Abstract
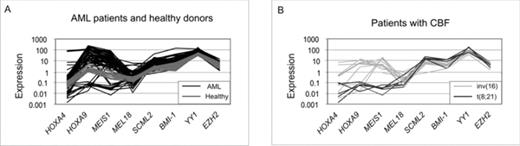
The Polycomb group (PcG) of genes is important for differentiation and X-chromosome silencing. Recently much attention has been afforded to the role of its aberrant expression in cancer, especially in relation to the inactivation of tumor suppressor genes. We hypothesized that a deregulation in the expression profile may contribute to the development of acute myeloid leukaemia (AML). To address this, we determined the RNA levels by RQ-PCR in diagnostic bone marrow samples from 126 patients and 20 healthy donors to delineate their expression profile of the PcG genes BMI-1, MEL18, SCML2, YY1 and EZH2. To address the interplay with downstream targets of PcG proteins, we also determined the expression of HOXA4, HOXA9 and MEIS1. These data were compared not only to the demographic and clinical data of the patients, but also to a large number of molecular assays already performed in these patients (Olesen LH et al. Br.J. Haematol. 2005 131(4):457–467; Rethmeier et al. Br.J. Haematol. 2006 133(3):276–283.). At first we noticed a striking heterogeneity in the expression profiles of the AML patients (Fig. 1). We also observed that HOXA9, MEIS1, SCML2, YY1, BMI-1 and EZH2 were significantly (p≤0.003) higher expressed in the patients compared to the healthy donors. Moreover, when patients were analyzed according to the three cytogenetic prognostic groups (normal, core-binding factor positive and complex), the expression profile of patients with the t(8,21) aberration was characterized by a significantly decreased expression of HOXA9 and MEIS1 and a higher one of SCML2, YY1 and BMI-1 than AML patients in general (p<0.003). When evaluating the impact of cytogenetic subgrouping, the expression levels of MEL18 and EZH2 significantly (p< 0.025) reflected highest expression in patients with adverse prognosis and lowest expression with patients exhibiting the most favourable prognosis. While the expression levels of the genes in focus did not correlate to course of disease, we observed that a direct relationship between transcript levels of PcG and PcG-related on the one hand and the DNA methyl transferases (DNMT’s), apoptosis and multidrug-resistance genes (p<0.001) on the other. In conclusion, in this study, which is the first to systematically analyze a series of PcG genes and genes regulated by PcG, we failed to demonstrate a correlation to the clinical outcome of patients with AML. On the other hand, our data strongly suggest that these genes might be involved in the leukaemogenic process by virtue of their relations to DNA methylation (DNMT1, DNMT3B), apoptosis (BAX, CASPASE 3) and multidrug resistance (MDR1, MRP1). Figure 1. Expression profiles of PcG or PcG-regulated genes in AML patients and healthy controls. A. Gene expression profile of all 126 AML patients included (black lines) compared to 20 healthy donors. B. Patients with CBF aberrations, t(8,21), n =7, or inv(16), n =12. The expression is calculated as 2−ΔCt *100), where ΔCt = CtTG−CtCG, CtTG is the Ct value of the target gene, and CtCG is the mean Ct value of the two control genes (B2M and ABL).
Author notes
Disclosure: No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal