Abstract
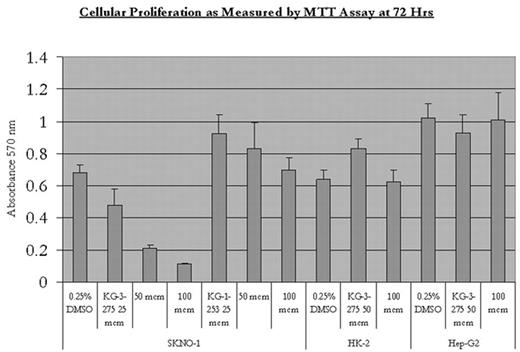
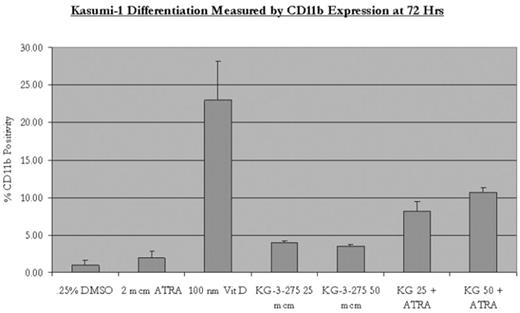
The protein-protein interaction between the subunits of the nuclear transcription factor core binding factor, RUNX1 (CBFα) and CBFβ, plays a critical role in hematopoiesis. Chromosomal rearrangements that target CBF genes are among the most common mutations in leukemia, including t(8;21) found in approximately 12% of cases of AML and resulting in the protein AML1-ETO. We describe the development of small molecule inhibitors of the interaction between RUNX1 and CBFβ. We used virtual screening to identify lead compounds. These leads were used to generate a library of compounds to explore the structure-activity relationships and optimize activity, resulting in the identification of low micromolar IC50 inhibitors of this protein-protein interaction. We confirmed by FACS FRET that inhibition also occurs in mammalian cells. HEK-293 cells were transfected with Cerulean-Runt domain and Venus-CBFβ. FRET emission from cells was monitored on a flow cytometer to assess binding of CBFβ and the Runt domain. Results with drug were compared to FRET between Runt and wild-type CBFβ and FRET between Runt and a mutant CBFβ with 2 point mutations abrogating binding to Runt. The small molecule inhibitor KG-3-275 inhibited CBFβ - Runt domain binding in a dose dependent manner (Table 1). Treatment of the t(8;21) leukemia cell lines Kasumi-1 and SKNO-1 results in inhibition of proliferation. SKNO-1 cells are inhibited in a dose-dependent manner by KG-3-275 but not by the weak inhibitor KG-1-253 (Fig 1). KG-3-275 does not inhibit the growth of renal tubular and hepatocellular cell lines (Fig 1). Kasumi-1 and SKNO-1 undergo apoptosis in a dose-dependent manner upon treatment with KG-3-275 (Table 2). Finally, KG-3-275 shows synergy with ATRA in increasing differentiation of Kasumi-1 cells as measured by CD11b expression, consistent with recent published results establishing a link between AML1-ETO and repression of RAR signaling (Fig 2). These data indicate drugs inhibiting CBF interactions hold promise as targeted agents in the treatment of CBF leukemias.
Fluorescence Resonance Energy Transfer Geometric Mean in HEK-293 Cells
| Cerulean-Runt domain + Venus-CBFb | 4.07 +/− 0.1 |
| Cerulean-RD + Venus-CBFb61/104 | 1.85 +/− .09 |
| C-RD + V-CBFb + KG-3-275 200 mcm | 3.05 +/− .15 |
| C-RD + V-CBFb + KG-3-275 100 mcm | 3.25 +/− 0.1 |
| C-RD + V-CBFb + KG-3-275 50 mcm | 3.45 +/− .25 |
| C-RD + V-CBFb + KG-3-275 25 mcm | 3.6 +/− 0.2 |
| Cerulean-Runt domain + Venus-CBFb | 4.07 +/− 0.1 |
| Cerulean-RD + Venus-CBFb61/104 | 1.85 +/− .09 |
| C-RD + V-CBFb + KG-3-275 200 mcm | 3.05 +/− .15 |
| C-RD + V-CBFb + KG-3-275 100 mcm | 3.25 +/− 0.1 |
| C-RD + V-CBFb + KG-3-275 50 mcm | 3.45 +/− .25 |
| C-RD + V-CBFb + KG-3-275 25 mcm | 3.6 +/− 0.2 |
% Cells Annexin V/PI Negative at 72 Hrs
| Kasumi-1 | 0.25% DMSO | 91.2 +/− 0.9 |
| KG-3-275 25 mcm | 91.2 +/− 1.2 | |
| KG-3-275 50 mcm | 85.9 +/− 2.4 | |
| KG-3-275 100 mcm | 66.7 +/− 6.7 | |
| SKNO-1 | 0.25% DMSO | 77.6 +/− 3.4 |
| KG-3-275 25 mcm | 77.6 +/− 2.3 | |
| KG-3-275 50 mcm | 63.7 +/− 3.6 | |
| KG-3-275 100 mcm | 13.7 +/− 6.8 |
| Kasumi-1 | 0.25% DMSO | 91.2 +/− 0.9 |
| KG-3-275 25 mcm | 91.2 +/− 1.2 | |
| KG-3-275 50 mcm | 85.9 +/− 2.4 | |
| KG-3-275 100 mcm | 66.7 +/− 6.7 | |
| SKNO-1 | 0.25% DMSO | 77.6 +/− 3.4 |
| KG-3-275 25 mcm | 77.6 +/− 2.3 | |
| KG-3-275 50 mcm | 63.7 +/− 3.6 | |
| KG-3-275 100 mcm | 13.7 +/− 6.8 |
Cellular Proliferation as measured by MTTAssay at 72 Hrs
Kasumi-1 Differentiation Measured by CD11b expression at 72 Hrs
Author notes
Disclosure: No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal