for the pathogenesis of impaired granulopoiesis in SDS is controversial. Dror and Freedman42 showed that both granulocytic cells and myeloid progenitors obtained from patients with SDS displayed increased susceptibility to Fas-mediated apoptosis. Likewise, in 32Dcl3 cells exhibiting lentiviral-mediated knockdown of Sbds, an increase in Annexin V+ cells was observed in culture.43 In contrast, Kuijpers et al11 reported no increase in apoptosis in circulating neutrophils in patients with SDS. In the present study, we show that reduced Sbds expression in myeloid progenitor cells results in a cell intrinsic delay, but not an absolute block, in granulocytic differentiation in vitro. No increase in surface expression of Annexin V was observed, suggesting that increased apoptosis is not responsible for this phenotype. Of note, however, the defect in granulocytic differentiation was more modest in vivo. This finding may relate to the reduced efficacy of Sbds RNAi over time, whereas Sbds protein expression was reduced 86% in cultured myeloid cells 7 days after transduction, only an approximately 68% reduction was observed in bone marrow cells 3 to 6 months after transplantation. Alternatively, signals generated in vivo by the bone marrow microenvironment may be able to compensate for the cell-intrinsic defect in granulocytic differentiation. The contribution of stromal cell dysfunction to SDS was not addressed in this study because Sbds was inhibited only in hematopoietic cells, within the context of wild-type bone marrow stroma. Collectively, these data show that reduced Sbds expression leads to a cell intrinsic defect in myeloid progenitor generation or survival and impaired granulocytic differentiation.
B-lymphocyte number, function, or both are often abnormal in patients with SDS. Dror et al5 reported that B-lymphocyte defects were present in approximately 50% of patients with SDS. These defects include low IgG or subclasses of IgG, absence of specific antibody or isohemagglutinin production, reduced in vitro proliferation, and low numbers of mature B lymphocytes in the circulation. We show that, in mice stably engrafted with Sbds RNAi-transduced cells, the number of circulating transduced B lymphocytes is significantly reduced. In contrast, the number of Sbds RNAi B lymphocytes in the bone marrow was comparable to controls. Moreover, the distribution of B-lymphocyte precursors in the bone marrow into pre- to pro-, pro-, and mature B-lymphocyte fractions, as assessed by CD43/B220 staining, was unaffected by Sbds inhibition (data not shown). Finally, the number of transduced B-lymphocyte progenitors (pre-B CFUs) was comparable to control cells. Together, these data suggest the B lymphopoiesis was normal in Sbds RNAi-transduced cells. Accordingly, we speculate that the reduction in circulating B lymphocytes is likely due to altered survival or trafficking or both.
Bone marrow failure is a common and devastating complication of SDS. In addition to neutropenia, anemia and thrombocytopenia are common in patients with SDS. Pancytopenia and, as noted earlier, bone marrow hypoplasia are present in a substantial number of patients with SDS (reviewed in Dror and Freedman42 ). Moreover, there is a very high rate of progression to MDS or AML; in a recent report, approximately 30% of patients with SDS developed one of these complications (reviewed in Dror6 ). As noted earlier, there is evidence for both a hematopoietic and a bone marrow stromal cell defect in SDS. Similar to other bone marrow failure syndromes, abnormal telomere shortening in leukocytes from patients with SDS has been reported.44 Despite these important clinical observations, the cellular and molecular pathogenesis of bone marrow failure in SDS remains unclear. In the present study, we show that short-term hematopoietic engraftment is severely reduced by the inhibition of Sbds in hematopoietic progenitors. Impaired homing of Sbds RNAi-transduced hematopoietic progenitors to the bone marrow makes a significant contribution to the overall engraftment defect. Furthermore, the more severe hematopoietic engraftment defect upon secondary transplantation of these transduced cells also suggests a defect in hematopoietic self-renewal.
There is accumulating evidence that cell motility, migration, or both defects might play an important role in the pathogenesis of SDS. It is well established that neutrophils isolated from patients with SDS exhibit cell motility and chemotactic deficiencies, probably because of cytoskeletal or microtubular abnormalities.5,18,20,21,23,24 Furthermore, a recent study of the social amoebae Dictyostelium discoideum has shown a potential role for the Sbds homolog in pseudopod-based cell locomotion.19 Although a role for Sbds in motility has not yet been directly described in neutrophils, it is interesting to note that a rare subset of patients with X-linked severe congenital neutropenia carry a mutation in the Wiskott-Aldrich syndrome gene that abolishes the autoinhibition of Wiskott-Aldrich syndrome protein (WASP). This deregulation of WASP leads to constitutive actin polymerization and the subsequent destabilization of the cytoskeleton, and it is thought to reduce cell motility or chemotaxis.45 In the present study, we examined the effect of Sbds inhibition on CXCL12α-induced chemotaxis of HPCs, because this property has been implicated in homing to the bone marrow.34,,–37 Surprisingly, CXCL12α-induced migration of Sbds RNAi-transduced HPCs was comparable to control cells. Further study will be required to determine whether Sbds regulates HPC migration in response to other stimuli or whether, in fact, defects in motility in the absence of Sbds are noncell intrinsic.
In summary, we show that inhibition of Sbds expression in primary murine hematopoietic cells by RNAi reproduces many of the hematopoietic defects observed in patients with SDS. Thus, these data provide the first experimental evidence that Sbds is required for normal hematopoiesis. We show for the first time that Sbds contributes to HPC homing to the bone marrow and subsequent hematopoietic reconstitution. Studies to define the molecular basis for this impaired homing may pro-vide insight into the pathogenesis of bone marrow failure in patients with SDS.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jeff Millbrandt for providing pFCYsi; Benjamin Capoccia, David Jin, and the Siteman Cancer Center Cell Sorting core for outstanding technical assistance; and Matthew Christopher, Monica Bessler, and Philip Mason for helpful scientific discussions.
This work was supported by National Institutes of Health grant HL079562 (D.C.L) and grant T32 HL07088–23 (A.S.R.).
National Institutes of Health
Authorship
Contribution: A.S.R., A.D.G., F.L., and J.R.W. performed experiments; A.S.R. and A.D.G. analyzed results and made the figures; A.S.R. and D.C.L. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Division of Oncology, Department of Medicine, 660 South Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail:dlink@im.wustl.edu.