Abstract
Severe deficiency of ADAMTS13, a von Willebrand factor (VWF)–cleaving metalloprotease, causes thrombotic thrombocytopenic purpura. When analyzed with VWF multimers, but not with an abbreviated VWF peptide (VWF73) as the substrate, the plasma ADAMTS13 activity levels of mouse strains segregated into a high and a low group that differed by approximately 10 fold. Low ADAMTS13 activity was detected in mice containing 2 alleles of intracisternal A-type particle (IAP) retrotransposon sequence in the ADAMTS13 gene. Molecular cloning of mouse ADAMTS13 identified 2 truncated variants (IAP-a and IAP-b) in the low-activity mice. Both of the IAP variants lacked the 2 carboxyl terminus thrombospondin type 1 repeat (TSR) and CUB domains of full-length ADAMTS13. The IAP-b variant also had splicing abnormalities affecting the spacer domain sequence and had miniscule enzymatic activity. Compared with full-length ADAMTS13, the IAP-a variant was approximately one ninth as active in cleaving VWF multimers but was only slightly less active in cleaving VWF73 peptide. Recombinant human ADAMTS13 was also less effective in cleaving VWF multimers than VWF73 when the C-terminal TSR sequence was deleted. In summary, the carboxyl terminus TSR sequence is important for cleaving VWF multimers. Assay results should be interpreted with caution when peptide substrates are used for analysis of variant ADAMTS13 proteins.
Introduction
Human ADAMTS13 (hADAMTS13), a member of the “a disintegrin and metalloprotease with thrombospondin type 1 motif (ADAMTS)” family, cleaves von Willebrand factor (VWF) at the peptide bond between Y1605 and M1606 of the A2 domain.1 Human ADAMTS13 gene contains 29 exons and spans approximately 37 kb on chromosome 9q34. The mRNA, 4.7 kb in size, is detected primarily in the stellate cells of the liver,2,3 although it may also be expressed in other cell types.4-7 Mutations of the ADAMTS13 gene are detected in patients with congenital thrombotic thrombocytopenic purpura (TTP), whereas autoimmune inhibitors of the enzyme are detected in patients with an acquired form of the disease.8
ADAMTS13 consists of several homologous domains: a signal peptide, a propeptide, a reprolysin-type metalloprotease domain, a disintegrin domain, a thrombospondin type 1 repeat (TSR), a cysteine-rich domain (Cys), a spacer domain (Spa), 7 additional TSRs, and 2 CUB domains. In vitro studies have revealed that deletion of the C-terminal TSR and CUB domains partially decreases the VWF cleaving activity by 50% to 75%, whereas deletion of the spacer domain markedly decreases the VWF binding and cleaving activity of the protease to less than 1% to 2%,9-12 indicating that the spacer domain plays a pivotal role in the cleavage of VWF. Nevertheless, the role of the sequence downstream of the spacer domain remains unresolved.
Intracisternal A-type particles (IAPs) are defective retroviruses that may undergo transpositions and act as endogenous mutagens. In Mus musculus, approximately 2000 full-length and truncated IAP elements are present in each haploid genome.13 Genomic IAP sequences have been associated with activation of various genes involved in oncogenesis.14-17 Inactivation of the affected genes has also been described.18-20 IAP may also be involved in the pathogenesis of certain autoimmune diseases.21
An IAP element of 6026 base pairs is located in intron 23 of the ADAMTS13 gene in some strains of mice.22 This IAP element is 92% homologous to the MIA14 IAP element (M17551) originally identified in the kappa light-chain gene of a mouse hybridoma cell line.23,24 It was previously reported that the IAP did not significantly affect the levels of ADAMTS13 in the affected mouse strains.22
Considerable variability in VWF levels has been noted in various strains of mice.25 In contrast, strain-dependent variation of ADAMTS13 has not been described. In our effort to develop animal models for investigation of ADAMTS13 function in vivo, we discovered that mouse strains differed markedly in their plasma ADAMTS13 activity levels. In this study, we report on how the IAP element in the ADAMTS13 gene affects ADAMTS13 activity and how its study provides new insights of the ADAMTS13 structure-function relation.
Materials and methods
Heparin-coated capillary tubes and Corning Transwell cell culture isets were purchased from Fisher Scientific (Pittsburgh, PA); HEK 293T cells and MDCK cells were from ATCC (Manassas, VA); PfuUltra DNA polymerase was from Stratagene (La Jolla, CA); Centricon YM-30 concentrators were from Millipore (Billerica, MA); horseradish peroxidase-conjugated goat anti-mouse IgG and SuperSignal chemiluminescent substrate were from Pierce (Rockford, IL); biotinylated monoclonal anti-His was from Qiagen (Valencia, CA); avidin-peroxidase was from Sigma-Aldrich (St Louis, MO); FRETS-VWF73 was from Peptides International (Louisville, KY); LightCycler was from Roche Applied Science (Indianapolis, IN); Easy-DNA kit, Total RNA Purification System, ThermoScript RT-PCR System for First-Strand cDNA Synthesis Kit, pCR4-TOPO vector, pCDNA3.1/V5-His vector, Lipofectamine 2000, and monoclonal anti-V5 antibody were from Invitrogen (Carlsbad, CA); and laboratory mice were from The Jackson Laboratory (Bar Harbor, ME). All of the oligonucleotides used in this study, as listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), were synthesized by Sigma-Aldrich.
Animals
Mice were maintained and bred under controlled light and temperature conditions with free access to pelleted food and water. The Animal Care and Use Committee approved the animal protocol in accordance with institutional and National Institutes of Health (NIH) guidelines.
Mouse plasma, DNA, and mRNA
Mouse blood samples were collected from the tail vein or retro-orbital sinus of 6- to 12-week-old mice into heparin-coated capillary tubes. The plasma fraction was obtained by centrifugation at 12 000g for 5 minutes and saved under −70°C. Genomic DNA was isolated from tail tips by using Easy-DNA kit. Mouse mRNA was isolated from the livers by using Total RNA Purification System and reverse-transcribed by using ThermoScript RT-PCR System for First-Strand cDNA Synthesis Kit.
PCR, RT-PCR, and real-time RT-PCR
Polymerase chain reaction (PCR), reverse transcription PCR (RT-PCR), and real-time RT-PCR using LightCycler were performed as previously described.9 The amplified DNA fragments were separated by agarose gel electrophoresis and visualized fluorescently in the presence of ethidium bromide. β-actin or 18s ribosomal RNA was amplified with primer pairs PP14 or PP15 (Table S1) as housekeeping controls for RT-PCR and real-time RT-PCR.
Determination of 3′ end sequence of ADAMTS13 cDNA
The 3′ end sequence of the mouse ADAMTS13 mRNA was analyzed with 3′ RACE (rapid amplification of cDNA 3′ end). Briefly, total RNA and cDNA were prepared from a C57BL/6J mouse as in the reverse transcription experiments, except that an oligo-dT anchor primer (Table S1) was used for reverse transcription. PCR was carried out with the primer pair PP5, followed by another round of PCR using primer pair PP6. PCR products were cloned into pCR4-TOPO vector for sequence analysis.
Cloning of ADAMTS13 cDNA
The complete cDNA sequences of mouse ADAMTS13 were amplified from the liver total RNA by reverse transcription and PCR with PfuUltra DNA polymerase. Primer pairs PP7 and PP8 were used for FVB/NJ and C57BL/6J mice, respectively. For C57BL/6J cDNA, a second round of PCR with PP9 was performed to yield sufficient amount of cDNA for cloning. The PCR products were cloned into vector pCDNA3.1/V5-His and the constructs were verified by sequencing. Human ADAMTS13 cDNA (AD2, AD5, AD6, and AD7) were cloned for expression as previously described.9
Transient in vitro expression of ADAMTS13 proteins
The pCDNA3.1/V5-His plasmids containing the ADAMTS13 cDNA or its variants were used to transfect HEK 293T cells in the presence of Lipofectamine 2000.9 The conditioned serum-free media were collected after 48 hours and concentrated 15-fold on Centricon YM-30 concentrators. The cells were collected in an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. All products were saved at −70°C.
Recombinant proteins were separated in 7.5% SDS-PAGE and visualized by probing with monoclonal anti-V5 antibody, horseradish peroxidase-conjugated goat anti-mouse IgG, and SuperSignal chemiluminescent substrate.9 The molar concentrations of recombinant ADAMTS13 proteins were estimated by comparing their respective optic densities with a reference curve constructed from serial dilutions of an AD2 protein, whose concentration had previously been calibrated.9
To analyze the polarity of ADAMTS13 secretion, MDCK cells were grown on 6-well Transwell filter insert units. Transfection with ADAMTS13 cDNA or its variants was performed in the presence of lipofectamine 2000. The secreted ADAMTS13 proteins in the apical and basolateral compartments were analyzed by SDS-PAGE and visualized with monoclonal anti-V5. The relative amount of the proteins in the apical and basolateral compartments was measured by densitometry.
Detection of the IAP element in the mouse ADAMTS13 gene
To determine the presence or absence of the IAP sequence in intron 23, mouse genomic DNA was amplified by PCR using oligonucleotide pairs PP12 or PP13 as the primers (Figure 3). The sense primer of both pairs was derived from a sequence in intron 23, upstream of the IAP element. The antisense oligonucleotide of PP12 was derived from the 5′ long-terminal repeat (LTR) sequence that was included in exon 24 of the IAP variants, whereas the antisense oligonucleotide of PP13 was derived from an intronic sequence downstream of the IAP element. The PCR analysis was expected to yield a 229-base pair product with PP12 for the genomic sequence containing the IAP element in intron 23 and a 304-base pair product with PP13 for the genomic sequence without the IAP insert.
Measurement of ADAMTS13 activity
The proteolytic activity of ADAMTS13 proteins was measured against purified human plasma VWF multimers pretreated with 1.5 M guanidine hydrochloride (protein concentration in reaction mixtures, 41 μg/mL) or a recombinant VWF73 fusion peptide consisting of GST-VWF73 (D1596-R1668)-His as the substrate (concentration in reaction mixtures, 3.9 nM). In some experiments, a murine counterpart of the human VWF73 (D1596-T1668) was also used as the substrate. Measurement of activity against VWF multimers was based on the generation of the dimer of a 176-kDa fragment resulting from cleavage of VWF at the Y1605-M1606 bond.9 The activity against VWF73 fusion peptides was determined by enzyme-linked immunosorbent assay (ELISA) as previously described,26 except that the VWF73 fusion peptides in the microwells were probed by using a biotinylated anti-6xHis and an avidin-conjugated horseradish peroxidase. The activity against a fluorogenic form of the VWF73 peptide (FRETS-VWF73; concentration in reaction mixtures, 3.7 nM) was measured according to the manufacturer's instructions. All activity assays were measured against a reference curve of serially diluted normal human plasma samples. The activity results were expressed in U/mL for plasma samples and in U/nmol for recombinant ADAMTS13 proteins. The activity in 1 mL of normal human plasma was defined as 1 unit (U). For calculation of VWF multimer-to-VWF73 activity ratios, the same recombinant proteins were used for measurement against both substrates.
Kinetic studies
Reactions were performed with different concentrations of VWF multimers, GST-VWF73-His, or FRETS-VWF73 as the substrate. For FRETS-VWF73, the initial rates of reaction were determined by the levels of fluorescent intensity measured at intervals of 5 minutes for up to 120 minutes. For VWF multimers or GST-VWF73-His, the initial rates of reaction were determined by sampling reaction mixtures at 10, 20, 30, 60, and 120 minutes. The optic density of the 350-kDa band resulting from digestion of VWF multimers or the optic density of the 30.4-kDa band resulting from digestion of GST-VWF73-His was measured against a reference curve constructed from serial dilutions of corresponding cleavage product in a 120-minute reaction mixture of the respective substrate and an FVB/NJ plasma, a recombinant mouse full-length ADAMTS13, or human ADAMTS13 AD7. Reaction rates were obtained by linear regression analysis and fitted to the Michaelis-Menten equation (V = [Vmax × substrate concentration]/[Km + substrate concentration]) to obtain the equivalent values of Michaelis constant (Km) and catalytic rate constant (kcat, ie, Vmax/enzyme concentration). For comparison, the results were expressed as fractions of the corresponding values of FVB/NJ plasma, a recombinant mouse full-length ADAMTS13, or AD7. For mouse plasma samples, the enzyme concentrations could not be reliably measured with existing techniques. Consequently, Vmax instead of kcat values were obtained. Since all plasma samples were measured at the same volume, the Vmax ratios should reflect the difference in the kcat ratios. Each type of sample was measured at least 3 times and the mean values were shown.
Statistics
All results were expressed as means ± standard deviations (SDs), except when indicated otherwise. The difference among experimental groups was analyzed by analysis of variance (ANOVA).
Results
Proteolytic activity of mouse plasma ADAMTS13 against VWF multimers
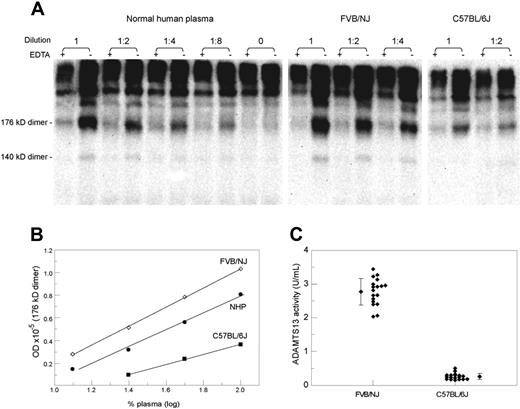
A representative analysis of mouse plasma ADAMTS13 activity is demonstrated in Figure 1A, which depicts VWF fragments generated after VWF multimers were incubated with normal human plasma, FVB/NJ strain plasma, or C57BL/6J plasma at various dilutions. For comparison, each incubation mixture (lane −) was accompanied with a control mixture containing EDTA to suppress the ADAMTS13 activity (lane +). Incubation of VWF multimers with plasma samples from either strain of mice generated different levels of the same VWF fragments. This difference was further illustrated by plotting the optic density of the 176-kDa fragment dimer against the plasma concentration (Figure 1B). The ADAMTS13 activity levels measured in 19 mice from each strain are illustrated in Figure 1C, confirming that the 2 strains were quite different in their plasma activity of cleaving VWF multimers (2.77 ± 0.40 U/mL for FVB/NJ vs 0.26 ± 0.09 U/mL for C57BL/6J; P < .001).
Comparison of ADAMTS13 activity in 2 mouse strains. (A) SDS-PAGE and immunoblotting depicting VWF proteolytic fragments (176-kDa and 140-kDa fragment dimmers) generated from cleavage of exogenous VWF multimers by ADAMTS13 in a normal human plasma, a FVB/NJ plasma, and a C57BL/6J plasma. VWF multimers were incubated with each plasma sample at the indicated dilution in the presence or absence of EDTA. The increased optic density in the absence of EDTA represents the product of proteolysis. (B) A plotting of the level of the 176-kDa fragment dimer against the plasma concentration. At each dilution, the FVB/NJ plasma was more active than the normal human plasma or the C57BL/6J plasma in generating the proteolytic fragment. (C) A dot plot of the ADAMTS13 activity levels in 18 FVB/NJ and 18 C57BL/6J mice. The mean and standard deviation are also shown for each group (P < .001).
Comparison of ADAMTS13 activity in 2 mouse strains. (A) SDS-PAGE and immunoblotting depicting VWF proteolytic fragments (176-kDa and 140-kDa fragment dimmers) generated from cleavage of exogenous VWF multimers by ADAMTS13 in a normal human plasma, a FVB/NJ plasma, and a C57BL/6J plasma. VWF multimers were incubated with each plasma sample at the indicated dilution in the presence or absence of EDTA. The increased optic density in the absence of EDTA represents the product of proteolysis. (B) A plotting of the level of the 176-kDa fragment dimer against the plasma concentration. At each dilution, the FVB/NJ plasma was more active than the normal human plasma or the C57BL/6J plasma in generating the proteolytic fragment. (C) A dot plot of the ADAMTS13 activity levels in 18 FVB/NJ and 18 C57BL/6J mice. The mean and standard deviation are also shown for each group (P < .001).
Analysis of mouse ADAMTS13 mRNA
To determine whether the 2 mouse strains differed in ADAMTS13 transcription, we compared the ADAMTS13 mRNA levels by real-time RT-PCR. Using primer pair PP1 (Document S1), the ADAMTS13 transcript level was not different between FVB/NJ and C57BL/6J mouse strains (0.88 ± 0.60, n = 14; vs 0.98 ± 0.52, n = 8).
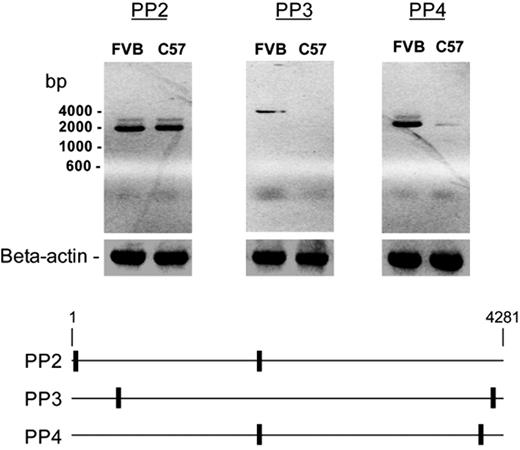
Further RT-PCR analysis using additional primer pairs (PP2, PP3, and PP4) to amplify different parts of the predicted ADAMTS13 coding sequence (NM_001001322) revealed that these 2 strains were quite different when primer pairs PP3 or PP4 were used (Figure 2). This discrepancy suggested that in the C57BL/6J strain, most of the ADAMTS13 cDNA ended upstream of the antisense sequence of primer pair PP4.
A composite of agarose gels depicting the products of mouse ADAMTS13 mRNA by RT-PCR. mRNA isolated from the livers of a FVB/NJ mouse and a C57BL/6J mouse was reverse transcribed and amplified with the indicated primer pairs. The analysis yielded similar levels of products with primer pair PP2 but much lower levels of products with primer pairs PP3 or PP4 in C57/BL/6J mice, suggesting that most of the mRNA was truncated. The approximate locations of the primers are illustrated in the bottom panel.
A composite of agarose gels depicting the products of mouse ADAMTS13 mRNA by RT-PCR. mRNA isolated from the livers of a FVB/NJ mouse and a C57BL/6J mouse was reverse transcribed and amplified with the indicated primer pairs. The analysis yielded similar levels of products with primer pair PP2 but much lower levels of products with primer pairs PP3 or PP4 in C57/BL/6J mice, suggesting that most of the mRNA was truncated. The approximate locations of the primers are illustrated in the bottom panel.
Mouse ADAMTS13 transcripts
To confirm the validity of this interpretation, we performed 3′ RACE to identify the 3′ end of ADAMTS13 transcripts from C57BL/6J mice. Sequence analysis of PCR products revealed that the mRNA of the C57BL/6J strain ended with an exon composed of 283 base pairs from the LTR of a previously reported retrotransposon of the IAP type as well as 8 base pairs from the intronic sequence immediately upstream of the LTR sequence. Based on this knowledge, we designed primer pairs PP8 and PP9 (Document S1) to amplify and clone the ADAMTS13 cDNA from C57BL/6J mice. ADAMTS13 cDNA was amplified and cloned from FVB/NJ with primer pair PP7.
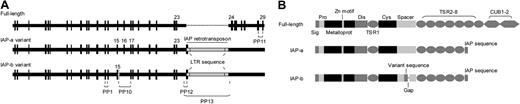
Alignment of the cloned cDNA sequences with the mouse genomic DNA sequence27 showed that the ADAMTS13 cDNA of the FVB/NJ strain was the same as that of NM_001001322 and was composed of 29 exons (Figure 3A). In contrast, 2 variants of ADAMTS13 cDNA were cloned from the C57BL/6J strain: IAP-a and IAP-b (Figure 3A). Both variants used a stop codon and a poly A signal derived from the LTR sequence of the IAP retrotransposon. IAP-a variant was found to be the same as AB071302 in the National Center for Biotechnology Information (NCBI) database.28 The IAP-b variant (Table S2) has not been described previously; it differed from the IAP-a variant in that its exon 15 contained an extra sequence of 153 base pairs from intron 15 but lacked 180 base pairs of predicted exon 16. This variation was not expected to alter the reading frame of the downstream mRNA sequence.
Mapping of coding sequences of mouse ADAMTS13 cDNA and the domain structures of the predicted proteins. (A) A full-length coding sequence consisting of 29 exons was cloned from a FVB/NJ mouse, whereas 2 coding sequences (IAP-a and IAP-b) were cloned from a C57BL/6J mouse. Both IAP variants ended with an exon consisting of sequences from the LTR of IAP and its 5′ flanking intronic sequence. IAP-b differed from IAP-a in that its exon 15 contained extra residues from intron 15 and lacked the exon 16 of IAP-a. As estimated by real-time RT-PCR using 2 primer pairs indicated in this panel, IAP-b accounted for 26% of the total ADAMTS13 mRNA. (B) The predicted domain structures of the 3 forms of ADAMTS13 cloned from mouse livers. Both IAP variants ended with a 16-residue sequence derived from the aberrant exon 24. A 61-residue sequence in the spacer domain of IAP-a was replaced by a novel sequence of 52 residues encoded by the extraneous sequence of exon 15.
Mapping of coding sequences of mouse ADAMTS13 cDNA and the domain structures of the predicted proteins. (A) A full-length coding sequence consisting of 29 exons was cloned from a FVB/NJ mouse, whereas 2 coding sequences (IAP-a and IAP-b) were cloned from a C57BL/6J mouse. Both IAP variants ended with an exon consisting of sequences from the LTR of IAP and its 5′ flanking intronic sequence. IAP-b differed from IAP-a in that its exon 15 contained extra residues from intron 15 and lacked the exon 16 of IAP-a. As estimated by real-time RT-PCR using 2 primer pairs indicated in this panel, IAP-b accounted for 26% of the total ADAMTS13 mRNA. (B) The predicted domain structures of the 3 forms of ADAMTS13 cloned from mouse livers. Both IAP variants ended with a 16-residue sequence derived from the aberrant exon 24. A 61-residue sequence in the spacer domain of IAP-a was replaced by a novel sequence of 52 residues encoded by the extraneous sequence of exon 15.
To determine the relative abundance of these 2 IAP variant transcripts, we performed real-time RT-PCR of mouse liver mRNA using primer pairs PP1 and PP10 (Figure 3A; Table S1). Serial dilutions of a plasmid DNA containing the cloned IAP-b variant were used as the reference. The results showed that the IAP-b variant accounted for 26% of the total ADAMTS13 cDNA. Similar real-time RT-PCR analysis using primer pairs PP1 and PP11 showed that the full-length mRNA accounted for less than 10% of the total ADAMTS13 transcripts in C57BL/6J mice.
Expression of ADAMTS13 proteins
Analysis of the translated sequences showed that the full-length mouse ADAMTS13 consisted of 1426-amino acid residues, whereas the 2 IAP variants consisted of 1037 and 1028 residues, respectively. Both variants ended with a novel 16-residue sequence (ALVWEAAPTFAVTRWR) not found in the full-length mouse ADAMTS13 (Figure 3B; Table S2). The IAP-b variant differed from the IAP-a variant in that residues A601-Q661 of the spacer domain of IAP-a were replaced by a sequence G601-K652 (Table S2).
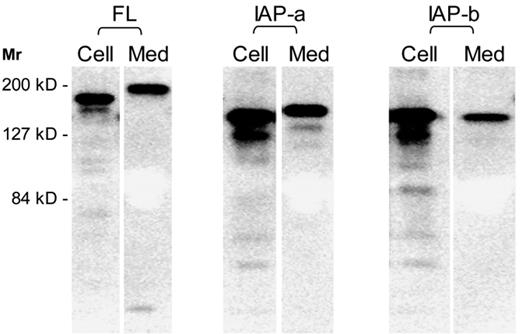
To compare the synthesis, secretion, and enzymatic activity of the 3 forms of mouse ADAMTS13, we cloned cDNA sequences into pCDNA3.1/V5-His-TOPO and expressed the proteins in HEK 293T cells, a human embryonic kidney cell line. SDS-PAGE and Western blotting showed that the full-length (FL) form was expressed as a 175-kDa protein but secreted as a larger 190-kDa band. Both IAP-a and IAP-b variants were approximately 150-kDa in size in the cells. The IAP-a variant was synthesized and secreted more efficiently as a 165-kDa band, whereas the IAP-b variant was secreted less efficiently and without an apparent increase in size (Figure 4; Table 1).
Synthesis and secretion of mouse ADAMTS13. Mouse ADAMTS13 cDNA constructs were used to transfect HEK 293T cells. The synthesized proteins were separated by SDS-PAGE and visualized by immunoblotting with ant-V5 tag sequence. A representative composite gel of ADAMTS13 proteins in the cell lysates and culture media for each cDNA. The cell lysates were diluted by 2-fold for the analysis.
Synthesis and secretion of mouse ADAMTS13. Mouse ADAMTS13 cDNA constructs were used to transfect HEK 293T cells. The synthesized proteins were separated by SDS-PAGE and visualized by immunoblotting with ant-V5 tag sequence. A representative composite gel of ADAMTS13 proteins in the cell lysates and culture media for each cDNA. The cell lysates were diluted by 2-fold for the analysis.
The percentages of protein secretion (medium/cells + medium), the estimated concentrations of the proteins in the culture media, and the VWF-cleaving activity levels of the secreted proteins
| Protein . | Secretion, % (n) . | Medium, nM (n) . | Activity, U/nmol* (n) . |
|---|---|---|---|
| Full-length | 23 ± 7 (5) | 1.6 ± 0.4 (4) | 578 ± 72 (3) |
| IAP-a | 33 ± 12 (5) | 6.7 ± 1.6† (4) | 65 ± 16† (4) |
| IAP-b | 5 ± 1‡ (3) | 2 ± 0.9 (3) | <0.05 U |
| Protein . | Secretion, % (n) . | Medium, nM (n) . | Activity, U/nmol* (n) . |
|---|---|---|---|
| Full-length | 23 ± 7 (5) | 1.6 ± 0.4 (4) | 578 ± 72 (3) |
| IAP-a | 33 ± 12 (5) | 6.7 ± 1.6† (4) | 65 ± 16† (4) |
| IAP-b | 5 ± 1‡ (3) | 2 ± 0.9 (3) | <0.05 U |
All values are expressed as mean ± SD. The numbers (n) of expressed proteins analyzed are also indicated for each cDNA.
Measured against VWF multimers as the substrate.
P < .01 (compared with full-length values).
P < .05.
Analysis of cleaving activity against VWF multimers showed that the IAP-a variant was approximately one ninth as active as the full-length protein, whereas the activity of the IAP-b variant was too low for further analysis (Table 1).
It has been suggested that ADAMTS13 is secreted with 95% apical polarity and that the CUB domain sequence is critical for this polarization of secretion.7 To determine whether the IAP-a variant protein was secreted with a different polarity, we transfected MDCK cells grown on Transwell filters with mouse ADAMTS13 cDNA plasmids. The results showed that 48.9% ± 10.1% (n = 5) of recombinant IAP-a and 48.2% ± 9.6% (n = 5) of recombinant mouse full-length ADAMTS13 were secreted apically (P > .5).
Linkage of low VWF cleaving activity to IAP genotype
To confirm that the difference in ADAMTS13 activity among the mouse strains was due to the presence of the IAP retrotransposon in the ADAMTS13 gene, we measured the protease activity in 4 additional mouse strains (CAST/EiJ, DBA/2J, 129/SvJ, and BALB/c) and correlated the results with the presence of the IAP sequence in the ADAMTS13 gene. The analysis showed that the IAP was detected in all 3 strains with low ADAMTS13 activity levels but in none of the strains with high protease activity (Table 2).
IAP genotype and ADAMTS13 activity levels
| Strain . | No. tested . | IAP genotype . | ADAMTS13 activity, mean ± SD, U/mL . |
|---|---|---|---|
| FVB/NJ | 23 | −/− | 2.95 ± 0.51 |
| 129x1/SvJ | 6 | −/− | 3.13 ± 0.42 |
| CAST/EiJ | 5 | −/− | 3.37 ± 0.65 |
| C57BL/6J | 21 | +/+ | 0.26 ± 0.09 |
| DBA/2J | 6 | +/+ | 0.24 ± 0.06 |
| BALB/c | 6 | +/+ | 0.30 ± 0.12 |
| F1* | 10 | +/− | 2.43 ± 0.43 |
| F2* | 11 | −/− | 2.51 ± 0.68 |
| F2* | 21 | +/− | 2.15 ± 0.50 |
| F2* | 4 | +/+ | 0.30 ± 0.13 |
| Strain . | No. tested . | IAP genotype . | ADAMTS13 activity, mean ± SD, U/mL . |
|---|---|---|---|
| FVB/NJ | 23 | −/− | 2.95 ± 0.51 |
| 129x1/SvJ | 6 | −/− | 3.13 ± 0.42 |
| CAST/EiJ | 5 | −/− | 3.37 ± 0.65 |
| C57BL/6J | 21 | +/+ | 0.26 ± 0.09 |
| DBA/2J | 6 | +/+ | 0.24 ± 0.06 |
| BALB/c | 6 | +/+ | 0.30 ± 0.12 |
| F1* | 10 | +/− | 2.43 ± 0.43 |
| F2* | 11 | −/− | 2.51 ± 0.68 |
| F2* | 21 | +/− | 2.15 ± 0.50 |
| F2* | 4 | +/+ | 0.30 ± 0.13 |
All ADAMTS13 activity levels were measured against VWF multimers.
F1 = FVB/NJ × C57BL/6J; F2 = F1 × F1.
To further analyze the association between the IAP genotype with ADAMTS13 activity, C57BL/6J mice were crossed with FVB/NJ mice. The resulting F1 mice were crossed with one another to create F2 mice with 0, 1, or 2 alleles of the IAP genotype. Activity analysis against VWF multimers revealed that only the mice with 2 alleles of the IAP had low ADAMTS13 levels (Table 2).
Mouse ADAMTS13 activity against VWF peptides
We compared the protease activity levels of mouse plasma samples against both human and murine versions of the VWF73 fusion peptide. The results showed that the FVB/NJ-to-C57BL/6J ratio decreased from approximately 11 when the activity was assayed against VWF multimers to 1.6 when the activity was against either human or murine VWF73 peptide (Figure 5A).
Substrate-dependent variation of VWF cleaving activity of mouse plasma samples and recombinant mouse ADAMTS13 proteins. (A) Activity levels of FVB/NJ and C57BL/6J plasma samples against VWF multimers (multimers), human VWF73 fusion peptide (hVWF73), and mouse VWF73 fusion peptide (mVWF73). Mouse plasma samples were assayed for activity of cleaving the indicated substrates, and all values were expressed using normal human plasma as the reference. Each column in the graph represents the mean and standard deviation of results from 23 FVB/NJ or 21 C57BL/6J different mouse plasma samples for VWF multimers and 5 different plasma samples each for the VWF peptide substrates. FVB/NJ plasma ADAMTS13 was much more effective than C57BL/6J plasma in cleaving VWF multimers (P < .001). This difference diminished from 11- to 1.6-fold when either human or mouse VWF73 was used as the substrate. (B) Activity levels (means and standard deviations) of recombinant full-length ADAMTS13 protein and its IAP-a variant against various types of substrates. FRETS indicates FRETS-VWF73. Recombinant proteins were assayed for enzymatic activity against the indicated substrates. All values were expressed against normal human plasma as the reference. Each column represents the mean and standard deviation of results obtained with 3 lots of the protein. The IAP-a protein was less effective than full-length mouse ADAMTS13 in cleaving VWF multimers (P < .001). The difference diminished from 9- to 1.4- to 2.0-fold when VWF73 peptides were used as the substrates.
Substrate-dependent variation of VWF cleaving activity of mouse plasma samples and recombinant mouse ADAMTS13 proteins. (A) Activity levels of FVB/NJ and C57BL/6J plasma samples against VWF multimers (multimers), human VWF73 fusion peptide (hVWF73), and mouse VWF73 fusion peptide (mVWF73). Mouse plasma samples were assayed for activity of cleaving the indicated substrates, and all values were expressed using normal human plasma as the reference. Each column in the graph represents the mean and standard deviation of results from 23 FVB/NJ or 21 C57BL/6J different mouse plasma samples for VWF multimers and 5 different plasma samples each for the VWF peptide substrates. FVB/NJ plasma ADAMTS13 was much more effective than C57BL/6J plasma in cleaving VWF multimers (P < .001). This difference diminished from 11- to 1.6-fold when either human or mouse VWF73 was used as the substrate. (B) Activity levels (means and standard deviations) of recombinant full-length ADAMTS13 protein and its IAP-a variant against various types of substrates. FRETS indicates FRETS-VWF73. Recombinant proteins were assayed for enzymatic activity against the indicated substrates. All values were expressed against normal human plasma as the reference. Each column represents the mean and standard deviation of results obtained with 3 lots of the protein. The IAP-a protein was less effective than full-length mouse ADAMTS13 in cleaving VWF multimers (P < .001). The difference diminished from 9- to 1.4- to 2.0-fold when VWF73 peptides were used as the substrates.
Similarly, analysis of the mouse recombinant ADAMTS13 protein activities using either human or mouse VWF73 fusion peptide as the substrate showed that the FL-to-IAP-a ratio of protease activity decreased from 9 against VWF multimers to 1.4 to 2.0 against human or mouse VWF73 in ELISA or fluorescence assays (Figure 5B).
Activity of recombinant human ADAMTS13
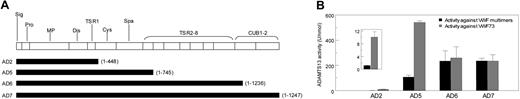
To determine whether the substrate-dependent variation also occurred with human ADAMTS13, we compared the activity of previously described recombinant human ADAMTS13 proteins AD2, AD5, AD6, and AD7 against VWF multimers and human VWF73 fusion peptide. As shown in Figure 6, human ADAMTS13 proteins lacking the TSR 2-8 domains (AD2 and AD5) were relatively less active in cleaving VWF multimers than in cleaving the VWF73 fusion peptide. In contrast, no such substrate-dependent difference was observed with AD6 or AD7.
Comparison of the activity levels of recombinant human ADAMTS13 and its truncated variants against VWF multimers or human VWF73 fusion peptide. (A) The lengths of the recombinant proteins used in the experiment are depicted against the domain structure of ADAMTS13. (B) The activity levels of each protein against VWF multimers (■) or human VWF73 fusion peptide (▩) are depicted. Recombinant proteins were assayed for activity of cleaving either VWF multimers or human VWF73 fusion peptide. All values were expressed using normal human plasma as the reference. Each column represents the mean and standard deviation of results obtained from 3 lots of the protein. The inset shows a magnification of the results for AD2. AD2 and AD5 were relatively less effective in cleaving VWF73 multimers than cleaving VWF73 peptide (P < .05 and < .001, respectively). No such substrate-dependent difference was observed for AD6 or AD7.
Comparison of the activity levels of recombinant human ADAMTS13 and its truncated variants against VWF multimers or human VWF73 fusion peptide. (A) The lengths of the recombinant proteins used in the experiment are depicted against the domain structure of ADAMTS13. (B) The activity levels of each protein against VWF multimers (■) or human VWF73 fusion peptide (▩) are depicted. Recombinant proteins were assayed for activity of cleaving either VWF multimers or human VWF73 fusion peptide. All values were expressed using normal human plasma as the reference. Each column represents the mean and standard deviation of results obtained from 3 lots of the protein. The inset shows a magnification of the results for AD2. AD2 and AD5 were relatively less effective in cleaving VWF73 multimers than cleaving VWF73 peptide (P < .05 and < .001, respectively). No such substrate-dependent difference was observed for AD6 or AD7.
Kinetic analysis
The equivalent values of Michaelis constant (Km) and maximal reaction (Vmax) or catalytic rate constant (kcat) were estimated from the levels of the cleavage products and expressed as ratios of the respective reference samples (FVB plasma or recombinant mouse or human full-length ADAMTS13 proteins; Table 3). For both C57BL/6J and recombinant IAP-a measured against VWF multimers as the substrate, the Km values were markedly elevated (14.0 and 16.5, respectively) compared with their respective reference samples (FVB plasma and recombinant full-length mouse ADAMTS13). The ratios were not substantially different from 1.0 (0.6 and 0.8, respectively) when these samples were measured against FRETS-VWF. The Vmax or kcat ratios were not affected by the substrates used. For the C57BL/6J and FVB/NJ plasma samples, we also measured the kinetic constants against the VWF73 fusion peptide by ELISA, and the results (Km ratio = 0.9; Vmax ratio = 0.6) were quite similar to those obtained with FRETS-VWF.
Comparison of kinetic values
| ADAMTS13 . | Ratio . | |||
|---|---|---|---|---|
| Km . | kcat . | |||
| VWF . | FRETS-VWF73 . | VWF . | FRETS-VWF73 . | |
| C57/FVB | 14.0 | 0.6 | NA | NA |
| IAP-a/FL | 16.5 | 0.8 | 0.34 | 0.31 |
| AD6/AD7 | 1.7 | 1.4 | 0.5 | 1.5 |
| AD5/AD7 | 5.2 | 1.0 | 1.1 | 1.9 |
| AD2/AD7 | 316 | 4.8 | 0.5 | 0.2 |
| ADAMTS13 . | Ratio . | |||
|---|---|---|---|---|
| Km . | kcat . | |||
| VWF . | FRETS-VWF73 . | VWF . | FRETS-VWF73 . | |
| C57/FVB | 14.0 | 0.6 | NA | NA |
| IAP-a/FL | 16.5 | 0.8 | 0.34 | 0.31 |
| AD6/AD7 | 1.7 | 1.4 | 0.5 | 1.5 |
| AD5/AD7 | 5.2 | 1.0 | 1.1 | 1.9 |
| AD2/AD7 | 316 | 4.8 | 0.5 | 0.2 |
The Vmax ratios for C57/FVB plasma ADAMTS13 were 0.6 and 0.9, respectively, against substrates VWF and FRETS-VWF73.
NA indicates not applicable.
For human ADAMTS13 proteins, the Km value of AD6 was similar to that of AD7, irrespective of the substrates used. For AD5 and AD2, this Km ratio increased to 5.2 and 316, respectively, with VWF multimers but was 1.0 and 4.8, respectively, with FRETS-VWF73 substrate. The substrates caused no or minimal difference in the kcat ratios.
Discussion
This study shows that the presence of a genomic IAP element is the main reason for the low ADAMTS13 activity levels that we detected in some mouse strains. Two lines of evidence support this interpretation. First, all 3 mouse strains with the IAP element had much lower ADAMTS13 activity than those without the element. Second, interspecies breeding of FVB/NJ (high ADAMTS13 activity) with C57BL/6J (low ADAMTS13 activity) mice showed that low plasma ADAMTS13 activity level is linked to the homozygous IAP genotype. Intriguingly, mice with 1 IAP allele had ADAMTS13 levels similar to those with no IAP alleles, indicating that the IAP allele behaved as a recessive trait. The basis for this recessive phenomenon remains to be elucidated.
Comparison of the IAP genotype with the published mouse family tree reveals that mouse strains with the IAP element belong to 3 (groups 3, 4, and 6) of the 7 different phylogenetic groups that are not directly related,29 suggesting intriguingly that integration of the IAP element into the ADAMTS13 genome might have occurred independently. In this study, we designed the primers such that both IAP-positive and IAP-negative genotypes were detected. Furthermore, the IAP element must have been inserted into the same intron-23 locus in all 3 positive strains of mice. Further investigation is required to understand how the IAP integrated at the same site of the ADAMTS13 gene if it occurred independently in various mouse strains.
In this study, as in a previous one by Banno et al,22 the IAP element of the mouse ADAMTS13 gene did not substantially affect the levels of total ADAMTS13 mRNA. Instead, it affected the structure of the mRNA and hence the predicted proteins. Part of the LTR sequence of the IAP element, including a stop codon and a poly (A) signal, was retained to become the last exon in more than 90% of the mRNA. This variant form of mRNA (IAP-a) is predicted to result in the synthesis of truncated forms of ADAMTS13 protein that lack both the CUB and the last 2 TSR domain sequences, ending with a novel sequence derived from intron 23 and LTR.
Sequencing analysis of the cloned cDNA showed that some of the IAP-type mouse mRNA had other splicing aberrations that affected the spacer domain sequence of the predicted protein. As estimated by real-time RT-PCR analysis, this IAP-b variant accounted for 26% of the total mRNA. In contrast, the presumed full-length mRNA accounted for less than 10%. This suggests that the presence of the IAP element affects the conformation of nuclear RNA, resulting in alternative intronic splicing of nonconsecutive introns. Presumably because of its low abundance, the full-length ADAMTS13 was not cloned from the liver cDNA of the C57BL/6J strain in our studies.
The IAP-b variant differed from the IAP-a variant in that part of the wild-type spacer domain was replaced by an entirely different sequence. The IAP-b variant not only was secreted less efficiently but also had miniscule enzymatic activity. This is consistent with the current view that integrity of the spacer domain is a major determinant of ADAMTS13 activity. It is not clear why the IAP-b variant was not detected in a previous study.22
The obviously lower plasma ADAMTS13 activity levels in mouse strains with an IAP element in the ADAMTS13 gene contrast with the lack of difference described in a previous study.22 Since that study used a mouse VWF73 fusion peptide as the substrate for activity assay, we suspected that difference in the assay substrates might account for the discrepant observations. To explore this possibility, we compared the activity levels using both VWF multimers and human or murine VWF73 peptides as the enzymatic substrates. The results showed that, when analyzed against either human or mouse VWF73 peptide, the difference between mouse strains with and without the IAP element was less than 2-fold. Thus the discrepancy between these 2 studies resulted primarily from the difference in the assay substrates.
This interpretation is further supported by analysis of recombinant mouse ADAMTS13 proteins. The full-length mouse ADAMTS13 was much more active than the truncated IAP-a variant protein when the proteases were analyzed against VWF multimers. The difference between these 2 proteins diminished when the activity assay was performed against VWF73 peptide. Since the VWF73 substrates were used in both ELISA and fluorescence formats, it is unlikely that assay formats caused the observed variations. In our expression studies, the IAP-a variant protein was synthesized and released in culture media more efficiently than the full-length protein, and no apical polarization of secretion was detected with either form of mouse ADAMTS13 in MDCK cells, a cell line known to exhibit polarized secretion of certain proteins. It is not clear why mouse ADAMTS13 did not exhibit apical polarity of secretion as previously reported with human ADAMTS13.7
A similar pattern of substrate-dependent divergence in activity was also observed with human ADAMTS13 and its recombinant variants. Compared with normal human plasma ADAMTS13, truncated proteins AD2 and AD5, but not AD6 or the full-length AD7, exhibited higher activity against VWF73 peptides than against VWF multimers. These findings suggest that the TSR 2-8 domains are important for efficient cleavage of VWF multimers but play little or no role in the cleavage of abbreviated peptide substrates. Interestingly we found that AD5 is more active than full-length AD7 in cleaving VWF73, suggesting that the TSR 2-8 and CUB domains may negatively impact the cleavage of VWF73. Similarly, hyperactivity has been observed in a flow system with truncated ADAMTS13 variant lacking the TSR 2-8 and CUB domains.30
The Km data of this study suggest that, compared with the full-length mouse ADAMTS13, the IAP-a variant has a much lower affinity for VWF multimers. Such a difference was not observed when FRETS-VWF73 was used as the substrate. This impact of the substrate form was similarly reflected in the difference of the Km ratio of C57BL/6J versus FVB/NJ plasma, since the VWF cleaving activity is expected to derive primarily from the IAP-a variant form of mouse ADAMTS13 in the C57BL/6J plasma and from full-length protease in FVB/NJ plasma. The type of substrate did not substantially affect the kcat or Vmax data, suggesting that the sequence difference between IAP-a and full-length mouse ADAMTS13 did not impact on the catalytic activity of the protease. Because we were unable to obtain sufficient mouse VWF multimers for analysis, only human VWF multimers were used in this study. Nevertheless, the divergence between VWF multimers and VWF73 peptide was also observed with AD5 and AD2, but not with AD6 or AD7, suggesting that the TSR 2-8 sequence enhanced the binding of human ADAMTS13 protein to VWF multimers but had no effect on the binding of the protease to VWF73.
Recent studies have demonstrated that truncation of human ADAMTS13 upstream of the spacer domain markedly decreases but does not abolish the enzymatic activity.9,11,12 This decrease of VWF cleavage was believed to result primarily from decreased substrate binding in the absence of the spacer domain sequence. The very low VWF cleaving activity and high Km ratio observed with AD2 in this study is consistent with the interpretation that the spacer domain promotes the binding of the protease to VWF. While TSR 2-8 promotes the binding of the protease to VWF multimers, the promoting effect of the spacer domain is observed with VWF in its multimeric or abbreviated peptide form.
The role of the CUB domain sequence at the carboxyl end of ADAMTS13 remains controversial. It has been suggested that the CUB domain sequence may promote ADAMTS13 binding to and cleavage of VWF multimers.11,12,31 Nevertheless, this observation has not been supported by data of VWF cleaving activity using either VWF multimers or abbreviated VWF peptides as the substrate.9,32,33 Our activity and kinetic data in this study also did not detect a substantial role of the CUB domain in promoting cleavage of conformationally unfolded VWF multimers. Difference in the design of the variant proteins and in the conditions used for cleavage analysis may affect the observed discrepancy, as suggested by a previous observation that truncation at the end of TSR7 results in increased binding with immobilized VWF multimers.11 Furthermore, because of inherent limitations of the assays, a minor role of the CUB domain sequence in promoting binding of ADAMTS13 to VWF multimers could not be excluded in our study.
Taking all of the available data into consideration, we propose that the spacer domain sequence enhances the binding of ADAMTS13 with VWF multimers or VWF73 peptide, whereas the TSR 2-8 sequence promotes the binding of the protease with VWF multimers but not with the VWF73 peptide. Further structural investigations are necessary to determine how the TSR sequence accomplishes this complex interaction. Our data obtained with the mouse IAP variants suggest that the sequence needed for effective multimer cleavage may be further narrowed to the TSR 7-8 domains. Nevertheless, since the mouse variants contained an extraneous 16-residue sequence at their carboxyl terminus, this interpretation requires confirmation with appropriate constructs in future studies.
Mouse strains have been shown to be quite heterogeneous in their VWF antigen levels, due to multiple genetic factors.25,34 Nevertheless, variability in VWF levels does not correlate with the risk of thrombosis in mice with ADAMTS13 deficiency.35 The present study reveals that mouse strains also differ considerably in their ADAMTS13 activity levels. In animal models, inactivation of the ADAMTS13 gene increases the size distribution of VWF multimers and causes TTP-like illness only in selected mouse strains.35,36 In this study, we found that despite marked difference in ADAMTS13 activity levels, the FVB/NJ and C57BL/6J mouse strains did not differ in their VWF multimer size distributions (data not shown). Thus, in these mouse strains, as in the C57BL6-129 × 1/Sv chimeric mice in a previous study,35 ADAMTS13 does not appear to play a major role in regulating the size of VWF multimers. Further investigation of the complex interplay between ADAMTS13, VWF, and other as yet unknown genes is necessary to elucidate the critical factors modulating the proteolysis of VWF and the propensity to thrombosis in ADAMTS13-deficient mice. Knowledge from such studies may be relevant for understanding why patients with the same mutations of ADAMTS13 occasionally present with vastly different disease severities. The present study also suggests that assay results should be interpreted with reservation when synthetic or recombinant peptides are used as the substrate for analysis of variant ADAMTS13 proteins.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs D. Ginsburg and D. Motto for kindly providing the construct of mouse VWF73 fusion peptide used in this study.
This study was supported in part by a grant (R01 HL62136) from the National Heart, Lung, and Blood Institute of the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: W.Z., E.E.B., and H.-M.T. contributed to conception, design, execution, data interpretation, and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Han-Mou Tsai, Montefiore Medical Center, Division of Hematology, 111 East 210th St, Bronx, NY 10467; e-mail: htsai@montefiore.org.