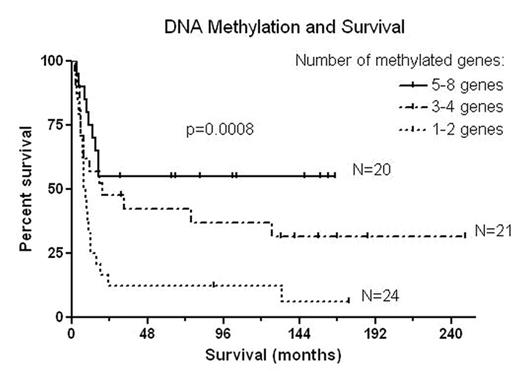
DNA methylation of CpG islands at transcriptional start sites (TSS) is an epigenetic modification that leads to permanent gene silencing and plays an important role in cancer development and progression. To better understand its significance in AML, we analyzed DNA methylation in bone marrow cells collected at diagnosis in two subsets of patients with AML distinguished by a long survival duration (group A, 33 patients, median, 90 months [range, 13–249+ months]) or short survival (group B, 32 patients, median, 7 months, [range, 2–12 months]). We quantitatively measured DNA methylation using bisulfite pyrosequencing of CpG islands at TSS of genes frequently methylated in leukemic cell lines: NOR1, CDH13, p15, NPM2, OLIG2, PGR, HIN1, and SLC26A4. We also studied FLT3-ITD and NPM1 mutations as the most frequent genetic changes in leukemia. We compared these results with cytogenetic abnormalities, clinical data, and the AML outcome, and statistically analyzed the data with nonparametric tests and the Cox multivariate analysis. The patients in group A were significantly younger (median, 46 years, [range, 18–78 years]) than those in group B (median, 61 years, [range, 17–77 years]; p=0.02). We noted no significant differences between these groups according to gender, FAB classification, blast count, WBC, Hgb, platelet count, LDH and the frequency of NPM1 mutations and FLT3-ITD. The patients in the longer living group A had significantly lower levels of β2-microglobulin, better cytogenetic classification, and less frequent deletions of chromosome 5 and/or 7 than did those in group B; p<0.01. Remarkably, the patients in group A had significantly higher levels of methylation in multiple studied genes: NOR1, NPM2, OLIG2, HIN1, and SLC26A4, p<0.05. We observed no significant differences in methylation of CDH13, p15, and PGR. The median number of genes simultaneously methylated over the threshold was significantly higher in group A than in group B (five vs. two; p=0.0009). We observed longer survival in FLT3-ITD positive patients who had 4–8 methylated genes compared to those with 1–3 methylated genes, p=0.005. Similarly, the patients with wildtype NPM1 showed a significantly longer survival if they had 4–8 methylated genes compared to those with 1–3 methylated genes, p=0.002. No significant survival differences were seen in terms of the number of methylated genes in patients with FLT3 wild-type or with NPM1 mutations. The multivariate analysis revealed deletions of chromosome 5/7 or complex karyotype, FLT3-ITD, and advanced age as negative prognostic factors, p<0.002. Surprisingly, increased methylation of NOR1, HIN1, SLC26A4 and PGRB, simultaneous methylation of multiple genes and a high methylation z score were positive prognostic factors, p<0.03. We hypothesize that frequent DNA methylation denotes a subset of patients with good prognosis where epigenetics plays an essential role. This “epigenetic leukemia” seems to be more amenable to treatment and may have greater potential for long-term survival or cure compared to AML with less DNA methylation.
DNA methylation and Survival
Disclosure:Research Funding: SPORE.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal