In allogenic hematopoietic stem-cell transplantation, an effect of HLA locus mismatch in allele level on clinical outcome has been clarified. However, the effect of each HLA allele mismatch combination is little known, and its molecular mechanism to induce acute graft-versus-host disease (aGVHD) remains to be elucidated. A total of 5210 donor-patient pairs who underwent transplantation through Japan Marrow Donor Program were analyzed. All HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles were retrospectively typed in all pairs. The impacts of the HLA allele mismatch combinations and amino acid substitution positions in 6 HLA loci on severe aGVHD were analyzed. A total of 15 significant high-risk HLA allele mismatch combinations and 1 HLA-DRB1-DQB1 linked mismatch combinations (high-risk mismatch) for severe aGVHD were identified, and the number of high-risk mismatches was highly associated with the occurrence of severe aGVHD regardless of the presence of mismatch combinations other than high-risk mismatch. Furthermore, 6 specific amino acid substitution positions in HLA class I were identified as those responsible for severe aGVHD. These findings provide evidence to elucidate the mechanism of aGVHD on the basis of HLA molecule. Furthermore, the identification of high-risk mismatch, that is, nonpermissive mismatch, would be beneficial for the selection of a suitable donor.
Introduction
Allogenic hematopoietic stem-cell transplantation (HSCT) from an HLA-matched unrelated (UR) donor has been established as a treatment for hematologic malignancies, when an HLA-identical sibling donor is unavailable.1,2 When a matched unrelated donor was not found in the donor registry, a partially HLA-matched unrelated donor was one of the candidates for alternative donor. But the higher risk of immunologic events, especially graft-versus-host disease (GVHD), was an important drawback. Extensive recent research has accumulated evidence of the role of each HLA locus mismatch on clinical outcome for UR-HSCT,3,,,,,–9 which has made it easy to search and select a partially matched donor. To further expand options for donor selection, our next challenge is to identify permissive and nonpermissive mismatch combinations of each HLA allele. Although there were some divisional trials with small populations,10,11 a large-scale cohort is essential for comprehensive analysis to identify nonpermissive mismatch combinations that are significant risk factors for severe acute graft-versus-host disease (aGVHD).
In this study, we identified nonpermissive HLA mismatch allele combinations of all major 6 HLA loci, and their responsible positions of amino acid substitution for aGVHD.
Patients, materials, and methods
Patients
A total of 5210 donor-patient pairs who underwent transplantation through the Japan Marrow Donor Program (JMDP) with T-cell–replete marrow from a serologically HLA-A, -B, and -DR antigen-matched donor between January 1993 and January 2006 were analyzed in this cohort study. Patients who received a transplant of harvested marrow outside Japan (n = 51) or were unavailable for blood sample (n = 428) were not eligible for this study of a total of 5689 consecutively registered patients.
Patient characteristics are shown in Table S1, available on the Blood website (see the Supplemental Materials link at the top of the online article). The final clinical survey of these patients was completed by June 1, 2006. Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki, and approval of the study was obtained from the Institutional Review Board of Aichi Cancer Center and JMDP.
HLA typing of patients and donors
Alleles at the HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 loci were identified by the methods described previously.4,5 Six HLA locus alleles were typed in all 5210 pairs. HLA genotypes of HLA-A, -B, -C, -DQB1, and -DPB1 allele of patient and donor were reconfirmed by the Luminex microbead method (Luminex 100 System; Luminex, Austin, TX). For convenience, we showed the frequency of HLA alleles that existed with more than a 5% allele frequency in the current Japanese data set and less than a 1% allele frequency in white populations12 in Table S2.
Matching of HLA allele between patient and donor
For the analysis of aGVHD, HLA allele mismatch among the donor-recipient pair was scored when the recipient's alleles were not shared by the donor (GVH vector). We also used GVH vectors for the analysis of overall survival (OS) to indicate OS of aGVHD high-risk or low-risk group.
Evaluation of acute GVHD
Occurrences of aGVHD were graded with grade 0, I, II, III, and IV according to established criteria.13 Grades III and IV were defined as severe aGVHD.
Definitions of amino acid substitution
Amino acid sequences of HLA-A, -B, -C, -DR, -DQ, and -DP molecules were obtained from IMGT/HLA sequence database.14 For example, Tyr9A-Phe9A indicated amino acid substitutions of position 9 in HLA-A molecule at which the donor had tyrosine and the patient phenylalanine. Substituted amino acids in HLA class I were summarized in Tables S3Table S4. Positions of amino acid substitution and substituted amino acids in HLA-B (PDF, 14 KB)–S5.
Definition of nonpermissive HLA combinations
Definition of hydropathy scale
The hydropathy scale proposed by Kyte and Doolittle17 evaluates the hydrophilicity and hydrophobicity of 20 amino acids to estimate the protein structure. Hydrophobic amino acid has a plus value and hydrophilic amino acid a minus value, and their absolute value indicates the grade of each property.
Statistical analysis
Cumulative incidences of aGVHD were assessed by the method described elsewhere to eliminate the effect of competing risk.18,19 The competing event regarding aGVHD was defined as death without aGVHD. A log-rank test was applied to assess the impact by the factor of interest. Multivariable Cox regression analyses20 were conducted to evaluate the impact of HLA allele mismatch combination, and the positions and types of amino acid substitution (for example, alanine, arginine, asparagines) of HLA molecules.
The HLA mismatch combination was evaluated for each locus separately, and the HLA match and HLA one-locus mismatch in every locus were analyzed. For example, A0206-A0201 mismatch combination meant that the donor has HLA-A*0206, recipient has HLA-A*0201, and another HLA-A allele of each donor and recipient was identical. This mismatch was compared with the HLA-A allele match. The mismatch combination of which the number of pairs was less than 10 was lumped together as “other mismatch.” This is because, according to the computer simulation by Peduzzi et al,21 it is generally accepted that regression analysis for a variable having fewer than 10 events might give an unreliable estimation. The model was constructed with mismatch combinations, mismatch status in other loci (match, 1 locus mismatch, and 2 locus mismatches as ordinal variable), and potential confounders. Confounders considered were sex (donor-recipient pairs), patient age (linear), donor age (linear), type of disease, risk of leukemia relapse (standard, high, and diseases other than leukemia), GVHD prophylaxis (cyclosporine [CSP] vs FK 506 [FK]), ATG (ATG vs no ATG), and preconditioning (total body irradiation [TBI] vs non-TBI). We used these confounders in all analyses in this paper to keep results comparable.
The impact of positions and types of amino acid substitution in HLA molecules was evaluated in pairs with HLA one-locus mismatch in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 separately. The amino acid positions we analyzed were all those at which amino acid was substituted in each locus. We analyzed the impact of each amino acid substitution on each position separately. Multivariable Cox models including positions and types of amino acid substitution, mismatch status in other loci (match, 1 locus mismatch, and 2 locus mismatches as ordinal variable), and confounders described in “Statistical analysis” were constructed.
We applied a P value of less than .005 as statistically significant to eliminate false-positive associations. All the analyses were conducted by STATA version 9.2 (Stata, College Station, TX).
Validation of statistical analysis
We validated the statistical analysis using 2 methods, traditional training-and-test method and bootstrap resampling method, in HLA-A analysis to confirm the usability of bootstrap resampling. In the traditional training-and-test method, donor-recipient pairs were divided at random in 2 equally scaled groups, group A and group B. When consistent results were obtained in both analyses, we considered the results as validated. In the bootstrap resampling method,22 we estimated the measure of association with the resampled data repeatedly drawn from the original data. Although around 100 to 200 bootstrapped samplings are generally sufficient,23 we explored 500, 1000, 5000, 10 000, and 50 000 bootstrappings in analysis of HLA-A mismatch combinations. We confirmed that an analysis using more than 5000 bootstrappings made the results stable. Because there was high concordance between these 2 methods (Table S6), we adopted bootstrap resampling using 10 000 bootstrap samples for all analyses in this paper as the method for validation. This is because traditional training-and-test methods do not work efficiently when small subgroups are considered as in this paper. Only when the results of base analysis and validating analysis using bootstrap resampling were significant concurrently were the results of the analysis judged to be statistically significant. When the result of base analysis was significant but the result of validating analysis using bootstrap resampling was not, we indicated this by adding an asterisk next to the P value of the base analysis.
Results
Impact of HLA allele mismatch combinations on severe aGVHD
Hazard ratios (HRs) of HLA allele mismatch combinations in HLA-A and -C on severe aGVHD are shown in Table 1 (HLA-B, -DR, -DQ, and -DP are available in Table S7).
Multivariable analysis of impact of mismatch pairs for sever aGVHD in HLA-A and -C
| Mismatch combination, donor-patient . | N . | HR (95% CI) . | P . |
|---|---|---|---|
| A locus match | 4510 | 1 | NA |
| A0201-A0206 | 138 | 1.23 (0.87-1.73) | .223 |
| A0206-A0201 | 131 | 1.78 (1.32-2.41) | < .001 |
| A0201-A0207 | 28 | 0.83 (0.34-2.03) | .699 |
| A0207-A0201 | 20 | 1.12 (0.42-3.02) | .809 |
| A0201-A0210 | 11 | 1.57 (0.58-4.23) | .367 |
| A0206-A0207 | 27 | 3.45 (2.09-5.70) | < .001 |
| A0207-A0206 | 22 | 0.71 (0.23-2.24) | .571 |
| A2402-A2420 | 60 | 0.64 (0.32-1.30) | .225 |
| A2420-A2402 | 30 | 1.18 (0.56-2.49) | .66 |
| A2601-A2602 | 24 | 0.64 (0.26-1.58) | .34 |
| A2602-A2601 | 21 | 3.35 (1.89-5.91) | < .001 |
| A2601-A2603 | 34 | 1.37 (0.73-2.57) | .326 |
| A2603-A2601 | 35 | 2.17 (1.29-3.64) | .003 |
| A2602-A2603 | 10 | 1.23 (0.30-4.98) | .763 |
| A2603-A2602 | 12 | 1.50 (0.48-4.68) | .485 |
| A other mismatch | 97 | 1.47 (1.00-2.15) | .047 |
| C locus match | 3685 | 1 | NA |
| C0102-C0303 | 30 | 2.83 (1.50-5.32) | .001* |
| C0303-C0102 | 38 | 1.05 (0.47-2.36) | .899 |
| C0102-C0304 | 12 | 1.85 (0.59-5.81) | .287 |
| C0304-C0102 | 19 | 0.89 (0.28-2.79) | .854 |
| C0102-C0401 | 14 | 1.87 (0.77-4.55) | .164 |
| C0102-C0803 | 24 | 1.97 (0.87-4.42) | .099 |
| C0803-C0102 | 10 | 1.66 (0.53-5.19) | .383 |
| C0102-C1402 | 16 | 3.86 (1.98-7.51) | < .001* |
| C1402-C0102 | 13 | 0.46 (0.06-3.33) | .45 |
| C0303-C0304 | 83 | 1.08 (0.63-1.85) | .761 |
| C0304-C0303 | 62 | 0.83 (0.41-1.68) | .614 |
| C0303-C0401 | 31 | 1.73 (0.89-3.36) | .103 |
| C0401-C0303 | 42 | 2.81 (1.72-4.60) | < .001 |
| C0303-C0702 | 25 | 1.16 (0.52-2.62) | .706 |
| C0702-C0303 | 18 | 2.16 (0.96-4.85) | .062 |
| C0303-C0801 | 76 | 1.07 (0.63-1.84) | .782 |
| C0801-C0303 | 80 | 2.32 (1.58-3.40) | < .001 |
| C0303-C1502 | 25 | 3.22 (1.75-5.89) | < .001 |
| C0304-C0401 | 15 | 3.02 (1.34-6.79) | .007 |
| C0401-C0304 | 12 | 6.22 (3.07-12.5) | < .001* |
| C0304-C0702 | 26 | 2.35 (1.16-4.76) | .017 |
| C0702-C0304 | 33 | 1.22 (0.58-2.59) | .59 |
| C0304-C0801 | 69 | 2.34 (1.55-3.52) | < .001 |
| C0801-C0304 | 47 | 1.64 (0.98-2.76) | .057 |
| C0304-C1402 | 28 | 3.06 (1.68-5.60) | < .001* |
| C1402-C0304 | 23 | 3.66 (2.00-6.68) | < .001 |
| C0304-C1502 | 53 | 1.82 (1.08-3.05) | .023 |
| C1502-C0304 | 27 | 3.77 (2.20-6.47) | < .001 |
| C0801-C0102 | 10 | 2.88 (0.92-9.03) | .068 |
| C0801-C0803 | 27 | 1.55 (0.69-3.48) | .284 |
| C0803-C0801 | 26 | 2.04 (1.04-3.99) | .037 |
| C0801-C1502 | 36 | 1.59 (0.79-3.21) | .19 |
| C1502-C0801 | 23 | 2.28 (1.07-4.85) | .031 |
| C1402-C1502 | 55 | 1.67 (1.01-2.77) | .043 |
| C1502-C1402 | 50 | 4.97 (3.41-7.25) | < .001 |
| C other mismatch | 347 | 1.69 (1.34-2.14) | < .001 |
| Mismatch combination, donor-patient . | N . | HR (95% CI) . | P . |
|---|---|---|---|
| A locus match | 4510 | 1 | NA |
| A0201-A0206 | 138 | 1.23 (0.87-1.73) | .223 |
| A0206-A0201 | 131 | 1.78 (1.32-2.41) | < .001 |
| A0201-A0207 | 28 | 0.83 (0.34-2.03) | .699 |
| A0207-A0201 | 20 | 1.12 (0.42-3.02) | .809 |
| A0201-A0210 | 11 | 1.57 (0.58-4.23) | .367 |
| A0206-A0207 | 27 | 3.45 (2.09-5.70) | < .001 |
| A0207-A0206 | 22 | 0.71 (0.23-2.24) | .571 |
| A2402-A2420 | 60 | 0.64 (0.32-1.30) | .225 |
| A2420-A2402 | 30 | 1.18 (0.56-2.49) | .66 |
| A2601-A2602 | 24 | 0.64 (0.26-1.58) | .34 |
| A2602-A2601 | 21 | 3.35 (1.89-5.91) | < .001 |
| A2601-A2603 | 34 | 1.37 (0.73-2.57) | .326 |
| A2603-A2601 | 35 | 2.17 (1.29-3.64) | .003 |
| A2602-A2603 | 10 | 1.23 (0.30-4.98) | .763 |
| A2603-A2602 | 12 | 1.50 (0.48-4.68) | .485 |
| A other mismatch | 97 | 1.47 (1.00-2.15) | .047 |
| C locus match | 3685 | 1 | NA |
| C0102-C0303 | 30 | 2.83 (1.50-5.32) | .001* |
| C0303-C0102 | 38 | 1.05 (0.47-2.36) | .899 |
| C0102-C0304 | 12 | 1.85 (0.59-5.81) | .287 |
| C0304-C0102 | 19 | 0.89 (0.28-2.79) | .854 |
| C0102-C0401 | 14 | 1.87 (0.77-4.55) | .164 |
| C0102-C0803 | 24 | 1.97 (0.87-4.42) | .099 |
| C0803-C0102 | 10 | 1.66 (0.53-5.19) | .383 |
| C0102-C1402 | 16 | 3.86 (1.98-7.51) | < .001* |
| C1402-C0102 | 13 | 0.46 (0.06-3.33) | .45 |
| C0303-C0304 | 83 | 1.08 (0.63-1.85) | .761 |
| C0304-C0303 | 62 | 0.83 (0.41-1.68) | .614 |
| C0303-C0401 | 31 | 1.73 (0.89-3.36) | .103 |
| C0401-C0303 | 42 | 2.81 (1.72-4.60) | < .001 |
| C0303-C0702 | 25 | 1.16 (0.52-2.62) | .706 |
| C0702-C0303 | 18 | 2.16 (0.96-4.85) | .062 |
| C0303-C0801 | 76 | 1.07 (0.63-1.84) | .782 |
| C0801-C0303 | 80 | 2.32 (1.58-3.40) | < .001 |
| C0303-C1502 | 25 | 3.22 (1.75-5.89) | < .001 |
| C0304-C0401 | 15 | 3.02 (1.34-6.79) | .007 |
| C0401-C0304 | 12 | 6.22 (3.07-12.5) | < .001* |
| C0304-C0702 | 26 | 2.35 (1.16-4.76) | .017 |
| C0702-C0304 | 33 | 1.22 (0.58-2.59) | .59 |
| C0304-C0801 | 69 | 2.34 (1.55-3.52) | < .001 |
| C0801-C0304 | 47 | 1.64 (0.98-2.76) | .057 |
| C0304-C1402 | 28 | 3.06 (1.68-5.60) | < .001* |
| C1402-C0304 | 23 | 3.66 (2.00-6.68) | < .001 |
| C0304-C1502 | 53 | 1.82 (1.08-3.05) | .023 |
| C1502-C0304 | 27 | 3.77 (2.20-6.47) | < .001 |
| C0801-C0102 | 10 | 2.88 (0.92-9.03) | .068 |
| C0801-C0803 | 27 | 1.55 (0.69-3.48) | .284 |
| C0803-C0801 | 26 | 2.04 (1.04-3.99) | .037 |
| C0801-C1502 | 36 | 1.59 (0.79-3.21) | .19 |
| C1502-C0801 | 23 | 2.28 (1.07-4.85) | .031 |
| C1402-C1502 | 55 | 1.67 (1.01-2.77) | .043 |
| C1502-C1402 | 50 | 4.97 (3.41-7.25) | < .001 |
| C other mismatch | 347 | 1.69 (1.34-2.14) | < .001 |
A0206-A0201 mismatch combination meant that the donor has HLA-A*0206, recipient has HLA-A*0201 and another HLA-A allele of each donor and recipient was identical. Each mismatch pair in HLA-A was compared with the HLA-A allele match, and each mismatch pair in HLA-C was compared with the HLA-C allele match. Confounders considered were sex (donor-recipient pairs), patient age (linear), donor age (linear), type of disease, risk of leukemia relapse (standard, high and diseases other than leukemia), GVHD prophylaxis, (CSP vs. FK), ATG (ATG vs. no ATG) and preconditioning (TBI vs non-TBI).
HR denotes hazard ratio; CI, confidence interval; NA, not applicable.
The result of base analysis was significant, but the result of validating analysis using bootstrap resampling was not. The results of the analysis were thus judged not to be statistically significant.
In HLA-A locus mismatch combinations, A*0206-A*0201 (HR: 1.78; CI, 1.32-2.41), A*0206-A*0207 (HR: 3.45; CI: 2.09-5.70), A*2602-A*2601 (HR: 3.35; CI: 1.89-5.91), and A*2603-A*2601 (HR: 2.17; CI: 1.29-3.64), were significant risk factors for severe aGVHD.
In HLA-C locus mismatch combinations, 7 combinations were significant risk factors for severe aGVHD; those were as follows: Cw*0401-Cw*0303 (HR: 2.81; CI: 1.72-4.60), Cw*0801-Cw*0303 (HR: 2.32; CI: 1.58-3.40), Cw*0303-Cw*1502 (HR: 3.22; CI: 1.75-5.89), Cw*0304-Cw*0801 (HR: 2.34; CI: 1.55-3.52), Cw*1402-Cw*0304 (HR: 3.66; CI: 2.00-6.68), Cw*1502-Cw*0,304 (HR: 3.77; CI: 2.20-6.47), and Cw*1502-Cw*1402 (HR: 4.97; CI: 3.41-7.25). To summarize, high-risk HLA allele mismatch combinations for severe aGVHD, that is, nonpermissive mismatch combinations, of all major 6 HLA loci were listed in Table 2. A total of 15 nonpermissive HLA allele mismatch combinations (4 in HLA-A, 1 in HLA-B, 7 in HLA-C, 1 in HLA-DRB1, and 2 in HLA-DPB1) and 1 HLA-DRB1-DQB1 linked mismatch combination (Table 2 legend) were identified.
Nonpermissive allele mismatch combinations for severe aGVHD
| Mismatch combination, donor-patient . | N . | HR (95% CI) . | P . |
|---|---|---|---|
| A0206-A0201 | 131 | 1.78 (1.32-2.41) | < .001 |
| A0206-A0207 | 27 | 3.45 (2.09-5.70) | < .001 |
| A2602-A2601 | 21 | 3.35 (1.89-5.91) | < .001 |
| A2603-A2601 | 35 | 2.17 (1.29-3.64) | .003 |
| B1501-B1507 | 19 | 3.34 (1.85-5.99) | < .001 |
| C0303-C1502 | 25 | 3.22 (1.75-5.89) | < .001 |
| C0304-C0801 | 69 | 2.34 (1.55-3.52) | < .001 |
| C0401-C0303 | 42 | 2.81 (1.72-4.60) | < .001 |
| C0801-C0303 | 80 | 2.32 (1.58-3.40) | < .001 |
| C1402-C0304 | 23 | 3.66 (2.00-6.68) | < .001 |
| C1502-C0304 | 27 | 3.77 (2.20-6.47) | < .001 |
| C1502-C1402 | 50 | 4.97 (3.41-7.25) | < .001 |
| DR0405-DR0403 | 53 | 2.13 (1.28-3.53) | .003 |
| (DR1403-DQ0301)-(DR1401-DQ0502) | 19 | 2.81 (1.44-5.51) | .002 |
| DP0301-DP0501 | 49 | 2.41 (1.49-3.89) | < .001 |
| DP0501-DP0901 | 71 | 2.03 (1.30-3.16) | .002 |
| Mismatch combination, donor-patient . | N . | HR (95% CI) . | P . |
|---|---|---|---|
| A0206-A0201 | 131 | 1.78 (1.32-2.41) | < .001 |
| A0206-A0207 | 27 | 3.45 (2.09-5.70) | < .001 |
| A2602-A2601 | 21 | 3.35 (1.89-5.91) | < .001 |
| A2603-A2601 | 35 | 2.17 (1.29-3.64) | .003 |
| B1501-B1507 | 19 | 3.34 (1.85-5.99) | < .001 |
| C0303-C1502 | 25 | 3.22 (1.75-5.89) | < .001 |
| C0304-C0801 | 69 | 2.34 (1.55-3.52) | < .001 |
| C0401-C0303 | 42 | 2.81 (1.72-4.60) | < .001 |
| C0801-C0303 | 80 | 2.32 (1.58-3.40) | < .001 |
| C1402-C0304 | 23 | 3.66 (2.00-6.68) | < .001 |
| C1502-C0304 | 27 | 3.77 (2.20-6.47) | < .001 |
| C1502-C1402 | 50 | 4.97 (3.41-7.25) | < .001 |
| DR0405-DR0403 | 53 | 2.13 (1.28-3.53) | .003 |
| (DR1403-DQ0301)-(DR1401-DQ0502) | 19 | 2.81 (1.44-5.51) | .002 |
| DP0301-DP0501 | 49 | 2.41 (1.49-3.89) | < .001 |
| DP0501-DP0901 | 71 | 2.03 (1.30-3.16) | .002 |
Analysis method is the same as in Table 1. We surveyed specific linked mismatches between nonpermissive mismatches elucidated. As a result, obvious specific linked mismatches exist only between DRB1*1403- DRB1*1401 and DQB1*0301- DQB1*0502. Therefore, we could not evaluate which mismatch combination impacted aGVHD, and we considered this linked mismatch did so. On the other hand, because other nonpermissive mismatch combinations had no specific link with the others, we judged other than DRB1*1403- DRB1*1401 and DQB1*0301- DQB1*0502 nonpermissive mismatches solely impacted aGVHD. (DR1403-DQ0301)-(DR1401-DQ0502) linked mismatch meant that the donor has HLA-DRB1*1403-HLADQB1*0301 and the recipient has HLA-DRB1*1401-HLADQB1*0502.
HR indicates hazard ratio; CI, confidence interval.
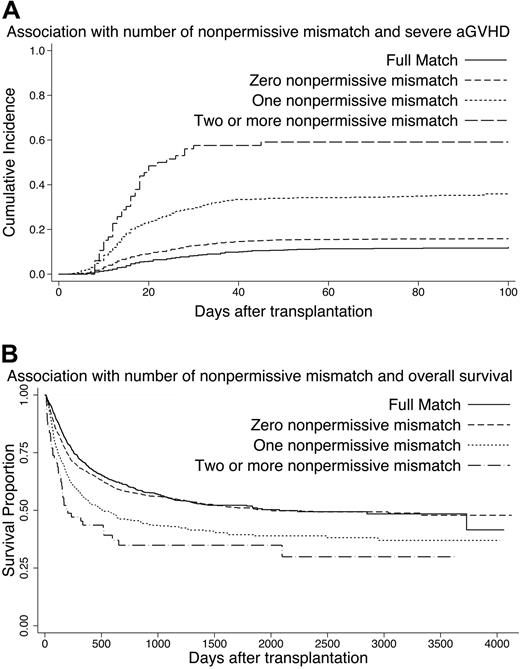
We divided donor-recipient pairs into 4 groups according to the number of nonpermissive mismatches: (1) full match (in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1) group; (2) zero nonpermissive mismatch (with mismatches other than nonpermissive mismatches) group; (3) 1 nonpermissive mismatch (with or without mismatches other than nonpermissive mismatches) group; and (4) 2 or more nonpermissive mismatches (with or without mismatches other than nonpermissive mismatches) group, and analyzed for association with severe aGVHD. This analysis excluded pairs with 2 locus mismatches in the same locus. Patient characteristics according to the number of nonpermissive mismatches are shown in Table 3. The curve of cumulative incidence of severe aGVHD is shown in Figure 1A. Multivariable analysis revealed that severe aGVHD occurred with almost equal frequency between the full match group and zero nonpermissive mismatch group, and was significantly associated with the number of nonpermissive mismatches (Table 4). Relative risk of significant factor for aGVHD and OS is shown in Table S8. In terms of the mortality due to aGVHD according to the number of nonpermissive mismatches, one nonpermissive mismatch group and 2 or more nonpermissive mismatch groups showed higher mortality (19.7% and 15.8%, respectively) than full match group and zero nonpermissive mismatch group (8.5% and 11.4%, respectively).
Patient characteristics according to number of nonpermissive mismatches
| Group . | Total . | Full match . | Zero nonpermissive mismatch . | One nonpermissive mismatch . | Two or more nonpermissive mismatches . |
|---|---|---|---|---|---|
| Total | 4050 | 712 | 2670 | 602 | 66 |
| Patient age, median y | 30 | 32 | 30 | 29 | 29 |
| Sex, donor/patient, no. patients | |||||
| Male/male | 1673 | 312 | 1096 | 237 | 28 |
| Male/female | 785 | 134 | 518 | 119 | 14 |
| Female/male | 769 | 115 | 524 | 117 | 13 |
| Female/female | 823 | 151 | 532 | 129 | 11 |
| Disease, no. patients | |||||
| ALL | 981 | 162 | 668 | 139 | 12 |
| ANLL | 1075 | 196 | 698 | 158 | 23 |
| CML | 703 | 119 | 453 | 115 | 16 |
| Hereditary disease | 85 | 14 | 56 | 15 | 0 |
| MDS | 476 | 91 | 304 | 72 | 9 |
| Malignant lymnphoma | 349 | 69 | 229 | 48 | 3 |
| Multiple myeloma | 42 | 8 | 29 | 4 | 1 |
| Severe aplastic anemia | 247 | 33 | 175 | 37 | 2 |
| Other disease | 92 | 20 | 58 | 14 | 0 |
| Risk of leukemia relapse,* no. patients | |||||
| Standard risk | 1308 | 249 | 857 | 181 | 21 |
| High risk | 1451 | 228 | 962 | 231 | 30 |
| Diseases other than leukemia | 1291 | 235 | 851 | 190 | 15 |
| GVHD prophylaxis, no. patients | |||||
| Cyclosporin-based | 2198 | 402 | 1444 | 319 | 33 |
| Tacrolimus-based | 1852 | 310 | 1226 | 283 | 33 |
| ATG, no. patients | |||||
| ATG | 323 | 48 | 215 | 53 | 7 |
| Non-ATG | 3727 | 664 | 2455 | 549 | 59 |
| Preconditioning, no. patients | |||||
| TBI regimen | 3117 | 539 | 2071 | 449 | 58 |
| Non-TBI regimen | 933 | 173 | 599 | 153 | 8 |
| Group . | Total . | Full match . | Zero nonpermissive mismatch . | One nonpermissive mismatch . | Two or more nonpermissive mismatches . |
|---|---|---|---|---|---|
| Total | 4050 | 712 | 2670 | 602 | 66 |
| Patient age, median y | 30 | 32 | 30 | 29 | 29 |
| Sex, donor/patient, no. patients | |||||
| Male/male | 1673 | 312 | 1096 | 237 | 28 |
| Male/female | 785 | 134 | 518 | 119 | 14 |
| Female/male | 769 | 115 | 524 | 117 | 13 |
| Female/female | 823 | 151 | 532 | 129 | 11 |
| Disease, no. patients | |||||
| ALL | 981 | 162 | 668 | 139 | 12 |
| ANLL | 1075 | 196 | 698 | 158 | 23 |
| CML | 703 | 119 | 453 | 115 | 16 |
| Hereditary disease | 85 | 14 | 56 | 15 | 0 |
| MDS | 476 | 91 | 304 | 72 | 9 |
| Malignant lymnphoma | 349 | 69 | 229 | 48 | 3 |
| Multiple myeloma | 42 | 8 | 29 | 4 | 1 |
| Severe aplastic anemia | 247 | 33 | 175 | 37 | 2 |
| Other disease | 92 | 20 | 58 | 14 | 0 |
| Risk of leukemia relapse,* no. patients | |||||
| Standard risk | 1308 | 249 | 857 | 181 | 21 |
| High risk | 1451 | 228 | 962 | 231 | 30 |
| Diseases other than leukemia | 1291 | 235 | 851 | 190 | 15 |
| GVHD prophylaxis, no. patients | |||||
| Cyclosporin-based | 2198 | 402 | 1444 | 319 | 33 |
| Tacrolimus-based | 1852 | 310 | 1226 | 283 | 33 |
| ATG, no. patients | |||||
| ATG | 323 | 48 | 215 | 53 | 7 |
| Non-ATG | 3727 | 664 | 2455 | 549 | 59 |
| Preconditioning, no. patients | |||||
| TBI regimen | 3117 | 539 | 2071 | 449 | 58 |
| Non-TBI regimen | 933 | 173 | 599 | 153 | 8 |
ALL indicates acute lymphoblastic leukemia; ANLL, acute non-lymphoblastic leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; and TBI, total body irradiation.
Standard risk for leukemia relapse was defined as the status of the 1st CR of AML and ALL and the 1st CP of CML at transplant, while high risk was defined as a more advanced status than standard risk in AML, ALL, and CML, and diseases other than leukemia was defined as other than ALL, ANLL, and CML.
Impact of number of nonpermissive mismatches on severe aGVHD and overall survival. (A) Cumulative incidence of severe aGVHD according to number of nonpermissive mismatches. –– indicates full match (in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1) group; ----, zero nonpermissive mismatch (with mismatches other than nonpermissive mismatches) group; · · · ·, one nonpermissive mismatch (with or without mismatches other than nonpermissive mismatches) group; and – – –, 2 or more nonpermissive mismatches (with or without mismatches other than nonpermissive mismatches) group. (B) Kaplan-Meier estimates of survival according to number of nonpermissive mismatches. Each group was divided as described for panel A.
Impact of number of nonpermissive mismatches on severe aGVHD and overall survival. (A) Cumulative incidence of severe aGVHD according to number of nonpermissive mismatches. –– indicates full match (in HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1) group; ----, zero nonpermissive mismatch (with mismatches other than nonpermissive mismatches) group; · · · ·, one nonpermissive mismatch (with or without mismatches other than nonpermissive mismatches) group; and – – –, 2 or more nonpermissive mismatches (with or without mismatches other than nonpermissive mismatches) group. (B) Kaplan-Meier estimates of survival according to number of nonpermissive mismatches. Each group was divided as described for panel A.
Multivariable analysis of impact of number of nonpermissive mismatches on severe aGVHD and overall survival
| . | N . | Event* . | Univariate analysis . | Multivariate analysis . | Bootstrap (10000) . | |||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |||
| For severe aGVHD | ||||||||
| Full match group | 972 | 129 | 1.00 | NA | 1.00 | NA | 1.00 | NA |
| Zero nonpermissive mismatch group | 2446 | 411 | 1.21 (0.95-1.54) | .111 | 1.00 (0.75-1.32) | .996 | 1.00 (0.74-1.33) | .996 |
| One nonpermissive mismatch group | 571 | 211 | 2.88 (2.20-3.78) | < .001 | 2.22 (1.62-3.04) | < .001 | 2.22 (1.63-3.02) | < .001 |
| Two or more nonpermissive mismatch group | 61 | 36 | 5.62 (3.77-8.39) | < .001 | 3.68 (2.33-5.80) | < .001 | 3.68 (2.33-5.80) | < .001 |
| For overall survival | ||||||||
| Full match group | 972 | 400 | 1.00 | NA | 1.00 | NA | 1.00 | NA |
| Zero nonpermissive mismatch group | 2446 | 1021 | 1.10 (0.98-1.23) | .091 | 1.06 (0.94-1.20) | .315 | 1.06 (0.94-1.20) | .299 |
| One nonpermissive mismatch group | 571 | 309 | 1.55 (1.34-1.78) | < .001 | 1.51 (1.30-1.76) | < .001 | 1.51 (1.29-1.77) | < .001 |
| Two or more nonpermissive mismatch group | 61 | 39 | 2.12 (1.54-2.90) | < .001 | 2.25 (1.65-3.08) | < .001 | 2.25 (1.65-3.08) | < .001 |
| . | N . | Event* . | Univariate analysis . | Multivariate analysis . | Bootstrap (10000) . | |||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |||
| For severe aGVHD | ||||||||
| Full match group | 972 | 129 | 1.00 | NA | 1.00 | NA | 1.00 | NA |
| Zero nonpermissive mismatch group | 2446 | 411 | 1.21 (0.95-1.54) | .111 | 1.00 (0.75-1.32) | .996 | 1.00 (0.74-1.33) | .996 |
| One nonpermissive mismatch group | 571 | 211 | 2.88 (2.20-3.78) | < .001 | 2.22 (1.62-3.04) | < .001 | 2.22 (1.63-3.02) | < .001 |
| Two or more nonpermissive mismatch group | 61 | 36 | 5.62 (3.77-8.39) | < .001 | 3.68 (2.33-5.80) | < .001 | 3.68 (2.33-5.80) | < .001 |
| For overall survival | ||||||||
| Full match group | 972 | 400 | 1.00 | NA | 1.00 | NA | 1.00 | NA |
| Zero nonpermissive mismatch group | 2446 | 1021 | 1.10 (0.98-1.23) | .091 | 1.06 (0.94-1.20) | .315 | 1.06 (0.94-1.20) | .299 |
| One nonpermissive mismatch group | 571 | 309 | 1.55 (1.34-1.78) | < .001 | 1.51 (1.30-1.76) | < .001 | 1.51 (1.29-1.77) | < .001 |
| Two or more nonpermissive mismatch group | 61 | 39 | 2.12 (1.54-2.90) | < .001 | 2.25 (1.65-3.08) | < .001 | 2.25 (1.65-3.08) | < .001 |
Each group was compared with Full match group. Confounders considered were sex (donor-recipient pairs), patient age (linear), donor age (linear), type of disease, risk of leukemia relapse (standard, highand diseases other than leukemia), GVHD prophylaxis, (CSP vs. FK), ATG (ATG vs. no ATG) and preconditioning (TBI vs. non-TBI).
HR indicates hazard ratio; CI, confidence interval; Boot strap (10000), bootstrap resampling using 10000 bootstrapping.
For severe aGVHD, “Event” refers to number of occurrences; for overall survival, number of deaths.
Impact of positions and types of amino acid substitutions of HLA molecules for severe aGVHD
One specific amino acid substitution at position 9 in HLA-A molecule and 6 specific amino acid substitutions at positions 9, 77, 80, 99, 116, and 156 in HLA-C molecule were significant risk factors for severe aGVHD: Tyr9A-Phe9A (HR: 1.66; CI: 1.19-3.32), Tyr9C-Ser9C (HR: 1.66; CI: 1.23-2.25), Asn77C-Ser77C (HR: 1.87; CI: 1.46-2.39), Lys80C-Asn80C (HR: 1.87; CI: 1.46-2.39), Tyr99C-Phe99C (HR: 1.64; CI: 1.21-2.22), Leu116C-Ser116C (HR: 3.40; CI: 2.20-5.25), and Arg156C-Leu156C (HR: 1.48; CI: 1.15-1.90) (Table 5). The amplitude of hydropathy scales were 4.1, 0.5, 2.7, 0.4, 4.1, 4.6, and 8.3, respectively. Although all amino acid positions substituted in each HLA locus were analyzed, amino acid substitutions of any other HLA-A and -C positions were not significant risk factors. As for HLA-B, DRB1, DQB1, and DPB1, there was no significant association between the positions of amino acid substitution and severe aGVHD. Impact for OS about positions and types of amino acid substitutions that were significant risk factors for aGVHD was shown in Table S9.
Multivariable analysis of impact of amino acid substitution on HLA class I molecules for severe aGVHD
| Posisiton and kind of amino acid substitution, donor-recipient . | HS . | N . | Event† . | HR (95% CI) . | P . |
|---|---|---|---|---|---|
| HLA-A locus | |||||
| Tyr9A-Phe9A | 4.1 | 163 | 64 | 1.66 (1.19-2.32) | .003 |
| Asn116A-Asp116A | 0 | 32 | 15 | 2.25 (1.26-4.01) | .005* |
| HLA-C locus | |||||
| Tyr9C-Ser9C | 0.5 | 146 | 59 | 1.66 (1.23-2.25) | .001 |
| Asn77C-Ser77C | 2.7 | 205 | 90 | 1.87 (1.46-2.39) | < .001 |
| Lys80C-Asn80C | 0.4 | 205 | 90 | 1.87 (1.46-2.39) | < .001 |
| Tyr99C-Phe99C | 4.1 | 146 | 59 | 1.64 (1.21-2.22) | .001 |
| Leu116C-Ser116C | 4.6 | 53 | 30 | 3.40 (2.20-5.25) | < .001 |
| Arg156C-Leu156C | 8.3 | 251 | 88 | 1.48 (1.15-1.90) | .002 |
| Posisiton and kind of amino acid substitution, donor-recipient . | HS . | N . | Event† . | HR (95% CI) . | P . |
|---|---|---|---|---|---|
| HLA-A locus | |||||
| Tyr9A-Phe9A | 4.1 | 163 | 64 | 1.66 (1.19-2.32) | .003 |
| Asn116A-Asp116A | 0 | 32 | 15 | 2.25 (1.26-4.01) | .005* |
| HLA-C locus | |||||
| Tyr9C-Ser9C | 0.5 | 146 | 59 | 1.66 (1.23-2.25) | .001 |
| Asn77C-Ser77C | 2.7 | 205 | 90 | 1.87 (1.46-2.39) | < .001 |
| Lys80C-Asn80C | 0.4 | 205 | 90 | 1.87 (1.46-2.39) | < .001 |
| Tyr99C-Phe99C | 4.1 | 146 | 59 | 1.64 (1.21-2.22) | .001 |
| Leu116C-Ser116C | 4.6 | 53 | 30 | 3.40 (2.20-5.25) | < .001 |
| Arg156C-Leu156C | 8.3 | 251 | 88 | 1.48 (1.15-1.90) | .002 |
HLA-B. -DRB1, -DQB1 -DPB1 locus had no significant substitutions. The impact of positions and types of amino acid substitution in HLA molecules was evaluated in pairs with HLA one-locus mismatch in HLA-A, -B, -C, -DRB1, -DQB1 and -DPB1 separately. For example, Tyr9A-Phe9A indicated amino acid substitutions of position 9 in HLA-A molecule at which donor had tyrosine and patient phenylalanine. The impacts of positions and kinds of amino acid substitutions in each HLA molecule were evaluated in pairs with HLA one locus mismatch in each HLA locus separately. Pairs which substituted specific amino acid at each position were compared with amino acid matched pairs at that position.
HS indicates hydropathy scale; HR, hazard ratio; CI, confidence interval; Tyr, tyrosine; Phe, phenylalanine; Asn, asparagine; Asp, asparatic acid; Ser, serine; Lys, lysine; Leu, leucine; and Arg, arginine.
Result of base analysis was significant but result of validating analysis using bootstrap resampling was not. Results of analysis were thus judged not to be statistically significant.
Measured in number of occurrences of severe acute GVHD.
Discussion
Extensive recent research has accumulated evidence of the role of each HLA locus mismatch on clinical outcome for UR-HSCT.3,,,,,–9 Our next concern is identifying the combinations of HLA allele mismatch and the positions of amino acid substitution of the HLA molecules responsible for aGVHD. In the present study, multivariable analysis revealed that 15 combinations of HLA allele mismatch and 1 HLA-DRB1-DQB1 haplotype mismatch significantly increase the occurrence of severe aGVHD (Table 2), and most of them increased the mortality rate after transplantation (data not shown). Thus, these mismatch combinations of HLA allele might be called nonpermissive clinically. We speculated that the effect of HLA locus mismatch was a reflection and summation of these HLA allele mismatch combinations. Discrepancies of responsible HLA locus for aGVHD between ethnically diverse transplantations might be explained by the proportions of nonpermissive mismatch combinations in each HLA locus. The same study in other populations would be needed to clarify this question as well as the severity of aGVHD. Interestingly, the full match group and zero nonpermissive mismatch group showed an almost equal occurrence of severe aGVHD, though pairs in zero nonpermissive mismatch group had one or more mismatches other than nonpermissive mismatches. And HR was elevated with the increase in the number of nonpermissive mismatches (Figure 1A; Table 4), while the number of nonpermissive mismatches also had a significant effect on OS after transplantation (Figure 1B; Table 4). These findings indicated at least that nonpermissive mismatches should be avoided in donor selection for UR-HSCT, and that the order of donor selection based on this nonpermissive mismatch would be useful, instead of that based on HLA locus mismatch. We also speculated that there are permissive mismatches in mismatches other than nonpermissive mismatches. It is therefore an important task in the future to identify permissive mismatches for partially HLA-matched donor selection. On the other hand, we do not deny the possibility that some mismatch combinations not classified as nonpermissive may actually be potential nonpermissive ones. Misclassification might happen because of insufficient statistical power due to the relatively small number of subjects in subcategories.
At present, there have been only a few reports indicating that the transplant-related immunologic reactions and clinical outcomes were caused by the HLA allele mismatch combinations. Macdonald et al24 reported that cytotoxic T lymphocytes (CTLs) discriminate between HLA-B*4402 and HLA-B*4403, and induce strong alloresponses, but the stronger T-cell alloreactivity is observed toward HLA-B*4403 compared with HLA-B*4402 in vitro. Zino et al10 and Fleischhauer et al11 attempted to develop an algorithm for prediction of nonpermissive HLA-DPB1 mismatches. The present report is the first to provide far more precise and detailed evidence for numerous HLA allele mismatch combinations for severe aGVHD.
In this study, substitutions of specific amino acids at positions 9, 77, 80, 99, 116, and 156 were elucidated as a significant risk factor for severe aGVHD. We speculated that the responsibility of positions 77 and 80 in HLA-C for severe aGVHD was associated with ligand matching of NK-cell receptor (KIR2DL). Although the role of KIR2DL in acute GVHD has been controversial,25 a recent JMDP analysis demonstrated that KIR2DL ligand mismatched pairs in GVH vector induced severe aGVHD in UR-HSCT with T-cell–replete marrow.9 The ligand of KIR2DL is located at positions 77 and 80, which are completely linked in HLA-C molecule. And almost all pairs in this study with Asn77C-Ser77C and Lys80C-Asn80C substitutions have a KIR2DL mismatch in GVH vector.
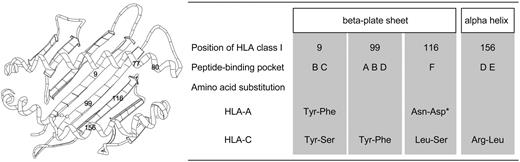
Except for positions 77 and 80, which are associated with KIR2DL ligand in HLA-C, positions 9, 99, 116, and 156 were elucidated. Positions 9, 99, and 116 are located in the beta-plated sheet, and position 156 is in the alpha helix of HLA class I molecule (Figure 2).26,27 Position 9 constitutes peptide-binding pockets B and C, position 99 constitutes A, B, and D pockets, position 116 constitutes F pocket, and position 156 constitutes D and E pockets.28 As a result, all amino acid positions elucidated in this study were important positions for peptide binding and T-cell recognition, although all substituted positions including positions at which residues are not accessible in the vicinity of peptide binding sites were analyzed.
Schematic presentation of HLA class I molecule and summary of features of significant amino acid substituted positions. Numbers in schema of HLA molecule indicate substituted amino acid positions that were elucidated as significant risk factor for severe aGVHD. Positions 9, 99, and 116 are located in the beta-plated sheet and positions 77, 80, and 156 in the alpha helix of HLA class I molecule (left). Positions 77 and 80 are associated with KIR2DL ligand in HLA-C molecule. Position 9 constitutes peptide-binding pockets B and C; position 99 constitutes A, B, and D pockets; position 116 constitutes F pocket; and position 156 constitutes D and E pockets (right). For example, Tyr-Phe indicated amino acid substitution at indicated position in HLA molecule at which donor had tyrosine and patient phenylalanine. Tyr indicates tyrosine; Phe, phenylalanine; Asn, asparagine; Asp, asparatic acid; Ser, serine; Lys, lysine; Leu, leucine; and Arg, arginine. *Result of base analysis was significant but result of validating analysis using bootstrap resampling was not. Results of analysis were thus judged not to be statistically significant.
Schematic presentation of HLA class I molecule and summary of features of significant amino acid substituted positions. Numbers in schema of HLA molecule indicate substituted amino acid positions that were elucidated as significant risk factor for severe aGVHD. Positions 9, 99, and 116 are located in the beta-plated sheet and positions 77, 80, and 156 in the alpha helix of HLA class I molecule (left). Positions 77 and 80 are associated with KIR2DL ligand in HLA-C molecule. Position 9 constitutes peptide-binding pockets B and C; position 99 constitutes A, B, and D pockets; position 116 constitutes F pocket; and position 156 constitutes D and E pockets (right). For example, Tyr-Phe indicated amino acid substitution at indicated position in HLA molecule at which donor had tyrosine and patient phenylalanine. Tyr indicates tyrosine; Phe, phenylalanine; Asn, asparagine; Asp, asparatic acid; Ser, serine; Lys, lysine; Leu, leucine; and Arg, arginine. *Result of base analysis was significant but result of validating analysis using bootstrap resampling was not. Results of analysis were thus judged not to be statistically significant.
To our knowledge, amino acid substitutions at position 9 (Tyr9A-Phe9A and Tyr9C-Ser9C) and position 99 (Tyr99C-Phe99C) were newly identified in the present study as responsible for severe aGVHD.
Ferrara et al reported that the amino acid substitution at position 116 in HLA class I molecule increased the risk for aGVHD, although the substituted amino acid was not taken into consideration.29 In our study, specific amino acid substitution at position 116 had a significant effect in HLA-C (Leu116C-Ser116C) and a marginal effect in HLA-A (Asn116A-Asp116A) for severe aGVHD (Table 5).
Position 156 of HLA molecule was certified to modify T-cell alloreactivity in vitro in HLA-A2,30,–32 HLA-B35,33 and HLA-B44.24 For example, in contrast to Asp156B in HLA-B*4402, the nonpolar nature of substituted Leu156B in HLA-B*4403 lost many interactions such as hydrogen bonds and van der Waals interactions with the other amino acid residues that constructed binding pockets. As a result, this substitution made the significant conformation change for alloreactivity.24 In the HLA-B*3501 and HLA-B*3508 combination, Leu156B in HLA-B*3501 with nonpolar residue was substituted for Asp156B in HLA-B*3508 with polar residue, and induced strong alloreactivity.33 In our study, the magnitude of the polar change of each substituted amino acid was calculated by “hydropathy scale,”17 because the influence of this scale on the amino acid interaction was much greater than the influence of the isoelectric point.34 Specific amino acid substitutions at position 9, 99, 116, and 156, which were not associated with KIR2DL ligand, were found to induce great polar changes except for Tyr9C-Ser9C. Generally speaking, the 3 major physicochemical properties of amino acids that play important roles in protein structure are the hydropathy scale, isoelectric point, and molecular weight, and molecular weight is reflected in the size of amino acids.34 Indeed, although tyrosine and serine in Tyr9C-Ser9C show few differences in hydropathy scale and isoelectric point, their molecular weights are quite different and may well induce an important conformation change in the HLA molecule. Thus, the change in the conformation by the polar change of the HLA molecule might be one of the mechanisms inducing alloreactivity. These data serve to clarify the mechanisms of aGVHD based on the HLA molecule.
The analysis of HLA-B, -DRB1, -DPB1, and -DQB1 mismatch for the substitution of amino acid elucidated no responsible position for severe aGVHD, and the analysis of HLA-A elucidated only one position. We speculate that the reason for the above result in HLA class I was that in this population there were fewer HLA-mismatched pairs in HLA-A and -B than in HLA-C. Although the findings are due mainly to the HLA-C molecule, specific amino acid substitution at positions 9, 99, 116, and 156 on the HLA class I molecule may induce strong alloreactivity because the structures of HLA class I molecules are quite similar.29 Indeed, position 9 is selected in HLA-A and -C concurrently, and position 116 had a significant effect on HLA-C and a marginal effect on HLA-A (Figure 2). In HLA class II, we speculated that the molecular base of aGVHD caused by the HLA class II mismatch might be different from that in HLA class I.
In conclusion, we clarified nonpermissive mismatch combinations of all major 6 HLA loci. These data would be beneficial for the selection of suitable donors and international donor exchange for UR-HSCT. Furthermore, we identified the positions and types of amino acid substitutions responsible for severe aGVHD and presented speculations for alloreactivity on the basis of the conformation change of the HLA molecule. These findings provide evidence to elucidate the mechanism of aGVHD on the basis of the HLA molecule.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by Health and Labor Science Research Grant from Ministry of Health, Labor and Welfare of Japan (Research on Human Genome, Tissue Engineering), Grant-in-Aid B from the Japan Society for the Promotion of Science, and a Grant from Third-Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor and Welfare, Japan.
We thank the staff members of the transplant center, donor centers, and JMDP office for their generous cooperation; Ms Ryouko Yamauchi for the data management; and Dr Toshitada Takahashi and Dr Setsuko Kawase for their expert technical assistance.
Authorship
Contribution: T.S., Y.M., T.K., T.J., and Y.K. participated in the conception of this study; K.K., H.I., and H.S. performed the execution for histocompatibility; Y.M. and S.K. performed the execution for transplantation; T.K. and K.M. performed statistical data analysis; T.K. and Y.M. wrote the paper; all authors checked the final version of the paper.
A complete list of the institutions participating and registering patients through the Japan Marrow Donor Program for the present study is available on the Blood website; see the Supplemental Appendix link at the top of the online article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yasuo Morishima, Department of Hematology and Cell Therapy, Aichi Cancer Center, 1-1 Kanokoden chikusa-ku Nagoya 464-8681, Japan; e-mail:ymorisim@aichi-cc.jp.