Abstract
Dasatinib and nilotinib are potent tyrosine kinase inhibitors (TKIs) with activity against many imatinib-resistant chronic myeloid leukemia (CML) clones with BCR-ABL kinase domain (KD) mutations, except T315I. We assessed for changes in the BCR-ABL KD mutation status in 112 patients with persistent CML who received a second-generation TKI after imatinib failure. Sixty-seven different KD mutations were detected before the start of therapy with a second TKI, with T315I seen in 15%. Equal numbers of patients received nilotinib or dasatinib following imatinib, and 18 received 3 TKIs. Response rates were similar for patients with and without mutations, regardless of mutation site except for T315I. Overall, 29 patients (26%) developed new KD mutations after therapy with a second (n = 24) or third (n = 5) TKI, but only 4 (4%) developed T315I. In 73% of cases, the KD mutations that persisted or developed following switch to new TKI were at sites also found in prior in vitro TKI mutagenesis assays. Although there is only a mild increase in mutation frequency with sequential TKI treatment, novel mutations do occur and mutation regression/acquisition/persistence generally reflects the in vitro differential sensitivity predicted for each TKI. In this study, there was no marked increase in development of T315I.
Introduction
Clinical resistance to imatinib in chronic myeloid leukemia (CML) has been attributed to several mechanisms.1-6 Among them, point mutations in the BCR-ABL kinase domain appear to be the most common, occurring in 30% to 90% of patients who develop resistance.7-11 Mutations in more than 40 different amino acids have been described, conferring differing levels of resistance to imatinib.10,11
To overcome imatinib resistance, more potent tyrosine kinase inhibitors (TKIs) such as nilotinib, dasatinib, and bosutinib have been developed with demonstrable activity against most of the BCR-ABL kinase domain mutations seen in patients treated with imatinib, with the notable exception of the T315I mutation.12-16 Experimental models of in vitro drug sensitivity have shown that specific mutations may develop after incubation with nilotinib or dasatinib, albeit at a decreased frequency compared with imatinib.17-19 Some of these mutations were novel, not being previously described after imatinib failure, and in some instances they did not confer resistance to imatinib.
With the increased use of these newer TKIs, it has been suggested that the spectrum of kinase domain (KD) mutations may change, possibly selecting for the pan-resistant T315I.20,21 We thus analyzed the patterns of mutations occurring in patients with CML treated sequentially with imatinib, and 1 or 2 second-generation TKIs. The goals of the study were to investigate the dynamics of the mutations and the emergence of new mutations in patients with persistent disease after sequential therapy with TKI.
Patients and methods
Between June 2003 and February 2006, 217 patients with CML who failed therapy with imatinib were treated with second-generation TKIs. Mutational analysis by direct sequencing was performed in all patients after imatinib failure and prior to the start of therapy with the second TKI. To date, mutational analysis has also been performed after treatment with another TKI in 112 patients who had failure to imatinib therapy. These patients with mutation analysis before and after treatment with second TKI constitute the focus of this report. Of these 112 patients treated with at least 2 TKIs, 18 received therapy with a third TKI after failing imatinib and one other inhibitor.
The criteria to trigger mutation analysis were based on clinical evidence of treatment failure. For this purpose, we used the recent recommendations of the European LeukemiaNet.22 Briefly, treatment failure was defined as transformation to accelerated or blast phase, loss of a cytogenetic or complete hematologic response (CHR), or failure to achieve a CHR (chronic phase [CP] only) or any hematologic response (for patients in accelerated phase [AP] or blast phase [BP]) after 3 months of therapy, or persistence of 100% Philadelphia chromosome (Ph)–positive metaphases after 6 months of therapy, or 35% or more after 12 months. Although these criteria were developed for patients treated with imatinib, in the absence of set criteria for second and subsequent line therapy, they were used for subsequent therapies in this study. Patients who had their treatment changed for other reasons (eg, TKI intolerance) also had a mutation analysis done. Patients were treated by M. D. Anderson Cancer Center IRB-approved protocols. Informed consent was obtained in accordance with the Declaration of Helsinki. Response criteria were as previously described.23 CHR was defined as a white blood cell (WBC) count of less than 10 × 109/L, a platelet count of less than 450 × 109/L, no immature cells (blasts, promyelocytes, myelocytes) in the peripheral blood, and disappearance of all signs and symptoms related to leukemia (including palpable splenomegaly). CHR was further categorized by the best cytogenetic remission as complete (0% Ph positive), partial (1%-35% Ph positive), minor (36%-65% Ph positive), and minimal (66%-95% Ph positive). A major cytogenetic remission (MCyR) included complete plus partial cytogenetic remissions (ie, ≤ 35% Ph positive). Cytogenetic remission was judged by standard cytogenetic analysis in 20 metaphases, done on bone marrow aspiration; fluorescence in situ hybridization using a dual-fusion probe, done on peripheral blood, was used only when routine cytogenetic analysis was unanalyzable (ie, insufficient metaphases). Major molecular response (MMR) was defined as a BCR-ABL/ABL ratio of less than 0.05% by real-time TaqMan-based quantitative polymerase chain reaction (PCR) done in peripheral blood samples, representing a 3-log reduction from the baseline for untreated patients in our laboratory; complete molecular response was (CMR) defined as undetectable levels of BCR-ABL with level of detection of at least 4.5 logs.24
Associations between categoric variables were assessed via cross-tabulation and Fisher exact test.
Mutation analysis
Total RNA was isolated from peripheral blood or bone marrow aspirate samples by Trizol solubilization (Gibco-BRL, Gaithersburg, MD) and cDNA synthesized by reverse transcriptase (Superscript II; Invitrogen, Carlsbad, CA). The kinase domain of the BCR-ABL fusion transcript was sequenced using a nested PCR strategy. BCR-ABL was first amplified followed by 2 separate PCR reactions that cover exons 221 to 394 and codons 381 to 500 of the ABL kinase domain. After purification of PCR products through DyeEx columns (Qiagen, Valencia, CA), standard dideoxy chain-termination DNA sequencing was performed using Big Dye chain terminator reagents on an automated 3130 genetic analyzer using Sequence Analysis V3.3 and SeqScape software V2.5 (ABI, Foster City, CA). All mutations were confirmed by sequencing of forward and reverse strands, with a sensitivity of 10% to 20% mutation-bearing cells in the analyzed population. For additional analysis of the T315I mutation, pyrosequencing was performed following first-round BCR-ABL PCR. Second round ABL KD PCR was performed using one biotin-tagged primer, and single-stranded product isolated on streptavidin sepharose beads (Amersham, Arlington Heights, IL) and sequenced using nucleotide dispensation tips and Pyro Gold reagents on a HSQ96 Pyrosequencer (Biotage, Uppsala, Sweden). The sensitivity of the pyrosequencing method was 5% mutation-bearing BCR-ABL transcripts.
Results
Patient characteristics
Table 1 summarizes the patients' characteristics. Their median age was 51 years (range, 17-96 years). Sixty-nine patients (62%) had received prior therapy with interferon-alpha before the start of imatinib and 43 (38%) had received imatinib as their first-line therapy for CML. At the start of imatinib, 88 (79%) patients were in CP, 18 (16%) in AP, and 6 (5%) in BP (3 myeloid and 3 lymphoid). Best response to imatinib was CHR in 58 (52%) patients and major cytogenetic response in 41 (37%; complete in 29 and partial in 11). Patients had received imatinib for a median of 32 months (range, 2-70 months) by the time they developed failure, and have been followed for a median of 64 months (range, 16-89 months) from the start of therapy with imatinib. Ninety-six percent of all patients were resistant to imatinib and 4% were intolerant (2 patients who lost their response after imatinib dose reduction due to side effects expressed KD mutations).
Patient characteristics, N = 112
| Parameter . | Median [range] or no. (%) . |
|---|---|
| Age, y | 51 [17-96] |
| Follow-up, mo | 64 [2-70] |
| Therapy before imatinib | |
| Interferon alpha | 69 (62) |
| None | 43 (38) |
| Stage at start of imatinib therapy | |
| CP | 88 (79) |
| AP | 18 (16) |
| BP | 6 (5) |
| Best response to imatinib | |
| CHR | 58 (52) |
| MCyR | 41 (37) |
| CCyR | 29 (26) |
| Duration of response, mo | 24 [2-68] |
| Time on imatinib, mo | 32 [2-70] |
| Mutation at imatinib failure | 61 (54) |
| Imatinib failure | |
| Resistance | 107 (96) |
| Primary resistance | 2 (2) |
| Secondary resistance | 105 (94) |
| Cytogenetic resistance | 29 (26) |
| Hematologic resistance | 76 (68) |
| Intolerance | 5 (4) |
| Follow-up after 2nd TKI, mo | 17 [4-31] |
| Stage at 2nd TKI | |
| CP | 38 (34) |
| AP | 54 (48) |
| BP | 20 (18) |
| 2nd TKI | |
| Dasatinib | 56 (50) |
| Nilotinib | 54 (48) |
| Bosutinib | 2 (2) |
| Stage at 3rd TKI | |
| CP | 5 (28) |
| AP | 9 (50) |
| BP | 4 (22) |
| 3rd TKI | |
| Dasatinib | 15 (83) |
| Nilotinib | 3 (17) |
| Parameter . | Median [range] or no. (%) . |
|---|---|
| Age, y | 51 [17-96] |
| Follow-up, mo | 64 [2-70] |
| Therapy before imatinib | |
| Interferon alpha | 69 (62) |
| None | 43 (38) |
| Stage at start of imatinib therapy | |
| CP | 88 (79) |
| AP | 18 (16) |
| BP | 6 (5) |
| Best response to imatinib | |
| CHR | 58 (52) |
| MCyR | 41 (37) |
| CCyR | 29 (26) |
| Duration of response, mo | 24 [2-68] |
| Time on imatinib, mo | 32 [2-70] |
| Mutation at imatinib failure | 61 (54) |
| Imatinib failure | |
| Resistance | 107 (96) |
| Primary resistance | 2 (2) |
| Secondary resistance | 105 (94) |
| Cytogenetic resistance | 29 (26) |
| Hematologic resistance | 76 (68) |
| Intolerance | 5 (4) |
| Follow-up after 2nd TKI, mo | 17 [4-31] |
| Stage at 2nd TKI | |
| CP | 38 (34) |
| AP | 54 (48) |
| BP | 20 (18) |
| 2nd TKI | |
| Dasatinib | 56 (50) |
| Nilotinib | 54 (48) |
| Bosutinib | 2 (2) |
| Stage at 3rd TKI | |
| CP | 5 (28) |
| AP | 9 (50) |
| BP | 4 (22) |
| 3rd TKI | |
| Dasatinib | 15 (83) |
| Nilotinib | 3 (17) |
CP indicates chronic phase; AP, accelerated phase; BP, blastic phase; CHR, complete hematologic response; MCyR, major cytogenetic response; and CCyR, complete cytogenetic response.
By the time therapy with the second TKI was initiated, 50 patients treated with imatinib while in CP had transformed to AP or BP, while one third of the patients still remained in CP. Fifty-four patients (13 CP, 31 AP, and 10 BP [8 myeloid and 2 lymphoid]) received nilotinib and 56 (24 CP, 22 AP, and 10 BP [7 myeloid and 3 lymphoid]) received dasatinib after developing failure to imatinib therapy. Their median follow-up was 18 months (range, 8-29 months) and 16 months (range, 4-31 months), respectively. Only 2 patients (CP and AP, one each) received bosutinib (SKI-606), another dual Src/Abl inhibitor,25 as their first salvage TKI. Among the 18 patients (5 CP, 9 AP, and 4 myeloid BP) treated sequentially with 3 TKIs, most were in the advanced phase of CML; 15 of them received dasatinib after having failed imatinib and nilotinib.
Response to therapy with a second and third TKI
The pattern of response to therapy with a second TKI is shown in Table 2. With a median follow-up of 17 months from the start of second TKI, the hematologic response rate was 82% (95% CI: 66-92) for patients in CP (79% CHR, 95% CI: 63-90), 78% (95% CI: 64-88) for patients in AP (74% CHR, 95% CI: 60-85), and 55% (95% CI: 32-77) for patients in BP (45% CHR, 95% CI: 23-68). Fifty-five percent (95% CI: 38-71) of patients treated in CP after imatinib failure achieved a cytogenetic response (complete in 26% and partial in 16%), while the cytogenetic response rate for patients treated in AP or BP was 33% (95% CI: 21-48) and 35% (95% CI: 15-59), respectively. Response rates to dasatinib and nilotinib were in line with what has been previously reported for these agents and were not statistically different between them. Best responses for patients treated with bosutinib were partial hematologic response for the patient in CP, and minor cytogenetic response for the one in AP. Among the 18 patients treated with a third TKI, 61% of patients responded to therapy, 39% achieving a cytogenetic response.
Patterns of response to therapy of imatinib-resistant CML
| . | CP . | AP . | BP/ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | Dasatinib . | Nilotinib . | P . | Total . | Dasatinib . | Nilotinib . | P . | Total . | Dasatinib† . | Nilotinib† . | |
| Response* | n=38 | n=24 | n=13 | n=54 | n=22 | n=31 | n=20 | n=10 | n=10 | ||
| Hematologic | 31 (82) | 21 (87) | 10 (77) | .64 | 42 (78) | 16 (73) | 26 (84) | .49 | 11 (55) | 6 (60) | 5 (50) |
| CHR | 30 (79) | 20 (84) | 10 (77) | .68 | 40 (74) | 14 (64) | 26 (84) | .11 | 9 (45) | 4 (40) | 5 (50) |
| Cytogenetic | 21 (55) | 15 (62) | 6 (46) | .49 | 18 (33) | 5 (23) | 13 (42) | .24 | 7 (35) | 4 (40) | 3 (30) |
| Complete | 10 (26) | 8 (32) | 2 (15) | .44 | 11 (20) | 3 (14) | 8 (26) | .33 | 7 (35) | 4 (40) | 3 (30) |
| Major | 16 (42) | 12 (50) | 4 (34) | .31 | 14 (26) | 4 (19) | 10 (33) | .35 | 7 (35) | 4 (40) | 3 (30) |
| . | CP . | AP . | BP/ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | Dasatinib . | Nilotinib . | P . | Total . | Dasatinib . | Nilotinib . | P . | Total . | Dasatinib† . | Nilotinib† . | |
| Response* | n=38 | n=24 | n=13 | n=54 | n=22 | n=31 | n=20 | n=10 | n=10 | ||
| Hematologic | 31 (82) | 21 (87) | 10 (77) | .64 | 42 (78) | 16 (73) | 26 (84) | .49 | 11 (55) | 6 (60) | 5 (50) |
| CHR | 30 (79) | 20 (84) | 10 (77) | .68 | 40 (74) | 14 (64) | 26 (84) | .11 | 9 (45) | 4 (40) | 5 (50) |
| Cytogenetic | 21 (55) | 15 (62) | 6 (46) | .49 | 18 (33) | 5 (23) | 13 (42) | .24 | 7 (35) | 4 (40) | 3 (30) |
| Complete | 10 (26) | 8 (32) | 2 (15) | .44 | 11 (20) | 3 (14) | 8 (26) | .33 | 7 (35) | 4 (40) | 3 (30) |
| Major | 16 (42) | 12 (50) | 4 (34) | .31 | 14 (26) | 4 (19) | 10 (33) | .35 | 7 (35) | 4 (40) | 3 (30) |
Data are number (percent in all but the P columns. For BP/ALL, all P values were .999.
CP indicates chronic phase; AP, accelerated phase; BP, blastic phase; ALL, Ph-positive acute lymphocytic leukemia; and CHR, complete hematologic response.
Two patients were treated with bosutinib: one in CP achieved partial hematologic response and one in AP achieved a minor cytogenetic response.
Eight patients with myeloid blast phase and 2 patients with lymphoid blast phase received nilotinib; 7 patients with myeloid blast phase and 3 patients with lymphoid blast phase received dasatinib.
Response rates, both hematologic and cytogenetic, were similar for patients without or with KD mutations, and were largely independent of mutation site. An exception was for patients with T315I of whom only 2 patients had a transient hematologic response (Table 3). There was no significant difference in the probability of achieving a CGCR between cohort with or without mutation (P = .11). The probability of achieving a CGCR at 6 and 12 months of therapy from the TKI switch was 26% (95% CI: 13-39) and 32% (95% CI: 19-46), respectively, for patients with mutations and 18% (95% CI: 6-30) and 28% (95% CI: 12-44), respectively, for those not harboring any mutation. The progression-free survival after a second TKI was similar whether patients had mutations or not (data not shown).
Patterns of clinical response to second TKI by KD mutation site
| Mutation type . | No. . | % Response . | ||
|---|---|---|---|---|
| Hematologic . | Cytogenetic . | |||
| Any . | Complete . | |||
| None | 51 | 78 | 43 | 27 |
| Any mutation site | 61 | 74 | 39 | 33 |
| P-loop | 19 | 84 | 47 | 34 |
| T315I | 10 | 20 | 0 | 0 |
| Other | 32 | 84 | 47 | 41 |
| Mutation type . | No. . | % Response . | ||
|---|---|---|---|---|
| Hematologic . | Cytogenetic . | |||
| Any . | Complete . | |||
| None | 51 | 78 | 43 | 27 |
| Any mutation site | 61 | 74 | 39 | 33 |
| P-loop | 19 | 84 | 47 | 34 |
| T315I | 10 | 20 | 0 | 0 |
| Other | 32 | 84 | 47 | 41 |
Mutation status after therapy with TKIs
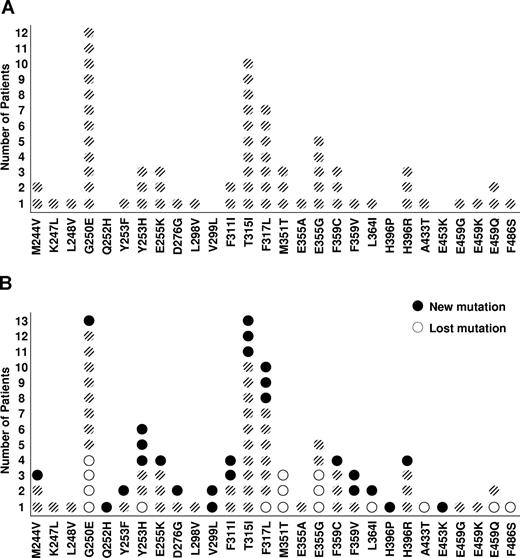
Sixty-seven mutations were identified in 61 patients with imatinib failure after a median of 32 months (range, 2-70 months) from the start of imatinib therapy and before the start of therapy with a second TKI. Five patients harbored more than one mutation (4 harboring 2 and 1 harboring 3). G250E was the most frequent mutation, present in 12 (20%) patients. As a group, P-loop mutations, defined as mutations in amino acids 248 to 255,22 accounted for 30% of the mutations. T315I was present in 10 patients, representing 15% of all mutations (Figure 1A).
The localization of mutations in the BCR-ABL kinase domain in imatinib-resistant CML. (A) Prior to starting a second tyrosine kinase inhibitor treatment. (B) Following treatment with a second tyrosine kinase inhibitor.
The localization of mutations in the BCR-ABL kinase domain in imatinib-resistant CML. (A) Prior to starting a second tyrosine kinase inhibitor treatment. (B) Following treatment with a second tyrosine kinase inhibitor.
After treatment with a second TKI, 11 patients showed regression of a KD mutation present before the start of therapy (1 patient lost 2 pre-existing mutations), most frequently G250E, M351T, and E355G. In contrast, 24 patients acquired a new KD mutation while on therapy with the new TKI (two patients acquired 2 new mutations) after a median of 9 months (range, 2-23 months) from the start of therapy with the second TKI resulting in a net gain of 10 mutations (Table 4). The rate of developing a new mutation after TKI switch was constant over time. Prior therapy with interferon-alpha did not impact the risk of developing second mutations (P = .23). Mutations most frequently involved codons 253, 317, and 359 (3 each). Overall, 6 new P-loop mutations were identified (Figure 1B). Thus, after a second TKI, 77 KD mutations were identified, up from 67 before therapy with the second TKI. Only 3 patients acquired a T315I mutation while on therapy with a second TKI.
Shift in overall distribution of mutations following sequential TKI therapies
| Mutation . | Amino acid change . | Nucleotide change (cDNA)* . | After imatinib, n = 67 . | After 2nd TKI, n = 77 . | After 3rd TKI . |
|---|---|---|---|---|---|
| M244V | Met to Val | c.1094A>G | 2 | 3 | — |
| K247L | Lys to Leu | c.1103A>G | 1 | 1 | — |
| L248V | Leu to Val | c.1106C>G | 1 | 1 | — |
| G250E | Gly to Glu | c.1113G>A | 12 | 9 | 2 |
| Q252H | Gln to His | c.1120G>T | — | 1 | — |
| Y253F | Tyr to Phe | c.1122A>T | 1 | 2 | — |
| Y253H | Tyr to His | c.1121T>C | 3 | 5 | — |
| E255K | Glu to Lys | c.1127G>A | 3 | 4 | 1 |
| D276G | Asp to Gly | c.1191A>G | 1 | 2 | — |
| L298V | Leu to Val | c.[1256C>G; 1258A>G] | 1 | 1 | — |
| V299L | Val to Leu | c.1259G>C/T | — | 2 | 1 |
| F311I | Phe to Ile | c.1294T>A | 2 | 4 | — |
| T315I | Thr to Ile | c.1308C>T | 10 | 13 | 2 |
| F317L | Phe to Leu | c.1315C>G/A; c.1313T>C | 7 | 9 | 3 |
| M351T | Met to Thr | c.1416T>C | 3 | — | — |
| E355A | Glu to Ala | c.1428A>C | 1 | 1 | — |
| E355G | Glu to Gly | c.1428A>G | 5 | 2 | 1 |
| F359C | Phe to Cys | c.1440T>G | 3 | 4 | 1 |
| F359V | Phe to Val | c.1439T>G | 1 | 3 | — |
| L364I | Leu to Ile | c.1455C>A | 1 | 1 | — |
| H396R | His to Arg | c.1551A>G | 3 | 4 | 2 |
| H396P | His to Pro | c.1551A>C | — | 1 | — |
| A433T | Ala to Thr | c.1658G>A | 1 | — | — |
| E453K | Glu to Lys | c.1721G>A | — | 1 | — |
| E459G | Glu to Gly | c.1740A>G | 1 | 1 | — |
| E459K | Glu to Lys | c.1739G>A | 1 | 1 | — |
| E459Q | Glu to Gln | c.1739G>C | 2 | 1 | — |
| F486S | Phe to Ser | c.1821T>C | 1 | — | — |
| T495R | Thr to Arg | c.1848C>G | — | — | 1 |
| Mutation . | Amino acid change . | Nucleotide change (cDNA)* . | After imatinib, n = 67 . | After 2nd TKI, n = 77 . | After 3rd TKI . |
|---|---|---|---|---|---|
| M244V | Met to Val | c.1094A>G | 2 | 3 | — |
| K247L | Lys to Leu | c.1103A>G | 1 | 1 | — |
| L248V | Leu to Val | c.1106C>G | 1 | 1 | — |
| G250E | Gly to Glu | c.1113G>A | 12 | 9 | 2 |
| Q252H | Gln to His | c.1120G>T | — | 1 | — |
| Y253F | Tyr to Phe | c.1122A>T | 1 | 2 | — |
| Y253H | Tyr to His | c.1121T>C | 3 | 5 | — |
| E255K | Glu to Lys | c.1127G>A | 3 | 4 | 1 |
| D276G | Asp to Gly | c.1191A>G | 1 | 2 | — |
| L298V | Leu to Val | c.[1256C>G; 1258A>G] | 1 | 1 | — |
| V299L | Val to Leu | c.1259G>C/T | — | 2 | 1 |
| F311I | Phe to Ile | c.1294T>A | 2 | 4 | — |
| T315I | Thr to Ile | c.1308C>T | 10 | 13 | 2 |
| F317L | Phe to Leu | c.1315C>G/A; c.1313T>C | 7 | 9 | 3 |
| M351T | Met to Thr | c.1416T>C | 3 | — | — |
| E355A | Glu to Ala | c.1428A>C | 1 | 1 | — |
| E355G | Glu to Gly | c.1428A>G | 5 | 2 | 1 |
| F359C | Phe to Cys | c.1440T>G | 3 | 4 | 1 |
| F359V | Phe to Val | c.1439T>G | 1 | 3 | — |
| L364I | Leu to Ile | c.1455C>A | 1 | 1 | — |
| H396R | His to Arg | c.1551A>G | 3 | 4 | 2 |
| H396P | His to Pro | c.1551A>C | — | 1 | — |
| A433T | Ala to Thr | c.1658G>A | 1 | — | — |
| E453K | Glu to Lys | c.1721G>A | — | 1 | — |
| E459G | Glu to Gly | c.1740A>G | 1 | 1 | — |
| E459K | Glu to Lys | c.1739G>A | 1 | 1 | — |
| E459Q | Glu to Gln | c.1739G>C | 2 | 1 | — |
| F486S | Phe to Ser | c.1821T>C | 1 | — | — |
| T495R | Thr to Arg | c.1848C>G | — | — | 1 |
TKI indicates tyrosine kinase inhibitor; and —, no data.
Nucleotide positions in coding DNA (c), according to GenBank accession number M14752.
The distribution of mutations by therapy for all patients is summarized in Figure 2. Of the 56 patients treated with dasatinib as second TKI, 8 (14%) developed new mutations: 5 in the mutation-naive group (one of them developed a new T315I mutation) and 3 among those who already had a mutation. Of 54 patients treated with nilotinib, 15 (28%) developed new mutations: 11 in the mutation-naive group, and 4 in patients with previous KD mutations (one of them developed a new T315I mutation). One patient treated with bosutinib developed a new T315I mutation.
Summary of the overall effects of particular second kinase inhibitors on the development and regression of BCR-ABL kinase domain mutations. After treatment with second TKI, 2 patients treated with nilotinib lost baseline mutations [(M35IT) and (G250E + F317L)] but acquired new mutations [(F359V) and (T315I + Y253H)], respectively; 1 patient treated with dasatinib lost baseline mutations (E459G + M351T + G250E) and acquired a new mutation (F317L). After treatment with a third TKI, 2 patients treated with dasatinib lost baseline mutations [(F311I + E453K) and (F359V)] but acquired new mutations [(F317L) and (V299L)], respectively.
Summary of the overall effects of particular second kinase inhibitors on the development and regression of BCR-ABL kinase domain mutations. After treatment with second TKI, 2 patients treated with nilotinib lost baseline mutations [(M35IT) and (G250E + F317L)] but acquired new mutations [(F359V) and (T315I + Y253H)], respectively; 1 patient treated with dasatinib lost baseline mutations (E459G + M351T + G250E) and acquired a new mutation (F317L). After treatment with a third TKI, 2 patients treated with dasatinib lost baseline mutations [(F311I + E453K) and (F359V)] but acquired new mutations [(F317L) and (V299L)], respectively.
Eighteen patients received a third TKI: 15 received dasatinib and 3, nilotinib. After treatment with a third TKI, 5 of the 18 patients acquired a new mutation, after a median of 4 months (range, 2-8 months) from the start of therapy with the third TKI: 2 at codon 317, and one each in codons 299, 315, and 495. Four patients lost pre-existing mutations. Four of the 15 patients treated with dasatinib and 1 of the 3 treated with nilotinib developed new mutations. The patient treated with nilotinib developed a new T315I mutation.
Correlation of mutation status with clinical resistance
Among the 29 patients who developed new mutations while on therapy with a second or third TKI, 24 had clinical evidence of resistance, including 5 who never responded and 19 who lost their response after a median of 11 months on therapy (range, 4 to 24 months). In 5 (17%) patients, responses were sustained for a median of 18+ months (range, 6+ to 24+ months) even in the presence of emerging mutations (2 on dasatinib who acquired a second mutation, 2 on nilotinib, 1 on bosutinib). Among these 5, those who acquired V299L, T495R, or F359C achieved only a CHR, 1 who gained H396R achieved a subsequent CGCR, and 1 who gained T315I had sustained minor cytogenetic response. Among 33 patients who had no detectable mutations throughout their therapy, 19 (58%) were resistant to a second or third TKI, including 5 who never responded.
Outcome was correlated with specific KD mutations present after sequential TKI therapy, including those that were acquired and those that persisted. Failure on dasatinib therapy was preferentially associated with mutations at codons 299 and 317. By contrast, in patients with nilotinib failure, the most frequent mutations were in the P-loop, particularly codons 253 and 255, as well as codons 311 and 359 (Figure 3). T315I and F359V developed after treatment with bosutinib.
Summary of the spectrum and frequency of BCR-ABL kinase domain that developed during treatment with a particular tyrosine kinase inhibitor. The solid color corresponds to the first amino acid change; the broken color corresponds to the second amino acid change if applicable.
Summary of the spectrum and frequency of BCR-ABL kinase domain that developed during treatment with a particular tyrosine kinase inhibitor. The solid color corresponds to the first amino acid change; the broken color corresponds to the second amino acid change if applicable.
Discussion
The availability of second-generation TKIs has provided new therapeutic options for patients with imatinib resistance. These agents have in vitro activity against imatinib-resistant kinase domain mutants, except T315I.15,16 Experiments exposing BCR-ABL–expressing cells to nilotinib or dasatinib under conditions that favor the development of mutations have shown that although their mutagenic potential is lower than that of imatinib, mutations may still emerge. These induced mutations included T315I, and novel mutations such as T315A or V299L that have never or only rarely been previously seen after imatinib exposure, including some that may not confer resistance to imatinib at all.17-19 As these agents are increasingly used, one concern is whether these new mutations may emerge in vivo and/or whether the incidence of T315I may increase.20,21
Acquisition of kinase domain mutations is the most frequently identified mechanism of resistance to imatinib. This was also true for patients in this study who had clinically resistant disease after receiving a second-generation TKI. The codons involved were diverse, and most frequently included those also seen after imatinib failure. However, some KD mutations occurred more frequently after specific second TKIs, such as F317L after dasatinib, and certain P-loop mutations after nilotinib. Few novel mutations also emerged, such as V299L after dasatinib.
We compared the spectrum of KD mutations that emerged following therapy with second TKIs to those that developed in previously published in vitro mutagenesis models.17-19 More than half of the KD mutations detected in our patient population were among those predicted from the in vitro models, representing 72% to 75% of all instances. One notable exception was T315I, which emerged in this study in only 3 patients treated with dasatinib or nilotinib (Table 5). More than half of the mutations emerging during nilotinib therapy were observed in the P-loop domain, and others included codons 311 and 359.18,19 These mutations were also responsible for nilotinib failure in 7 of 8 patients in a previous report.26 Half of the mutations encountered after therapy with dasatinib were those seen with in vitro exposure to intermediate and high dasatinib concentrations.17-19 These mutations are predicted to be contact residues that would impair dasatinib binding. F317L/V mutations have been reported in another series to be associated with dasatinib failure.27 V299L mutation, also involving a contact point, was reported to be responsible for failure in in vitro models,17,19 and was reported in 4 of 15 patients who failed dasatinib.21 This is a mutation rarely encountered with imatinib therapy.7 The mutations emerging after nilotinib and dasatinib also correlate to some extent with those that have the least in vitro sensitivity to these agents.14 Understanding the mechanisms for decreased sensitivity of these mutations and how they emerge after therapy could lead to rationale ways to select between different TKIs, and to develop novel agents.
The relationship of BCR-ABL KD mutations developing in patients compared with those predicted from in vitro treatment with TKIs
| . | In vitro models* . |
|---|---|
| Dasatinib, n=12 | |
| G250E (1) | – |
| V299L (3) | + |
| T315I (1) | + |
| F317L (5) | + |
| L364I (1) | – |
| T495R (1) | – |
| Correlation | 75% of patients; 50% of mutations |
| Nilotinib, n=18 | |
| M244V (1) | – |
| Q252H (1) | + |
| Y253H/F (4) | + |
| E255K (1) | + |
| D276G (1) | – |
| F311I (2) | + |
| T315I (2) | + |
| F359C (1) | + |
| F359V (2) | + |
| H396P (1) | – |
| H396R (1) | – |
| E453K (1) | – |
| Correlation | 72% of patients; 58% of mutations |
| . | In vitro models* . |
|---|---|
| Dasatinib, n=12 | |
| G250E (1) | – |
| V299L (3) | + |
| T315I (1) | + |
| F317L (5) | + |
| L364I (1) | – |
| T495R (1) | – |
| Correlation | 75% of patients; 50% of mutations |
| Nilotinib, n=18 | |
| M244V (1) | – |
| Q252H (1) | + |
| Y253H/F (4) | + |
| E255K (1) | + |
| D276G (1) | – |
| F311I (2) | + |
| T315I (2) | + |
| F359C (1) | + |
| F359V (2) | + |
| H396P (1) | – |
| H396R (1) | – |
| E453K (1) | – |
| Correlation | 72% of patients; 58% of mutations |
– indicates not induced in vitro; +, induced in vitro.
Despite the concern about the possible selection of T315I because this mutation is insensitive to second-generation TKIs and can be induced in vitro,15,16 we did not observe in this study a significant increase in the incidence of this mutation after the sequential use of TKI. This mutation was observed in 10 patients after imatinib failure (4 CP, 5 AP, and 1 BP), and 4 after second or third TKI: 1 after dasatinib, 2 after nilotinib, and 1 after bosutinib. The latter patient surprisingly achieved a minor cytogenetic response (Ph-positive metaphases, 40%) that was ongoing at the time of this report (6+ months). This is in contrast with other recent reports.20,21 In one series, T315I was detected after dasatinib failure in 12 (80%) of 15 patients.21 In a second series, T315I was detected after dasatinib failure in 8 patients.20 In addition, T315I mutation was detected in 2 of 147 mutation-naive patients treated with dasatinib and was responsible for treatment failure.28 In these 3 series, T315I occurred mostly in patients with lymphoid blast phase CML or Ph+ acute lymphoblastic leukemia (ALL). In our series, few patients had lymphoid blast crisis at the time of failure, likely explaining the lower rate of T315I detection. In addition, the possibility of an increase in occurrence of this mutation with further follow-up of our patients needs to be considered a possibility.
In conclusion, the types of mutations that occur with persistent/recurrent disease after treatment with sequential TKIs are diverse, including those seen in patients resistant to imatinib therapy and others that are novel and only seen after introduction of a new TKI. Overall frequency of mutation appears to increase with sequential TKI use and the mutations observed mostly coincide with what would be predicted from in vitro mutagenesis studies, except that an increase in the frequency of T315I is not observed. Acquisition of new KD mutations is usually associated with therapy resistance, but treatment failure can occur in many instances in the absence of KD mutations, emphasizing the complexity of the mechanisms of resistance to TKIs.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by Leukemia Specialized Program of Research Excellence [SPORE] grant P50CA100632.
Authorship
Contribution: J.C. and E.J. wrote the paper, analyzed data, and approved the paper; H.K., S.O., and F.G. analyzed data and approved the paper; C.C.Y. performed sequencing and analyzed data; J.S. performed statistical analysis; G.G.-M., M.B., and N.R. analyzed data; W.G.W. approved the paper; D.J. wrote the paper, performed the sequencing, analyzed data, and approved the paper. J.C. and E.J. contributed equally to the project.
Conflict-of-interest disclosure: J.C. and H.K. have received research grants from Novartis and BMS. F.G. has received research grants from Novartis. All other authors declare no competing financial interests.
Correspondence: Jorge Cortes, Professor of Medicine, Department of Leukemia, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Box 428, Houston, TX 77030; e-mail: jcortes@mdanderson.org.

![Figure 2. Summary of the overall effects of particular second kinase inhibitors on the development and regression of BCR-ABL kinase domain mutations. After treatment with second TKI, 2 patients treated with nilotinib lost baseline mutations [(M35IT) and (G250E + F317L)] but acquired new mutations [(F359V) and (T315I + Y253H)], respectively; 1 patient treated with dasatinib lost baseline mutations (E459G + M351T + G250E) and acquired a new mutation (F317L). After treatment with a third TKI, 2 patients treated with dasatinib lost baseline mutations [(F311I + E453K) and (F359V)] but acquired new mutations [(F317L) and (V299L)], respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-03-080838/4/m_zh80010810660002.jpeg?Expires=1769110596&Signature=ufsSxsEQNJ0XljKCIHFm8GcKIHv3kgp7FVzuCaWNdYjrukyv-71HMvdmLU~O18gdWkZEnvHLrnQgCfj5Fw9Hdw3ws~qARiXSffU0UoiGdBScLirRN3MIJ4dmF9hD8LWN4aGQDInwq6mHvwifNXc43ysBMvUmRtrFNuFzDkSkG5~GncJChoseOdQgBboGW1TIRds6LqympJUSUjafIfrKlPcyHw0DUwBpKuGEU8~GIbXxZ-3z~fgftiiz9IuSobb0ddEsBDH-902QKeBB4ceVwDfMufR8QKmrS1aHsVKQJINeBoNFhHTxH7w5bdZnWa3lu4i4CYBTpV~L1QO67MdG-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
