Abstract
Background: Imatinib resistance (IR) is a well known cause of treatment failure in chronic myeloid leukemia (CML) patients being treated with imatinib mesylate (IM). Several cellular and genetic mechanisms of IR have been proposed including amplification and overexpression of the BCR-ABL gene, the presence of specific point mutations and MDR1 gene overexpression.
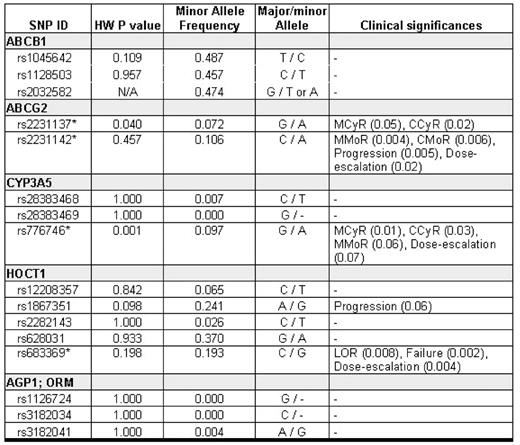
Methods: We investigated the impact of 16 single nucleotide polymorphisms (SNPs) in 5 genes potentially associated with pharmacogenetics of IM (ABCB1, multidrug resistance 1; ABCG2, breast-cancer resistance protein; CYP3A5, cytochrome P450 3A5; HOCT1, human organic cation transporter 1; AGP1, alpha-1-acid glycoprotein-1, plasma protein binding to IM). The major endpoints included:
response to IM: cytogenetic (CyR) or molecular response (MoR);
resistance to IM: loss of response (LOR), treatment-failure (including primary resistance or LOR);
progression to accelerated phase (AP) or blast crisis (BC), or death; and
need for IM dose escalation to overcome resistance or LOR. The DNAs from peripheral blood samples were genotyped using the Sequenom MassARRAY system based on MALDI-TOF technique.
The study population included 229 patients whose clinical outcomes following IM therapy were evaluated from January 2000 to January 2007 (male:female 96:133; median age at start of IM, 53 years-old; white:non-white 170:59; chronic phase:AP:BC, 199:23:3).
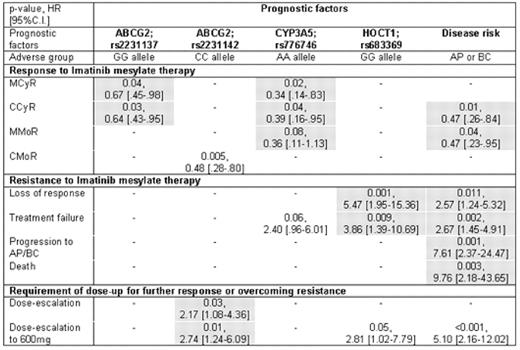
Results: The frequencies of genotypes in 16 SNPs are summarized in Table 1. The GG allele in ABCG2 (rs2231137), AA allele in CYP3A5 (rs776746) and advanced stage were significantly associated with poor response to IM, while GG allele at HOCT1 (rs683369) and advanced stage correlated with high rate of LOR or treatment failure. The CC allele in ABCG2 (rs2231142) was also identified as an independent predictor of more frequent need for IM dose escalation. The results of multivariate analyses are summarized in Table 2.
Conclusion: Using a novel, multiple candidate gene approach based on the pharmacogenetics of Imatinib mesylate, we identified several SNP candidates in patients with CML that are potential predictors of clinical response to IM (rs2231137, ABCG2 or rs776746, CYP3A5), resistance to IM (rs683369, HOCT1) and need for IM dose escalation (rs2231142, ABCG2). Further studies are warranted to validate the role of these SNPs in the early identification of individuals with CML who may not respond optimally to standard IM therapy.
Multivariate regression model based on Cox's proportional hazard model for each-point
Author notes
Disclosure: No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal