The mechanisms underlying granulocyte-colony stimulating factor (G-CSF)–induced mobilization of granulocytic lineage cells from the bone marrow to the peripheral blood remain elusive. We provide evidence that the transcriptional repressor growth factor independence-1 (Gfi-1) is involved in G-CSF–induced mobilization of granulocytic lineage cells from the bone marrow to the peripheral blood. We show that in vitro and in vivo G-CSF promotes expression of Gfi-1 and down-regulates expression of CXCR4, a chemokine receptor essential for the retention of hematopoietic stem cells and granulocytic cells in the bone marrow. Gfi-1 binds to DNA sequences upstream of the CXCR4 gene and represses CXCR4 expression in myeloid lineage cells. As a consequence, myeloid cell responses to the CXCR4 unique ligand SDF-1 are reduced. Thus, Gfi-1 not only regulates hematopoietic stem cell function and myeloid cell development but also probably promotes the release of granulocytic lineage cells from the bone marrow to the peripheral blood by reducing CXCR4 expression and function.
Introduction
The generation of neutrophils from hematopoietic precursors and their release to the peripheral circulation are highly regulated processes that ensure the maintenance of homeostatic neutrophil levels in the blood and their rise in response to bacterial infections and other signals. Granulocyte-colony stimulating factor (G-CSF) has emerged as a critical regulator of granulopoiesis because mice carrying homozygous deletions of G-CSF or its receptor are severely neutropenic,1,2 and dominant-negative mutations of granulocyte colony-stimulating factor receptor (G-CSFR) have been linked to severe defects of granulopoiesis.3,4 In addition, administration of G-CSF induces an expansion of myeloid lineage cells in the bone marrow and promotes the release of neutrophils and hematopoietic progenitor cells from the bone marrow to the peripheral blood.5 On the basis of these properties, G-CSF is widely used to induce granulopoiesis and to mobilize hematopoietic progenitors to the peripheral blood.
The biologic activities of G-CSF are solely mediated by its activation of the G-CSFR that is expressed on myeloid lineage progenitor cells.6 Compelling evidence from genetic studies and other studies showed that G-CSF indirectly promotes hematopoietic cell and neutrophil mobilization to the peripheral blood by modulating the activities of the chemokine SDF-1, its receptor CXCR4, or both, which are essential for the retention of hematopoietic cells to the bone marrow cavity.7,,,,–12 AMD3100, a competitive inhibitor of SDF-1 binding to its receptor, and a mutant form of SDF-1β, which induces prolonged down-regulation of the CXCR4 surface receptor, promote the mobilization of neutrophils and hematopoietic cells to the peripheral blood.13,14
Osteoblasts, stromal cells, and endothelial cells constitutively express SDF-1 in the bone marrow; hematopoietic cells express CXCR4.15,16 During stem-cell mobilization with G-CSF, SDF-1 and CXCR4 protein levels decrease in the bone marrow.7,,,–11 To explain such reductions, some studies have supported a role for enzymatic cleavage of SDF-1 or CXCR4 or both by metalloproteinase-9 neutrophil elastase, and cathepsin-G,10,17,18 but mice deficient of these enzymes responded normally to G-CSF mobilization.8 Other studies indicated that G-CSF transcriptionally down-regulates SDF-1 expression in the bone marrow, acting indirectly on osteoblasts that do not express G-CSFR.19 We have recently reported that G-CSF reduces CXCR4 expression in bone marrow Gr-1+ myeloid cells, which express G-CSFR.20 This is consistent with earlier observations that CXCR4 levels are reduced on neutrophils and CD34+ hematopoietic progenitor cells recently released from the bone marrow to the peripheral blood.21,22
In the current study, we show that G-CSF promotes the expression of the transcriptional repressor growth factor independence-1 (Gfi-1) in cells of myeloid lineage in vitro and in vivo and that Gfi-1 represses CXCR4 transcription. The transcription factor Gfi-1 is essential for granulocytic lineage maturation during development23,–25 and for maintenance of the hematopoietic stem- cell pool postnatally.26,27 Thus, the results described here increase understanding of the molecular mechanisms responsible for G-CSF–induced mobilization of myeloid cells from the bone marrow to the peripheral blood and extend the spectrum of activities of the Gfi-1 repressor.
Materials and methods
Cells
The murine IL-3–dependent 32Dcl3 cell line28 (a gift of Dr Alan D. Friedman, Johns Hopkins University) was maintained in Iscove-modified Dulbecco medium (Mediatech; Cellgro, Herndon, VA) containing 10% heat-inactivated fetal bovine serum (FBS; Biosource, Camarillo, CA) plus 100 pg/mL IL-3 (PeproTech, Rocky Hill, NJ) with biweekly subculture. For induction, 32Dcl3 cells were cultured (0.5–1.5 × 106/mL) with G-CSF (100 ng/mL) added to maintenance medium. Bone marrow cells were obtained by flushing femurs and tibias of C57BL/6Ncr female mice (6-9 weeks old; The Jackson Laboratory, Bar Harbor, ME). Gr1+ cells were selected by positive selection using FITC-conjugated rat anti–mouse Gr1 antibodies followed by anti-FITC microbeads (Miltenyi Biotech, Auburn, CA).
Virus packaging and infection
The retroviral constructs GFi-1–GFP-RV and GFP-RV were previously described.29 32Dcl3 cells were suspended (1 × 106 cells/mL) in culture medium with filtered virus-containing supernatant (dilution 1:2) supplemented with Polybrene (8 μg/mL; Sigma, St Louis, MO), centrifuged (1 hour at 1000 g), incubated (3 hour, 37°C), washed, and replated in culture medium. After 24 to 48 hours of incubation, 20% to 40% of cells were GFP positive; homogeneous populations of transduced cells (≥ 90% GFP-positive cells) were selected by electronic cell sorting (FACSVantage SE; BD Biosciences, San Jose, CA).
Mobilization of hematopoietic cells
All animal experiments were approved by the NCI-Bethesda Animal Care and Use Committee. C57BL/6NCr female mice, 6 to 10 weeks old were treated intraperitoneally with 5 μg human G-CSF (filgrastim; Amgen, Thousand Oaks, CA) daily for 5 days.20 Neutrophil mobilization was documented by peripheral blood cell counts showing leukocytosis. Control mice received daily intraperitoneal injections of PBS containing 2% human albumin for 5 days. On day 6 the mice were killed; unfractionated bone marrow cell populations were harvested from the femurs and tibias.
Immunoblotting
For intracytoplasmic and surface proteins, cells were lysed in 1% Triton X-100 TNE buffer (25mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA on ice with protease inhibitor cocktail [Complete, Roche, Indianapolis, IN]). After centrifugation, supernatants were For nuclear proteins, cells were lysed in 50 mM Tris (pH 7.4) buffer with 1% SDS and 150 mM NaCl, 1 mM NaVO4, and protease inhibitor cocktail. Samples were incubated (30 minutes, 37°C) or boiled (10 minutes), centrifuged, resolved by SDS–polyacrylamide gel electrophoresis (NuPage 10% Bis-Tris or 10%-20% Tricine gels; Invitrogen, Carlsbad, CA), and blotted onto nitrocellulose membranes (protran BA-83; Whatman Schleicher and Schuell, Florham Park, NJ). After blocking (PBS with 5% milk), membranes were incubated overnight with primary antibodies (myeloperoxidase, C-16 affinity-purified goat antibody; neutrophil elastase, C-17 affinity-purified goat antibody; Gfi-1, N-20 affinity-purified goat antibody; actin, C-11 goat IgG antibody [all from Santa Cruz Biotechnology, Santa Cruz, CA]); phospho-STAT1, Tyr701 rabbit monoclonal antibody; phospho-STAT, Tyr705 rabbit IgG antibody (both from Cell Signaling Technology, Beverly, MA); phospho-STAT5, Tyr694 rabbit monoclonal antibody; STAT3, clone 84 mouse monoclonal antibody; CXCR4, 2B11 rat antimouse monoclonal antibody (BD Biosciences). Secondary antibodies included horseradish peroxidase–conjugated goat anti–rat and donkey anti–goat (Santa Cruz Biotechnology); donkey antimouse and donkey antirabbit (Amersham Pharmacia Biotech, GE Healthcare, Piscataway, NJ). Bound antibodies were detected by chemiluminescence; Amersham Pharmacia Biotech, GE Healthcare).
RNA isolation, semiquantitative and quantitative RT-PCR
Total RNA was isolated using TRIsol reagent (Invitrogen). Semiquantitative polymerase chain reaction (PCR) was performed as described,30 using primers for amplification of mouse CXCR4, GAPDH,20 and Gf1-1.29 PCR products were visualized in 1.8% agarose gels (NuSieve agarose; FMC, Rockland, ME). For quantitative reverse transcription (RT)–PCR, we used One-step RT-PCR kit (Qiagen, Chatworth, CA) with SYBR Green PCR mix (Applied Biosystems, Foster City, CA). Reactions were performed in an Abi Prism 7900HT sequence detection system (Applied Biosystems). Primers used for quantitative PCR of mouse Gfi-1,29 CXCR4, and GAPDH20 mRNAs were described.
Flow cytometry and cytochemistry
Flow cytometric analysis was performed as described.20 Cells were stained for surface CXCR4 with FITC-conjugated rat anti–mouse 2B11 monoclonal antibody (BD Biosciences) and for surface Gr1 with the APC-conjugated rat anti–mouse Gr1 monoclonal antibody (BD Biosciences). Isotype-matched FITC and APC-conjugated immunoglobulin (BD Biosciences) served as controls. Nontransduced parental cells served as controls for GFP-expressing cells. Cells were analyzed with a FACSCalibur cytofluorimeter (BD Biosciences). Data were collected from 10 × 103 viable cells, and results were analyzed with CELLQuest software (BD Biosciences). Cells were cytocentrifuged and stained with Wright-Giemsa solutions.
Cell proliferation, migration, and adhesion assays
32Dcl3 cells were incubated (0.1-1 × 106/mL; 1-8 days) in triplicate 96-well plates in maintenance culture medium (containing 100 pg/mL IL-3) or with the addition of G-CSF (100 ng/mL). Proliferation was measured by 3H-thymidine deoxyribose incorporation (0.5 μCi/well [0.022 MBq]; New England Nuclear, Waltham, MA) during the last 6 hours of culture. Results are expressed as mean (± SEM) cpm/culture. Cell migration and adhesion were evaluated as described.20 For cell migration we used 24-well transwell chambers (pore size, 3 μm; Costar, Corning, NY), incubating 32Dcl3 cells (1 × 106 cells) at 37°C for 4 hours in migration medium (Iscove-modified Dulbecco medium containing 0.5% BSA and 10 mM HEPES) in the upper chamber of the transwell. The lower chamber contained migration medium alone, with FBS (25% final concentration), or with recombinant human SDF-1α (100 ng/mL; PeproTech). For cell adhesion we coated 96-well plates (Immunon 4HBX; Thermo Labsystems, Franklin, MA) with 4 μg/mL SDF-1α or diluent (PBS with 1% BSA) at 4°C for 18 hours. After blocking (PBS with 5%BSA) at room temperature for 1 hour, 32Dcl3 cells (1 × 105 in 50 μL complete culture medium) were added and incubated (37°C for 30 minutes). After washing in PBS, attached cells were removed (5 mM EDTA in PBS) and counted.
Transient transfection and luciferase assay
CXCR4 promoter constructs (gifts of Dr Wilhelm Krek31 ; provided by Dr Rhoda Alani, John Hopkins University) included pGL2 −2632/86 (full-length CXCR4 promoter), pGL2 −1368/86, pGL2 −920/86, and pGL2 −832/86. Transfections were performed using Amaxa nucleofector system (Amaxa Biosystems, Gaithersburg, MD) optimized for 32D cells (program E-32), with Cell Line Nucleofector Solution V (100 μL/reaction). Cells were cotransfected with 2.5 μg phRL-SV40 (Renilla luciferase reference control plasmid; Promega, Madison, WI) to control for transfection efficiency. After transfection, cells were incubated (4-24 hours; 6-well culture plates) at 37°C in complete culture medium (0.5 mL). Firefly luciferase and Renilla luciferase activities from transfected cells were measured in triplicate microtiter wells using Dual-Glo Luciferase Assay System following the manufacturer's instructions (Promega). Relative luciferase activity is defined as the ratio of firefly luciferase to Renilla luciferase activity.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed as described.32 Briefly, 32Dcl3 cells (5 × 106) were fixed with 1% formaldehyde (10 minutes at 37°C), treated with glycine (125 mM, 5 minutes at room temperature), and sonicated (10 rounds of 30 seconds with 30-second breaks) at 4°C in Bioruptor (4.5 setting; Cosmo Bio, Carlsbad, CA). Soluble chromatin (0.2-0.5 mL, from 2–5 × 106 cells) was incubated overnight (4°C) with 3 μg goat anti–GFi-1 antibody (N-20; Santa Cruz Biotechnology) or goat IgG (Organon Technica, Durham, NC) and 20 μL protein G/A agarose (1:1 slurry, blocked with 0.5% BSA and 0.1 mg/mL salmon sperm DNA). Immunoprecipitates were incubated (65°C for 15 minutes), and supernatants were incubated at 55°C for 2 hours and 65°C overnight. After phenol/chloroform and chloroform extraction and ethanol precipitation (with glycogen carrier) samples (2-3 μL) were used per PCR reaction. Primer pairs were designed (Primer3 software, Whitehead Institute, Cambridge, MA) for amplification of 150 to 250 base pair (bp) surrounding putative Gfi-1–binding sites33 within 4-kilobase (kb) sequences upstream the murine CXCR4 transcription start site. Primers and location of target Gfi-1–binding site (counting from transcription start site) included sense, 5-GCT TTC CTC GAC CAA AGA GA-3, and antisense, 5-AGA GCG CTG GTG TTC AAG AT-3 (−1958/−1972); sense, 5-TAA AAC AGC TCC CCA GCA CT-3, and antisense, 5-TCT GAT GAT CCC GTT TGT CA-3 (−1427/−1441); sense, 5-CTG CCA CAC CTG ACC CTA AT-3, and antisense, 5-CTG GAG TTT ACG GGT GGA GA-3 (−545/−559); sense, 5-CTC GCC GCT CAA TTC TTT AT-3, and antisense, 5-TAG GGT CAG GTC TGG CAG A-3 (−390/−404). Conditions for PCR were optimized for sonicated chromatin and included 30 to 35 cycles of amplification.
Results
G-CSF reduces levels of CXCR4 protein in 32Dcl3 granulocytic lineage cells
We examined whether the murine 32D clone 3 (32Dcl3) granulocytic precursor cell line could serve as a surrogate for primary granulocytic lineage cells to study regulation of CXCR4 by G-CSF. With IL-3 supplementation and appropriate feeding, the 32Dcl3 cells grow continuously in culture as undifferentiated myeloblasts and can be induced to differentiate along the granulocytic lineage on withdrawal of IL-3 and exposure to G-CSF.28 Because removal of IL-3 from 32Dcl3 cell cultures results in cell death, we tested the effects of G-CSF (100 ng/mL) in the presence of IL-3. At 100 pg/mL, IL-3 maintains 32Dcl3 cell viability at 90% or greater for 3 days when the cells are cultured (0.5-1.0 × 106/mL) in complete culture medium alone or with G-CSF, and viability subsequently declines (30%-50% viability by day 6) if the medium is not changed and IL-3 is not repleted. Proliferation of 32Dcl3 cells is highest after 24-hour culture and then steadily declines with IL-3 alone or IL-3 and G-CSF (Figure 1A), and the cells maintain the morphology of myeloblast/promyelocytic cells after 3- and 5-day culture with G-CSF in the presence of IL-3 (Figure 1B). By day 8, cell death is prominent with a minority (10%-15%) of cells incubated with IL-3 plus G-CSF showing morphologic evidence of granulocytic differentiation (not shown). These results indicate that under the conditions used here, which include IL-3, G-CSF induces a modest morphologic granulocytic differentiation in 32Dcl3 cells, consistent with previous observations showing that IL-3 impairs G-CSF–induced granulocytic differentiation in 32Dcl3 cells.28
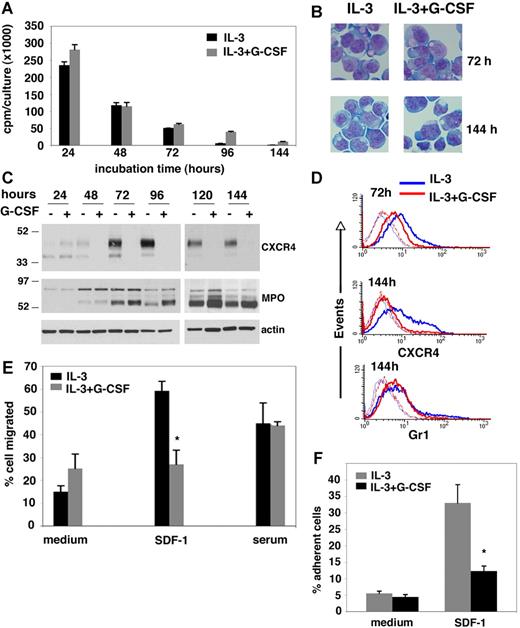
Effects of G-CSF on the growth, differentiation, and CXCR4 expression in the IL-3–dependent 32Dcl3 myeloid cells. (A) DNA synthesis in 32Dcl3 cells cultured for 24 to 144 hours in the presence of IL-3 alone or IL-3 plus G-CSF measured by 3H thymidine incorporation. The error bars represent standard deviation (SD) of the mean (triplicate cultures). (B) Microscopic morphology of 32Dcl3 cells cultured for 72 and 144 hours with IL-3 alone or with IL-3 plus G-CSF. Cytospin preparations were stained with May-Grünwald-Giemsa and mounted with PBS/1% glycerol. Cells were visualized through an Eclipse 6600 Nikon microscope (Melville, NY) equipped with a 60×1.40 oil objective (Nikon), and photographed with a Cool Snaps fx Roper Camera (Roper Scientific, Duluth, GA). Images were acquired with IPlab software (Biovision Technologies, Fairfax, VA) and imported into Adobe Photoshop 7.0 software (Adobe, San Jose, CA). (C) Western blot analysis of CXCR4, MPO, and actin content in 32Dcl3 cells cultured for 24 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. The blot was stained with antibodies to CXCR4 then stripped and stained for actin first and then for MPO. (D) Flow cytometric analysis of CXCR4 and Gr-1 surface levels in 32Dcl3 cells cultured with IL-3 alone or IL-3 plus G-CSF for 72 and 144 hours. Background fluorescence with control antibodies is also shown (dotted line). (E) Transwell migration of 32Dcl3 cells in response to recombinant SDF-1α or serum. Cells were precultured for 3 days with IL-3 alone or with IL-3 plus G-CSF. The results (expressed as the percentage of input cells) reflect the mean of 3 experiments, each performed in triplicate. The error bars represent SD of the mean (*P < .05). (F) Attachment of 32Dcl3 cells to wells coated with SDF-1α or medium only. Cells were precultured for 3 days with IL-3 alone or with IL-3 plus G-CSF. The results reflect the means of 3 experiments, each performed in 5 replicates. Results are expressed as the percentage of input cells; error bars represent SD of the mean (*P < .05).
Effects of G-CSF on the growth, differentiation, and CXCR4 expression in the IL-3–dependent 32Dcl3 myeloid cells. (A) DNA synthesis in 32Dcl3 cells cultured for 24 to 144 hours in the presence of IL-3 alone or IL-3 plus G-CSF measured by 3H thymidine incorporation. The error bars represent standard deviation (SD) of the mean (triplicate cultures). (B) Microscopic morphology of 32Dcl3 cells cultured for 72 and 144 hours with IL-3 alone or with IL-3 plus G-CSF. Cytospin preparations were stained with May-Grünwald-Giemsa and mounted with PBS/1% glycerol. Cells were visualized through an Eclipse 6600 Nikon microscope (Melville, NY) equipped with a 60×1.40 oil objective (Nikon), and photographed with a Cool Snaps fx Roper Camera (Roper Scientific, Duluth, GA). Images were acquired with IPlab software (Biovision Technologies, Fairfax, VA) and imported into Adobe Photoshop 7.0 software (Adobe, San Jose, CA). (C) Western blot analysis of CXCR4, MPO, and actin content in 32Dcl3 cells cultured for 24 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. The blot was stained with antibodies to CXCR4 then stripped and stained for actin first and then for MPO. (D) Flow cytometric analysis of CXCR4 and Gr-1 surface levels in 32Dcl3 cells cultured with IL-3 alone or IL-3 plus G-CSF for 72 and 144 hours. Background fluorescence with control antibodies is also shown (dotted line). (E) Transwell migration of 32Dcl3 cells in response to recombinant SDF-1α or serum. Cells were precultured for 3 days with IL-3 alone or with IL-3 plus G-CSF. The results (expressed as the percentage of input cells) reflect the mean of 3 experiments, each performed in triplicate. The error bars represent SD of the mean (*P < .05). (F) Attachment of 32Dcl3 cells to wells coated with SDF-1α or medium only. Cells were precultured for 3 days with IL-3 alone or with IL-3 plus G-CSF. The results reflect the means of 3 experiments, each performed in 5 replicates. Results are expressed as the percentage of input cells; error bars represent SD of the mean (*P < .05).
Using this system, we examined the effects of G-CSF (100 ng/mL) on CXCR4 protein levels in 32Dcl3 cells (1.0 × 106/mL) cultured in the presence of 100 pg/mL IL-3. By immunoblotting with specific antibodies (2B11), CXCR4 is identified as 2 bands with approximate relative molecular weight of 47 and 42 kDa; the relative intensities of the 2 bands vary.20 In the presence of IL-3 alone, CXCR4 was detected at low levels in 32Dcl3 cells during the initial 48 hours of incubation, but levels increased after 72 to 96 hours of culture, when cell proliferation was reduced, and decreased somewhat after 144 hours (Figure 1C). G-CSF reduced levels of CXCR4 in 32Dcl3 cells (in the presence of IL-3) beginning 48 hours after addition to culture, and this CXCR4 reduction persisted at the subsequent time points (72, 96, 120, and 144 hours) (Figure 1C). This difference in CXCR4 protein levels was not attributable to uneven loading as documented by membrane reprobing with antiactin antibodies (Figure 1A,C), providing evidence that G-CSF can significantly reduce CXCR4 expression levels in 32Dcl3 cells. Consistent with previous observations,28,34 we found that 32Dcl3 cells incubated with G-CSF plus IL-3 expressed greater levels of the approximate 60-kDa heavy subunit of the mature myeloperoxidase (MPO) protein, compared with cells incubated with IL-3 alone, beginning after 72 hours of incubation and continuing through the 144-hour time point (Figure 1C). These results show that increased MPO heavy subunit expression by G-CSF followed temporally the decreased CXCR4 expression and provide evidence that G-CSF can differentially regulate protein expression in this system. Dose-response experiments (not shown) indicated that, in the presence of a fixed IL-3 concentration (100 pg/mL), G-CSF at 1 ng/mL was ineffective at reducing CXCR4 levels in 32Dcl3 cells; G-CSF at the concentration of 10 ng/mL was effective at partially reducing CXCR4 levels; and G-CSF at concentrations of 100 ng/mL or higher produced maximal CXCR4 reductions (not shown). Consistent with the results of immunoblotting, fluorescence-activated cell sorting (FACS) analysis showed that cell-surface CXCR4 expression in 32Dcl3 cells is reduced after 72-hour and more prominently after 144-hour culture with IL-3 (100 pg/mL) and G-CSF (100 ng/mL) compared with cultures with IL-3 alone. Instead, surface expression Gr-1 is similar in the presence or absence of G-CSF (Figure 1D).
The 32Dcl3 cells migrate in response to serum in transwell migration assays.35 We now tested whether reduced surface CXCR4 levels induced by G-CSF lead to specific changes in the chemotactic response of 32Dcl3 cells to the CXCR4 ligand SDF-1α. After 3-day culture with IL-3 (100 pg/mL) alone, approximately 4-fold more 32Dcl3 cells migrated in response to SDF-1α (50 ng/mL) compared with cells incubated in medium alone, consistent with their significant expression of surface CXCR4 (Figure 1E). By contrast, 32Dcl3 cells cultured for 3 days in the presence of IL-3 (100 pg/mL) plus G-CSF (100 ng/mL) displayed minimal specific migration to SDF-1α, consistent with their reduced expression of surface CXCR4 (Figure 1E). By contrast, 32Dcl3 cell migration to serum-containing medium was similar after culture with IL-3 alone or with G-CSF (Figure 1E), providing evidence for SDF-1 specificity. In additional experiments, we found that 32Dcl3 cells displayed a reduced capacity to specifically attach to SDF-1–coated wells after 3-day culture in the presence of IL-3 (100 pg/mL) plus G-CSF (100 ng/mL) compared with cells cultured with IL-3 alone (Figure 1F). Migration and attachment to SDF-1 were markedly impaired in 32Dcl3 cells despite their continued expression of cell surface CXCR4 after 3-day culture with G-CSF, suggesting either a threshold effect or accompanying defects in CXCR4 internalization or signaling. These results show that G-CSF reduces CXCR4 protein expression and function in 32Dcl3 cells cultured with IL-3.
G-CSF reduces CXCR4 mRNA expression in 32Dcl3 cells
G-CSF reduces the intensities of the approximately 47-kDa and approximately 42-kDa CXCR4-related bands in 32Dcl3 granulocytic lineage cells after 3- and 6-day culture, without evidence for induction of higher or lower molecular weight forms of the protein attributable to CXCR4 processing or degradation detected by immunoblotting (Figure 2A). We therefore measured levels of CXCR4 and GAPDH mRNAs in 32Dcl3 cells cultured with or without G-CSF. By using quantitative PCR, we found that levels of CXCR4 mRNA were specifically reduced by 74% ± 5.8% of 3 experiments in 32Dcl3 cells cultured for 3 days with G-CSF (100 ng/mL) plus IL-3 (100 pg/mL) compared with cells cultured with IL-3 alone (Figure 2B). To determine whether this mRNA reduction was attributable to an effect of G-CSF on mRNA synthesis or degradation, we first measured the rate of spontaneous CXCR4 decay in the presence of the transcriptional inhibitor actinomycin-D (5 μg/mL). By quantitative PCR (Figure 2C), levels of CXCR4 mRNA decreased time dependently as a function of GAPDH in the presence of actinomycin-D; by 1 hour, CXCR4 mRNA levels were reduced by approximately 60% compared with the levels measured at time 0. We then tested whether G-CSF accelerated the rate of CXCR4 mRNA decay. On the basis of the time course of spontaneous CXCR4 decay, we selected the 1- and 2-hour time points to measure the effect of G-CSF (100 ng/mL) on CXCR4 levels in 32Dcl3 cells cultured with IL-3 (100 pg/mL). In the presence of actinomycin-D, we found levels of CXCR4 to be similarly reduced in cells exposed to IL-3 plus G-CSF as opposed to IL-3 alone (Figure 2D). When mRNA synthesis was not blocked, CXCR4 levels were not reduced or only slightly reduced by G-CSF at the 1-hour and 3-hour time points (Figure 2D). These results provide evidence that under the conditions used, G-CSF does not accelerate the rate of CXCR4 mRNA decay in 32Dc cells.
G-CSF regulation of CXCR4 mRNA synthesis. (A) Appearance of CXCR4-related bands evaluated by Western blotting. 32Dcl3 cells were cultured for 3 and 6 days with IL-3 alone or with IL-3 plus G-CSF. (B) CXCR4 mRNA levels were measured by quantitative PCR. 32Dcl3 cells were cultured for 3 days with IL-3 alone or IL-3 plus G-CSF. The results reflect the means of 3 experiments; the error bars reflect SD of the mean (*P < .05). (C) Kinetic of CXCR4 mRNA decay was measured by quantitative PCR. 32Dcl3 cells were cultured for 1 to 6 hours in the presence of actinomycin-D. The results are from a representative experiment and reflect the means of triplicate determinations; the error bars represent SD of the mean. (D) Effects of G-CSF on CXCR4 mRNA decay measured by quantitative PCR. 32Dcl3 cells were cultured for 1 hour or 2 hours with or without actinomycin-D in the presence of IL-3 alone or with IL-3 plus G-CSF. The results (expressed as percentage of CXCR4 mRNA in cells cultured with IL-3 alone) reflect the means of triplicate determinations; the error bars represent SD of the mean.
G-CSF regulation of CXCR4 mRNA synthesis. (A) Appearance of CXCR4-related bands evaluated by Western blotting. 32Dcl3 cells were cultured for 3 and 6 days with IL-3 alone or with IL-3 plus G-CSF. (B) CXCR4 mRNA levels were measured by quantitative PCR. 32Dcl3 cells were cultured for 3 days with IL-3 alone or IL-3 plus G-CSF. The results reflect the means of 3 experiments; the error bars reflect SD of the mean (*P < .05). (C) Kinetic of CXCR4 mRNA decay was measured by quantitative PCR. 32Dcl3 cells were cultured for 1 to 6 hours in the presence of actinomycin-D. The results are from a representative experiment and reflect the means of triplicate determinations; the error bars represent SD of the mean. (D) Effects of G-CSF on CXCR4 mRNA decay measured by quantitative PCR. 32Dcl3 cells were cultured for 1 hour or 2 hours with or without actinomycin-D in the presence of IL-3 alone or with IL-3 plus G-CSF. The results (expressed as percentage of CXCR4 mRNA in cells cultured with IL-3 alone) reflect the means of triplicate determinations; the error bars represent SD of the mean.
G-CSF transcriptionally regulates CXCR4 gene expression in 32Dcl3 cell
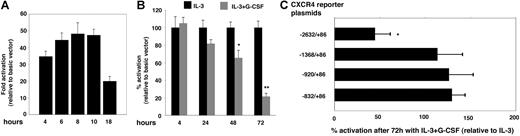
To investigate the possibility that G-CSF transcriptionally regulates CXCR4 gene expression, we used a luciferase reporter containing the human CXCR4 promoter, including the first intronic region (−2632/86,31 a gift of Dr W. Krek). In preliminary experiments, we determined that the PGL-2–CXCR4 promoter (−2632/86) vector induced significant luciferase expression in transiently transfected 32Dcl3 cells compared with the control PGL-2–Basic (empty) vector and that peak activity was measured 6 to 10 hours after transfection followed by a decline after 18 hours (Figure 3A). Using a 6- to 10-hour time point after transfection, we evaluated changes in PGL-2–CXCR4 promoter activity in 32Dcl3 cells cultured over time with IL-3 alone (100 pg/mL) or with G-CSF (100 ng/mL) (Figure 3B). We found that after 4 hours of incubation the levels of luciferase activity induced by the reporter containing the CXCR4 promoter (−2632/86) were similar in 32Dcl3 cells cultured with IL-3 alone or IL-3 plus G-CSF (Figure 3B). However, after 24, 48, and 72 hours of incubation, the levels of luciferase activity in cells incubated with IL-3 plus G-CSF were only 81.8%, 65.6%, and 31.5%, respectively, compared with cells incubated with IL-3 alone (Figure 3B). These results indicate that after 72-hour exposure to G-CSF, 32Dcl3 cells are significantly less effective at supporting CXCR4 promoter activity than control cells cultured without G-CSF, a result we confirmed in additional experiments (IL-3 + G-CSF/IL-3 = 44.77% ± 16.6%; 7 independent experiments) (Figure 3C). To identify which region of the CXCR4 promoter is necessary to observe a reduction in promoter activity by G-CSF, 3 luciferase reporters containing progressive deletions of the CXCR4 promoter (−1368/86, −920/86, −832/86; gifts of W. Krek) were transfected into 32Dcl3 cells (incubated for 3 days with either IL-3 alone or with G-CSF). Our results indicate that sequences located between −1.3 kb and −2.6 kb of the transcriptional start site are required for G-CSF to reduce reporter activity (Figure 3C).
Effects of G-CSF on CXCR4 promoter activity. (A) Time course of luciferase activity after transfection of 32Dcl3 cells with the CXCR4-promoter reporter plasmid or with the control basic (empty) vector. Cells were cotransfected with Renilla luciferase reference control plasmid to account for variation in transfection efficiencies. The results are expressed as fold activation relative to basic vector after correction for Renilla luciferase activity. The representative (n = 3) results reflect the means of triplicate determinations; the error bars represent SD of the mean. (B) Effects of G-CSF preculture on the luciferase activity induced by CXCR4-promoter reporter plasmid. 32Dcl3 cells were cultured (4-72 hours) with IL-3 alone or with IL-3 plus G-CSF and then transfected with the CXCR4-promoter reporter plasmid or with the basic vector. The results are expressed as the percentage of activation of luciferase activity relative to basic vector after correction for transfection efficiencies as measured by cotransfection with Renilla luciferase control plasmid. Luciferase activation in cells cultured with IL-3 alone was set at 100%. The results reflect the means of 5 experiments; the error bars represent SD of the mean (*P < .05; **P < .02). (C) Luciferase activity after transfection with full-length CXCR4-promoter reporter plasmid and deletion mutants. The results are expressed as the percentage of activation of luciferase activity in 32Dcl3 cells precultured with IL-3 plus G-CSF relative to luciferase activity in cells precultured with IL-3 alone (set at 100%). The results reflect the means of 3 experiments; the error bars represent SD of the mean (*P < .05).
Effects of G-CSF on CXCR4 promoter activity. (A) Time course of luciferase activity after transfection of 32Dcl3 cells with the CXCR4-promoter reporter plasmid or with the control basic (empty) vector. Cells were cotransfected with Renilla luciferase reference control plasmid to account for variation in transfection efficiencies. The results are expressed as fold activation relative to basic vector after correction for Renilla luciferase activity. The representative (n = 3) results reflect the means of triplicate determinations; the error bars represent SD of the mean. (B) Effects of G-CSF preculture on the luciferase activity induced by CXCR4-promoter reporter plasmid. 32Dcl3 cells were cultured (4-72 hours) with IL-3 alone or with IL-3 plus G-CSF and then transfected with the CXCR4-promoter reporter plasmid or with the basic vector. The results are expressed as the percentage of activation of luciferase activity relative to basic vector after correction for transfection efficiencies as measured by cotransfection with Renilla luciferase control plasmid. Luciferase activation in cells cultured with IL-3 alone was set at 100%. The results reflect the means of 5 experiments; the error bars represent SD of the mean (*P < .05; **P < .02). (C) Luciferase activity after transfection with full-length CXCR4-promoter reporter plasmid and deletion mutants. The results are expressed as the percentage of activation of luciferase activity in 32Dcl3 cells precultured with IL-3 plus G-CSF relative to luciferase activity in cells precultured with IL-3 alone (set at 100%). The results reflect the means of 3 experiments; the error bars represent SD of the mean (*P < .05).
G-CSF promotes expression of the transcriptional repressor Gfi-1 in 32Dcl3 cells
To gain insight in the mechanisms by which G-CSF transcriptionally down-regulates CXCR4 expression in 32Dcl3 cells, we examined G-CSF signaling in 32Dcl3 cells under conditions leading to CXCR4 down-regulation. Previous studies in various systems have identified STAT3, STAT1, and STAT5 as mediators of signaling by the activated G-CSF receptor.36,37 We cultured 32Dcl3 cells in medium supplemented with IL-3 alone (100 pg/mL) or with IL-3 (100 pg/mL) plus G-CSF (100 ng/mL) for 6 days and examined STAT phosphorylation at different time points. STAT3 phosphorylation was noticeable 1 hour after 32Dcl3 cells were exposed to G-CSF (in the presence of IL-3) and was more apparent at the subsequent time points (24, 48, 72, and 144 hours). By contrast, we found no evidence for sustained STAT1 and STAT5 phosphorylation induced by G-CSF in the current system (Figure 4A).
G-CSF induced STAT3 signaling and Gfi-1 expression. (A) Western blot analysis of STAT1, STAT3, and STAT5 phosphorylation in cell lysates of 32Dcl3 cells cultured for 1 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. Blots were first stained for phosphorylated Stat1 and then stripped and reprobed for P-STAT3, PSTAT5, and total STAT3. (B) Semiquantitative RT-PCR analysis of CXCR4, Gfi-1, and GAPDH mRNAs in 32Dcl3 cells before subculture (time 0) or after incubation with IL-3 alone or with IL-3 plus G-CSF for 24 and 96 hours. Representative experiment (n = 5). (C) Real-time RT-PCR analysis of CXCR4 and Gfi-1 mRNA in 32Dcl3 cells after 24 and 96 hours of incubation with IL-3 alone or with IL-3 pus G-CSF. The results reflect the mean fold (± SD) mRNA change in cells cultured with IL-3 plus G-CSF relative to cells cultured in IL-3 alone. (D) Kinetics of G-CSF–induced Gfi-1 protein expression evaluated by Western blotting. 32Dcl3 cells were cultured for 24 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. The blots were stripped and restained for actin. Relative ratios of Gfi-1 to actin are shown in the lower bar graph.
G-CSF induced STAT3 signaling and Gfi-1 expression. (A) Western blot analysis of STAT1, STAT3, and STAT5 phosphorylation in cell lysates of 32Dcl3 cells cultured for 1 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. Blots were first stained for phosphorylated Stat1 and then stripped and reprobed for P-STAT3, PSTAT5, and total STAT3. (B) Semiquantitative RT-PCR analysis of CXCR4, Gfi-1, and GAPDH mRNAs in 32Dcl3 cells before subculture (time 0) or after incubation with IL-3 alone or with IL-3 plus G-CSF for 24 and 96 hours. Representative experiment (n = 5). (C) Real-time RT-PCR analysis of CXCR4 and Gfi-1 mRNA in 32Dcl3 cells after 24 and 96 hours of incubation with IL-3 alone or with IL-3 pus G-CSF. The results reflect the mean fold (± SD) mRNA change in cells cultured with IL-3 plus G-CSF relative to cells cultured in IL-3 alone. (D) Kinetics of G-CSF–induced Gfi-1 protein expression evaluated by Western blotting. 32Dcl3 cells were cultured for 24 to 144 hours with IL-3 alone or with IL-3 plus G-CSF. The blots were stripped and restained for actin. Relative ratios of Gfi-1 to actin are shown in the lower bar graph.
The time course of CXCR4 down-regulation and CXCR4 promoter modulation by G-CSF indicated a requirement for prolonged G-CSF exposure, extending longer than 24 hours (Figure 3B). This suggested that a number of intermediate steps were needed for G-CSF to reduce CXCR4 promoter activity in 32D cells and that G-CSF did not directly regulate CXCR4 promoter activity. We analyzed (using Genomatrix software Genomatrix Software, Ann Arbor, MI) sequences (4 kb) upstream of the murine CXCR4 gene transcription start site for the presence of putative binding sites for transcriptional repressors. We identified the presence of 4 putative binding sites for the transcriptional repressor Gfi-1 (growth factor independence-1) based on the presence of the previously identified core sequence AA(T/G)C for Gfi-1 DNA binding.33,38 These include gcaAATCattttact (−1958/−1972 from the transcription start site), ttaAATCagagcttt (−1427/−1441), ccaAATCtcttcacg (−545/−559), and agtAATCactcctga (−390/−404). Further evidence pointing to involvement of Gfi-1 comes from studies linking Gfi-1 to regulation of granulopoiesis because Gfi-1 knock-out mice are neutropenic,23,25 and mutations of Gfi-1 cause neutropenia in humans.39
We tested whether G-CSF induces Gfi-1 in 32Dcl3 cells and looked for Gfi-1 mRNA and protein expression in 32Dcl3 cells cultured with IL-3 (100 pg/mL) plus G-CSF (100 ng/mL) and compared with cells cultured with IL-3 alone. By semiquantitative PCR, we found levels of Gfi-1 mRNA to be higher after 24-hour incubation with IL-3 plus G-CSF compared with IL-3 alone, at a time when levels of CXCR4 mRNA were similar (Figure 4B). After 96 hours, levels of Gfi-1 mRNA continued to be higher in cells incubated with IL-3 plus G-CSF compared with IL-3 alone, whereas levels of CXCR4 were now significantly reduced (Figure 4B). We confirmed by quantitative PCR that levels of Gfi-1 mRNA were higher in 32Dcl3 cells after 24 or 96 hours of culture with IL-3 plus G-CSF compared with IL-3 alone, whereas levels of CXCR4 mRNA were lower after 24 and 96 hours of culture with IL-3 plus G-CSF compared with IL-3 alone (Figure 4C). By immunoblotting with specific antibodies (N20 antibody), we found higher levels of Gfi-1 in cells cultured with G-CSF plus IL-3 compared with cells cultured with IL-3 alone. The difference was most prominent after 120 to 144 hours of culture (Figure 4D). Thus, G-CSF promotes the expression of the Gfi-1 repressor in 32Dcl3 cells, and the kinetics of mRNA and protein expression suggests that increased Gf1–1 expression precedes CXCR4 down-modulation in these cells.
Identification of Gfi-1 binding to CXCR4 upstream sequences
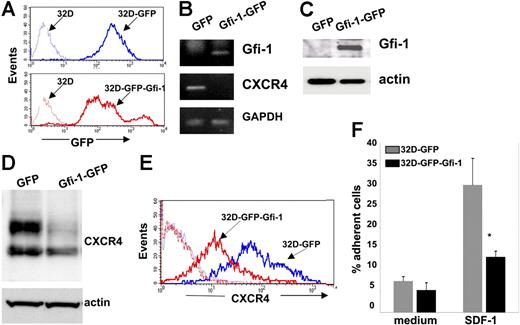
We stably overexpressed Gfi-1–GFP or GFP in 32Dcl3 cells by using retroviral expression constructs29 and examined CXCR4 expression in these cells. After infection with either control GFP or Gfi-1–GFP retroviruses, we selected 32Dcl3 cells that were more than 90% GFP positive by cell sorting (Figure 5A). Cell proliferation, viability, and morphology (mostly myeloblast and promyelocytes) were similar in 32Dc13 cells transduced with control or Gfi-1 retroviruses (not shown). By semiquantitative PCR (Figure 5B) and Western blotting (Figure 5C), Gfi-1–transduced cells expressed significantly more Gfi-1 mRNA and protein than the control cells transduced with control retrovirus. Importantly, CXCR4 mRNA levels were reduced in 32Dcl3 cells transduced with the Gfi-1 suppressor compared with control cells (Figure 5B). By Western blotting, 32Dcl3 cells transduced with Gfi-1–GFP retrovirus consistently expressed significantly lower levels of CXCR4 protein than control cells transduced with GFP retrovirus (Figure 5D), whereas levels of the proapoptotic protein Bax were not changed (not shown). In addition, 32Dcl3 cells transduced with Gfi-1–GFP expressed lower levels of cell-surface CXCR4 compared with control cells transduced with GFP alone as measured by flow cytometry (Figure 5E). Consistent with their reduced expression of surface CXCR4, significantly fewer 32Dcl3 cells transduced with Gfi-1–GFP retrovirus bound to SDF-1–coated plates than control cells (Figure 5F). Thus, constitutive expression of Gfi-1 causes a reduction of CXCR4 mRNA and protein expression in 32Dcl3 cells.
Gfi-1 transduction in 32Dcl3 cells and its effect on CXCR4 expression. (A) GFP expression in 32Dcl3 cells infected with control and Gfi-1 retrovirus was detected by flow cytometry. The profile of noninfected 32Dcl3 cells is also shown (--------). (B) Gfi-1, CXCR4, and GAPDH mRNAs in 32Dcl3 cells infected with control and Gfi-1 retroviruses were detected by semiquantitative RT-PCR. (C) Gfi-1 and actin were detected by Western blotting in cell lysates of 32Dcl3 cells infected with control and Gfi-1 retroviruses; representative results. (D) Western blot analysis of CXCR4 and actin in cell lysates of 32Dcl3 cells infected with control and Gfi-1 retroviruses; representative results. (E) Surface CXCR4 expression in 32Dcl3 cells infected with control and Gfi-1 retroviruses detected by flow cytometry. Background staining with control antibody is shown (--------). (F) Attachment of 32Dcl3 cells infected with control and Gfi-1 retroviruses to wells coated with SDF-1α or medium only. Results are expressed as the percentage of cell input; error bars represent SD of the mean (*P < .05).
Gfi-1 transduction in 32Dcl3 cells and its effect on CXCR4 expression. (A) GFP expression in 32Dcl3 cells infected with control and Gfi-1 retrovirus was detected by flow cytometry. The profile of noninfected 32Dcl3 cells is also shown (--------). (B) Gfi-1, CXCR4, and GAPDH mRNAs in 32Dcl3 cells infected with control and Gfi-1 retroviruses were detected by semiquantitative RT-PCR. (C) Gfi-1 and actin were detected by Western blotting in cell lysates of 32Dcl3 cells infected with control and Gfi-1 retroviruses; representative results. (D) Western blot analysis of CXCR4 and actin in cell lysates of 32Dcl3 cells infected with control and Gfi-1 retroviruses; representative results. (E) Surface CXCR4 expression in 32Dcl3 cells infected with control and Gfi-1 retroviruses detected by flow cytometry. Background staining with control antibody is shown (--------). (F) Attachment of 32Dcl3 cells infected with control and Gfi-1 retroviruses to wells coated with SDF-1α or medium only. Results are expressed as the percentage of cell input; error bars represent SD of the mean (*P < .05).
To examine the effects of Gfi-1 overexpression on the CXCR4 promoter activity in 32D cells, we transfected the luciferase reporter constructs containing the full-length and deletion mutants (−1368/86, −920/86, −832/86) of human CXCR4 promoter into 32Dcl3 cells stably transduced with Gfi-1–GFP or control GFP retrovirus. Six to 8 hours after transfection, levels of luciferase activity driven by the full-length promoter were significantly lower in Gfi-transduced 32D cells compared with the control-transduced cells (Figure 6A). By contrast, in the 32Dcl3 cells transduced with the deletion mutants, levels of luciferase activity were similar in Gfi-1– and control-transduced 32D cells (Figure 6A), providing further evidence that sequences located between −1.3kb and −2.6 kb of the transcriptional start site are required for Gfi-1 regulation of the human CXCR4 promoter activity. Note, the previously identified core sequence AA(T/G)C for Gfi-1 DNA binding38 is found at position −1616/−1631 (taaAATCaattctaa) from the transcription start site in the human CXCR4 promoter. To evaluate whether Gfi-1 binds to upstream sequences of the murine CXCR4 gene transcription start site, we used chIP testing chromatin extracts from 32Dcl3 cells in which Gfi-1 was overexpressed by retroviral infection or was induced by 3-day stimulation of cells with G-CSF and looked for the presence of DNA/Gfi-1 complexes recognized by N-20 goat anti–Gfi-1 antibody, as described.38 The immunoprecipitated DNA was tested for amplification by primers designed for each of the 4 putative Gfi-1–binding sites identified within the 4-kb sequences upstream of the murine CXCR4 start site (−1958/−1972, −1427/−1441, −545/−559, and −390/−404). As shown in Figure 6B, the specific goat antibody to Gfi-1, but not control goat IgG, immunoprecipitated DNA that was amplified by the primer set designed for amplification of the putative Gfi-1–binding site ttaAATCagagcttt located at −1427/−1441 from the transcription start site. Instead, this primer set did not amplify DNA sequences from immunoprecipitates of control 32Dcl3 cells transduced with GFP (Figure 4B). In addition, no specific amplification was derived from the use of the other 3 sets of primers for amplification of the 3 other putative Gfi-1–binding sites within sequences upstream of the murine CXCR4 gene transcription start site (Figure 6C). Similar results were derived from chromatin immunoprecipitation of 32Dcl3 cells cultured with G-CSF in the presence of IL-3. As shown (Figure 6D), specific antibodies to Gfi-1, but not control IgG, immunoprecipitated DNA that was amplified by primers for the putative Gfi-1 binding site at −1427/−1441 from the transcription start site, but no such specific amplification was observed when the 32Dcl3 cells were cultured in the absence of G-CSF. In addition, no specific amplification was derived from using the other 3 sets of primers for amplification of the other 3 putative Gfi-1–binding sites. Thus, Gfi-1 induced by treatment with G-CSF or virally transduced in 32Dcl3 cells is specifically detected in association with sequences upstream of the murine CXCR4 gene.
CXCR4 promoter activity with Gfi-1 overexpression and Gfi-1 binding to CXCR4 upstream sequences. (A) Luciferase activity after transfection of CXCR4 reporter plasmids in 32Dcl3 cells overexpressing Gfi-1. The results reflect the percentage of the mean activation of luciferase activity in 32Dcl3 cells transduced with Gfi-1 relative to luciferase activity in control-transduced cells (set at 100%). The results reflect the means of 4 experiments; the error bars represent SD of the mean (*P < .05). (B) Representative ChIP results from cell lysates of 32Dcl3 cells overexpressing Gfi-1 (Gfi-1–GFP) or control (GFP). Anti–Gfi-1 antibodies and control IgG were used for immunoprecipitation. The results reflect PCR amplification of genomic sequences upstream of the CXCR4 gene transcription start site (−1427/−1441). (C) PCR results from the indicated primer sets after ChIP of cell lysates from 32Dcl3 cells transduced with Gfi-1; representative results. (D) Cell lysates from 32Dcl3 cells cultured for 3 days with IL-3 alone or with IL-3 plus G-CSF were used for ChIP with antibodies to Gfi-1 or control IgG. Representative PCR results from the indicated primer sets.
CXCR4 promoter activity with Gfi-1 overexpression and Gfi-1 binding to CXCR4 upstream sequences. (A) Luciferase activity after transfection of CXCR4 reporter plasmids in 32Dcl3 cells overexpressing Gfi-1. The results reflect the percentage of the mean activation of luciferase activity in 32Dcl3 cells transduced with Gfi-1 relative to luciferase activity in control-transduced cells (set at 100%). The results reflect the means of 4 experiments; the error bars represent SD of the mean (*P < .05). (B) Representative ChIP results from cell lysates of 32Dcl3 cells overexpressing Gfi-1 (Gfi-1–GFP) or control (GFP). Anti–Gfi-1 antibodies and control IgG were used for immunoprecipitation. The results reflect PCR amplification of genomic sequences upstream of the CXCR4 gene transcription start site (−1427/−1441). (C) PCR results from the indicated primer sets after ChIP of cell lysates from 32Dcl3 cells transduced with Gfi-1; representative results. (D) Cell lysates from 32Dcl3 cells cultured for 3 days with IL-3 alone or with IL-3 plus G-CSF were used for ChIP with antibodies to Gfi-1 or control IgG. Representative PCR results from the indicated primer sets.
Effects of G-CSF on CXCR4 and Gfi-1 expression in primary myeloid cells
Previously, G-CSF was found to reduce CXCR4 expression in primary bone marrow granulocytic lineage cells in vitro,20 and systemic administration of G-CSF was found to decrease CXCR4 expression in cells mobilized from the bone marrow to the peripheral blood.20,–22 To evaluate whether the results from our in vitro model system of 32Dcl3 cells are applicable to primary cells, we examined Gfi-1 expression in primary bone marrow–derived granulocytic lineage cells (Gr-1+ cells) activated by G-CSF in vitro and in vivo. By quantitative RT-PCR, purified bone marrow Gr-1+ cells incubated with G-CSF (100 ng/mL) displayed increased levels of Gfi-1 mRNA after 1 hour of incubation and decreased levels of CXCR4 mRNA after 6 hours of incubation (Figure 7A). By immunoblotting, levels of Gfi-1 protein were similar in Gr-1+ cells cultured with or without G-CSF for up to 6 hours, but levels of CXCR4 protein were lower beginning at the 1.5-hour time point (Figure 7B). After 18 hours of culture with G-CSF (100 ng/mL), bone marrow Gr-1+ cells consistently displayed increased protein levels of Gfi-1 and decreased levels of CXCR4 compared with cells incubated in medium alone (Figure 7C).
In vitro and in vivo effects of G-CSF on CXCR4 and Gfi-1 expression in primary granulocytic lineage cells. (A) CXCR4 and Gfi-1 mRNA levels measured by real-time RT-PCR in Gr1+ bone marrow cells cultured in vitro with G-CSF for 1 hour to 6 hours. The results reflect the relative change in mRNA levels after culture compared with before culture; representative of 3 experiments. (B) CXCR4, Gfi-1, and actin levels detected by immunoblotting in Gr1+ bone marrow cells cultured in vitro with or without G-CSF for 1.5 to 6 hours; representative of 3 experiments. (C) CXCR4, Gfi-1, and actin content in cell lysates of Gr1+ bone marrow-derived cells after 18 hours of culture with G-CSF detected by Western blotting. The results reflect 3 independent experiments. Relative ratios of CXCR4/actin and Gfi-1/actin are shown in the bottom bar graph. (D) RNA was extracted from bone marrow Gr1+ cells from control mice, and mice were injected once with G-SCF 5 and 18 hours earlier. CXCR4 and Gfi-1 mRNA levels detected by real-time RT-PCR. The results reflect the relative change in mRNA levels in Gr1+ cells from mice treated with G-CSF compared with control. (E) Cells lysates were prepared from Gr1+ cells of control mice, and mice were injected once with G-CSF 5 and 18 hours earlier. CXCR4, Gfi-1, and actin levels were detected by immunoblotting. (F) After the mice were treated with G-CSF or diluent daily for 5 days, Gr1+ cells were purified from the bone marrow, and their content of CXCR4, Gfi-1, and actin were evaluated by Western blotting. The results are from 3 independent experiments. Relative ratios of CXCR4/actin and Gfi-1/actin are shown in the bottom bar graph.
In vitro and in vivo effects of G-CSF on CXCR4 and Gfi-1 expression in primary granulocytic lineage cells. (A) CXCR4 and Gfi-1 mRNA levels measured by real-time RT-PCR in Gr1+ bone marrow cells cultured in vitro with G-CSF for 1 hour to 6 hours. The results reflect the relative change in mRNA levels after culture compared with before culture; representative of 3 experiments. (B) CXCR4, Gfi-1, and actin levels detected by immunoblotting in Gr1+ bone marrow cells cultured in vitro with or without G-CSF for 1.5 to 6 hours; representative of 3 experiments. (C) CXCR4, Gfi-1, and actin content in cell lysates of Gr1+ bone marrow-derived cells after 18 hours of culture with G-CSF detected by Western blotting. The results reflect 3 independent experiments. Relative ratios of CXCR4/actin and Gfi-1/actin are shown in the bottom bar graph. (D) RNA was extracted from bone marrow Gr1+ cells from control mice, and mice were injected once with G-SCF 5 and 18 hours earlier. CXCR4 and Gfi-1 mRNA levels detected by real-time RT-PCR. The results reflect the relative change in mRNA levels in Gr1+ cells from mice treated with G-CSF compared with control. (E) Cells lysates were prepared from Gr1+ cells of control mice, and mice were injected once with G-CSF 5 and 18 hours earlier. CXCR4, Gfi-1, and actin levels were detected by immunoblotting. (F) After the mice were treated with G-CSF or diluent daily for 5 days, Gr1+ cells were purified from the bone marrow, and their content of CXCR4, Gfi-1, and actin were evaluated by Western blotting. The results are from 3 independent experiments. Relative ratios of CXCR4/actin and Gfi-1/actin are shown in the bottom bar graph.
We also treated groups of mice systemically with G-CSF (5 μg/mouse intraperitoneally for 1-5 days) to induce mobilization of myeloid cells from the bone marrow to the peripheral blood. We purified bone marrow Gr1+ cells 5 and 18 hours after a single G-CSF injection. By quantitative RT-PCR, levels of Gfi-1 mRNA were higher in Gr1+ cells obtained 18 hours after injection compared with control Gr1+ from untreated mice, whereas levels of CXCR4 mRNA were lower (Figure 7D). By immunoblotting, levels of Gfi-1 and CXCR4 proteins obtained at the same 5- and 18-hour time points after in vivo administration of G-CSF were similar to those detected in control Gr1+ cells from untreated mice (Figure 7E). We also examined the effects of 5-day systemic G-CSF administration on Gfi-1 and CXCR4 protein levels in Gr1+ bone marrow cells. In 3 experiments (Figure 7F), Gr1+ cells from groups of mice treated in vivo with G-CSF expressed higher levels of Gfi-1 compared with the control mice. By contrast, Gr1+ bone marrow cells from G-CSF–treated mice expressed lower levels of CXCR4 compared with the controls (Figure 7F). Thus, in vitro and in vivo G-CSF stimulates Gfi-1 expression in myeloid lineage cells and reduces CXCR4 expression in these cells.
Discussion
In this study we describe a novel role for the nuclear suppressor protein Gfi-1 as an inhibitor of CXCR4 expression in cells of granulocytic lineage and link G-CSF–induced mobilization of granulocytic lineage cells from the bone marrow to the peripheral blood to its up-regulation of Gfi-1, which down-regulates CXCR4 expression. Gfi-1 was previously identified as a regulator of neutrophil maturation from hematopoietic precursor cells and of quiescence in hematopoietic progenitor cells.23,25,–27 We now show that Gfi-1 regulates the release of myeloid lineage cells from the bone marrow to the peripheral blood acting through CXCR4, thus extending the previously identified roles of Gfi-1 as a key regulator of hematopoiesis. The identification of CXCR4 as a new target of Gfi–1 regulation raises the distinct possibility that this nuclear suppressor plays important and currently uncharacterized roles in cancer, HIV infection, reproduction, angiogenesis, and immunity in which CXCR4 is a well-established participant.30,40,,,–44
Much evidence supports a role of Gfi-1 as an inhibitor of CXCR4 expression induced by G-CSF. In 32Dcl3 cells and in primary myeloid lineage cells, G-CSF induces Gfi-1 expression, which temporally precedes or coincides with the decline of CXCR4. Overexpression of Gfi-1 causes a decline of CXCR4 expression and reduces CXCR4 promoter activity. When induced by G-CSF or overexpressed in myeloid cells, Gfi-1 physically associates with upstream sequences of the murine CXCR4 transcription start site.
Mutant mice lacking Gf1-1 (Gfi1−/−) are neutropenic because of a severe defect in differentiation of granulocytic precursors into mature neutrophils,23,25 and mutations in the GFI1 gene have been described in rare patients with congenital neutropenia.39 G-CSF treatment failed to induce neutrophilia in GFi1−/− mice, a result expected from the severe reduction of the very cell targets for G-CSF mobilization.11,37 Nonetheless, we suspect that a mobilization defect may have contributed to the high incidence of bacterial infections and early lethality of young (6 weeks old) Gfi1−/− mice at a time when bone marrow myelopoiesis was still relatively normal, and steady state white blood cell counts were similar to those in wild-type mice.23 In response to endogenous G-CSF induced by bacterial infections, Gfi1-null mice would have not reduced levels of CXCR4 expression in granulocytic lineage cells and would have not mobilized myeloid cells. CXCR4 was found normally expressed in Gfi1−/− hematopoietic progenitor cells,27 and would have served to retain myeloid cells within the bone marrow.
Besides having defective neutrophil maturation, patients with GFI1 mutations had reduced T and B lymphocytes in their blood and GFI1-null mice had reduced T cells in the thymus and reduced B cells in the bone marrow,23,45 revealing complex roles of the repressor in lymphoid cells maturation.24 The combined defect in myelopoiesis and lymphopoiesis suggested a striking similarity between patients with GFI1 mutations and patients with WHIM (warts, hypogammaglobulinemia, immunodeficiency, and myelokathexis) syndrome harboring CXCR4 gain-of-function mutations.46,47 This raises the possibility that Gfi-1 may serve as an important regulator of CXCR4 expression in the immune system.
STAT3 is a major target of G-CSF signaling,48 but it is not required for neutrophil differentiation from bone marrow progenitors,37 suggesting that STAT3 is probably not involved in the G-CSF–Gfi-1 pathway of neutrophil differentiation.24 However, administration of G-CSF to STAT3-deficent mice failed to induce an increase in peripheral blood neutrophils,37 raising the possibility that STAT3 is required for G-CSF–induced mobilization of myeloid lineage cells. We observed sustained STAT3 phosphorylation, increased expression of Gfi-1, and decreased expression of CXCR4 in 32Dcl3 cells after exposure to G-CSF and have identified a putative STAT-binding site (attttaTTCCtaaaaattt) within the human Gfi-1 promoter.49 Yeast 2-hybrid screens detected physical interaction between the STAT3 inhibitor PIAS3 and Gfi-1,50 raising the possibility that the STAT3–Gfi-1 axis may participate in modulation of G-CSF–induced CXCR4 expression and neutrophil mobilization.
Previous analyses of the human CXCR4 promoter identified putative consensus sequences for binding of different transcription factors,51 and subsequent studies have documented the binding of certain transcription factors to the CXCR4 promoter, including YY1,52,53 NF-κB,54 p53,54,55 and the von Hippel-Lindau tumor suppressor protein pVHL.31 Here, we identify CXCR4 as a target gene for Gfi-1. CXCR4 is a chemokine receptor of critical importance in the hematopoietic, nervous, and immune systems that has been implicated in cancer, autoimmunity, HIV infection, and angiogenesis. Thus, Gfi-1 defects could contribute to a variety of disease states.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs William E. Paul, Masashi Narazaki, Cassin Kimmel-Williams, Ombretta Salvucci, A. Virginia Gulino, Helen Kim, William Krek, Rhoda Alani, David Levens, Juhong Liu, Fang Dong, Jan Marcus, Alan D. Friedman, Stefania Pittaluga, Beverly Mock, Valery Bliskovski, and Mark Yarchoan for their help on various aspects of this work.
This work was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
National Institutes of Health
Authorship
Contribution: M.D.L.L.S. performed experiments; P.G. performed experiments and provided insightful comments; P.M. helped plan and performed some experiments; J.Z. provided critical reagents, performed some experiments, and helped in the design of some of the experiments; and G.T. designed the research, interpreted the results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giovanna Tosato, Laboratory of Cellular Oncology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; e-mail:tosatog@mail.nih.gov.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal