Abstract
TCR signal strength is involved in many cell fate decisions in the T-cell lineage. Here, we show that transcriptional events induced by Hedgehog (Hh) signaling reduced TCR signal strength in mice. Activation of Hh signaling in thymocytes in vivo by expression of a transgenic transcriptional-activator form of Gli2 (Gli2\#916;N2) changed the outcome of TCR ligation at many stages of thymocyte development, allowing self-reactive cells to escape clonal deletion; reducing transgenic TCR-mediated positive selection; reducing the ratio of CD4/CD8 single-positive (SP) cells; and reducing cell surface CD5 expression. In contrast, in the Shh\#8722;/\#8722; thymus the ratio of CD4/CD8 cells and both positive and negative selection of a transgenic TCR were increased, demonstrating that Shh does indeed influence TCR repertoire selection and the transition from double-positive (DP) to SP cell in a physiological situation. In peripheral T cells, Gli2\#916;N2 expression attenuated T-cell activation and proliferation, by a mechanism upstream of ERK phosphorylation.
Introduction
The Hedgehog (Hh) family of secreted intercellular signaling molecules specifies cell fate and patterning during embryonic development and is involved in homeostasis and renewal of adult tissues, including skin, gut, lung, blood, and thymus.1\#8211;9
The specialized environment of the thymus supports the maturation of lymphocyte precursors into functional T cells through stages defined by cell surface expression of developmentally regulated markers: CD4\#8722;CD8\#8722; double-negative (DN) thymocytes differentiate to CD4+CD8+ double-positive (DP) cells that become mature CD4 or CD8 single-positive (SP) T cells. The DN population can be subdivided by expression of CD44 and CD25. The earliest CD44+CD25\#8722; cells (DN1) acquire CD25 (DN2), then lose CD44 expression (DN3) and become CD44\#8722;CD25\#8722;DN (DN4) cells before differentiating to DP. The transition from DN to DP cell requires a functional pre-T-cell receptor (TCR) complex,10 whereas maturation from DP to mature SP involves positive selection of the \#945;\#946;TCR repertoire to ensure appropriate MHC restriction and negative selection of potentially self-reactive clones.11,12 This developmental program is regulated by the stroma, and one way in which the epithelium signals to developing thymocytes is by production of Sonic Hh (Shh).5
The 3 mammalian Hh proteins (Shh, Indian Hh, and Desert Hh) share a common signaling pathway. Hh proteins bind their cell surface receptor Patched, releasing the signal transducer Smoothened (Smo) to transmit the Hh signal into the cell. At the end of this pathway are the transcription factors Gli1, Gli2, and Gli3.1,2 Gli1 acts exclusively as an activator of transcription and, although not essential for mouse development or Hh signaling, is itself a Hh target gene, so measurement of its transcription can be used as a readout of Hh signaling in a given population of cells.13 Gli2 and Gli3 undergo processing to function as positive or negative regulators of transcription, depending on the presence or absence of Hh signaling, respectively.14 Both are essential for mouse development and have distinct but partially overlapping functions.15,16 Gli2 functions primarily as a transcriptional activator downstream of Hh signaling, and is necessary to initiate the first transcriptional changes induced by the Hh signal.17 In contrast, in vivo Gli3 functions primarily as a transcriptional repressor in the absence of the Hh signal.18
Analysis of Shh\#8722;/\#8722;, Gli3\#8722;/\#8722;, and Smo-deficient (conditional, T lineage) thymi has shown that Hh signaling is a positive regulator of the early stages of thymocyte development, controlling homeostasis of DN progenitors and differentiation from DN1 to DN2.5,7,8 Both the analysis of the Shh\#8722;/\#8722;5 and Gli3\#8722;/\#8722;7 thymus and in vitro experiments using wild-type mouse thymus explants4 have also suggested that Hh signaling is involved after TCR-\#946; gene rearrangement at the transition from DN to DP cell. Surprisingly, however, conditional deletion of Smo from T-lineage cells failed to show any influence of loss of Hh signaling after the DN2 stage.8 A role for Hh signaling has also been described in the activation of peripheral lymphocytes,19,20 although no defect in T-cell activation was detected in Smo-deficient lymphocytes.8
Here, we investigated the function of Hh signaling at late stages of thymocyte development and in T-cell activation. By analysis of the Shh\#8722;/\#8722; thymus, we show that Shh influences TCR repertoire selection and the transition from DP to SP. We also show that T-cell\#8211;autonomous activation of the Hh pathway in developing thymocytes inhibited TCR repertoire selection, altering differentiation to SP and the CD4/CD8 SP ratio. In addition, we demonstrate that in mature T cells the transcriptional changes induced by Hh reduce the ability of the TCR to signal for activation and proliferation, by a mechanism upstream of ERK phosphorylation.
Materials and methods
Construction of lck-Gli2\#916;N2 transgenic
Mice
lck-Gli2\#916;N2 transgene-positive mice were backcrossed for more than 7 generations with C57BL/6 (B&K Universal, Grimston, England). Gli2\#916;N2 and Shh+/\#8722; mice were crossed with HY-TCR transgenic22 and B6.21.6\#946;-TCR transgenics.23 Shh+/\#8722; mice24 were on a C57BL/6 background.5 Mice were maintained under United Kingdom Home Office regulations.
Genotyping
DNA was extracted using DNeasy tissue kit (Qiagen). Real-time polymerase chain reaction (PCR) for genomic Gli2\#916;N2 was carried out on an iCycler (Bio-Rad Laboratories, Hercules, CA) using the iQSYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions. Primers were Gli2F AGAACCTGAAGACACACCTGCG and Gli2R GAGGCATTGGAGAAGGCTTTG. To enable comparison between samples, Gli2 levels were normalized using levels of the J1.1 germ-line gene (primers were 5\#8242;J1.1GL CCTCATCCTATGGCACTG and 3\#8242;J1.1GL GCTTTGTCCGAAGAGAGAC). Embryo sex was determined as described.25
Cell sorting
Thymocytes were sorted on a MoFlo (Cytomation, Fort Collins, CO). DN1-4 populations were sorted using antibodies against CD25FITC, CD3/4/8PE, and CD44Cychrome. Staining with CD3FITC, CD4PE, and CD8Cychome allowed sorting of DPs and SPs and CD8FITC, CD4PE, and B220Cychrome allowed sorting of peripheral SPs and B cells. Cells collected fell within FSC/SSC live gate.
Quantitative RT-PCR for Gli2\#916;N2 transgene expression
RNA was extracted using Absolutely RNA Miniprep kit (Stratagene, La Jolla, CA) from fluorescence-activated cell sorter (FACS) cells. cDNA was synthesized using Superscript II (Invitrogen, Carlsbad, CA) and quantitative reverse-transcription (RT)\#8211;PCR carried out as described.7 Primers were Gli2\#916;N2F AGAACCTGAAGACACACCTGCG and Gli2\#916;N2R GAGGCATTGGAGAAGGCTTTG.
Antibodies and flow cytometry
Cells were stained as described7 using directly conjugated antibodies from BD PharMingen (San Diego, CA) and analyzed on a FACScan (BD Biosciences, Basel, Switzerland). Live cells were gated according to FSC/SSC profiles. Data are representative of more than 3 experiments.
CFSE (Sigma-Aldrich) labeling was carried out in PBS and 10 \#956;M CFSE for 10 minutes at 25°C in the dark.
DbSmcyPE tetramer (ProImmune, Oxford, United Kingdom) staining was prior to other antibody staining in PBS for 15 minutes at 25°C in the dark.
In vitro T-cell activation
Splenic T cells were cultured at 5 × 106/mL in AIM-V (Life Technologies), 10\#8722;5 M 2ME (Sigma-Aldrich) at 37°C, 5% CO2. Anti-CD3 and anti-CD28 (azide-free [NA/LE; BD PharMingen]) were at 0.01 \#956;g/mL of each, unless otherwise stated.
Fetal thymus organ culture (FTOC)
Analysis of Erk1 and Erk2 phosphorylation
Splenic T cells at 5 × 106 in OptiMEM (Invitrogen) media were activated with 1 \#956;g/mL anti-CD3\#1013; and anti-CD28 antibodies and incubated on ice for 30 minutes. For stimulation, cells were transferred to 37°C for 5, 15, 30, 45, and 60 minutes in 5% CO2. After a short spin, cells were lysed on ice for 40 minutes and analyzed by immunoblotting using anti-Erk1, anti-Erk2, anti\#8211;phospho-Erk, and anti\#8211;phospho-Erk2 (Cell Signaling, Beverly, MA) as described.26
For FACS analysis of intracellular phospho-Erk activation and staining were performed as described.27
Results
Expression of Gli1 in SP thymocytes
The Hh signaling pathway has previously been shown to be involved in regulation of thymocyte development at the transition from DN1 to DN25,7,8 and at the transition from DN to DP cell.4,5,7 Genes encoding components of the signaling pathway, including Smo and Gli1, are expressed by DN thymocytes, and anti-Smo staining and RT-PCR analysis of transcription in sorted DN subsets have both shown that their expression is highest in the DN2 subset.4,8 To assess activation status of the Hh signaling pathway at later stages of thymocyte development, we analyzed transcription of the Hh-target gene Gli1 in sorted thymocyte populations. We found that Gli1 transcription was down-regulated in the DN4 subset and DP population (Figure 1A). We did, however, detect an 11-fold increase in Gli1 transcription from the DP to CD4SP population, and a 4-fold increase from the DP to CD8SP population, indicating active Hh signaling at later stages of T-cell development, in SP cells (Figure 1A). The detection of Gli1 transcription in the CD4 and CD8 SP subsets is consistent with high cell surface expression of Smo on about 9% of CD4SP and 20% of CD8SP thymocytes.4 Shh is expressed by epithelial cells in the medulla and corticomedullary junction,4,6,8 and therefore expression of Gli1 and Smo in SP thymocytes is consistent with their location in the thymus, as like the early DN populations,28 they are located near the source of Shh.
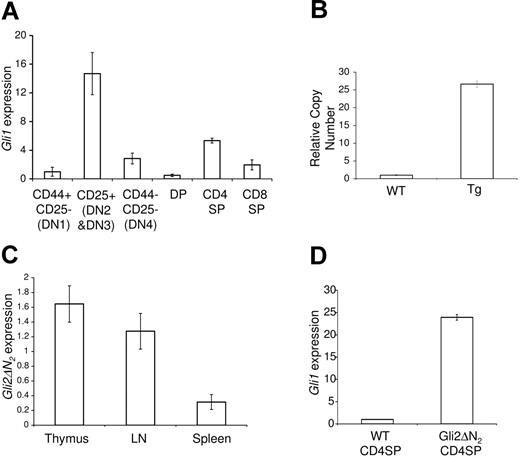
Expression of Gli1 and Gli2\#916;N2 in WT and Gli2\#916;N2 transgenic mice. (A) Hh activation status, shown by quantitative RT-PCR analysis of gli1 mRNA in FACS-sorted populations of the adult murine thymus. (B) Quantitative PCR of genomic Gli2\#916;N2, showing high copy number of the Gli2\#916;N2 transgene. (C) Relative Gli2\#916;N2 expression, by quantitative RT-PCR analysis, in thymus, lymph node, and spleen (D) and up-regulation of Gli1 transcription in Gli2\#916;N2 transgenic CD4SP thymocytes. Error bars show standard deviation (SD). Data are representative of at least 3 experiments.
Expression of Gli1 and Gli2\#916;N2 in WT and Gli2\#916;N2 transgenic mice. (A) Hh activation status, shown by quantitative RT-PCR analysis of gli1 mRNA in FACS-sorted populations of the adult murine thymus. (B) Quantitative PCR of genomic Gli2\#916;N2, showing high copy number of the Gli2\#916;N2 transgene. (C) Relative Gli2\#916;N2 expression, by quantitative RT-PCR analysis, in thymus, lymph node, and spleen (D) and up-regulation of Gli1 transcription in Gli2\#916;N2 transgenic CD4SP thymocytes. Error bars show standard deviation (SD). Data are representative of at least 3 experiments.
Transgenic Gli2 expression in thymocytes
Given that the Hh signaling pathway is active in SP thymocytes, we investigated the function of Hh signaling at later stages of thymocyte development. We made a transgenic mouse that expressed an N-terminally truncated form of Gli2 under the control of the lck promoter. This truncated form of Gli2 (Gli2\#916;N2) behaves as an activator of transcription independent of upstream effects on the pathway, thus constitutively activating Hh-dependent transcription in cells that express it,14 and allowing study of the effect of pathway activation specifically in thymocytes. Use of this lck cassette restricts transgene expression to thymocytes without affecting the thymic stroma.21 Two independent studies using the lck cassette to drive expression of a GFP reporter gene have shown transgenic expression first in the DN2 and DN3 subsets, and that full expression is achieved at DN4. Expression is maintained in DP, SP, and peripheral T-cell populations.29,30
Transgenic mice were produced and quantitative PCR of genomic DNA using primers that amplify both transgenic and genomic Gli2 gene demonstrated high copy number (Figure 1B). Quantitative RT-PCR demonstrated Gli2\#916;N2 expression in transgenic thymocytes, splenocytes, and lymph node cells, with varying expression levels corresponding to the proportion of T-lineage cells (Figure 1C). To confirm that the Gli2\#916;N2 protein was functional and that it activated transcription of Hh-target genes, we analyzed the transcription of Gli1 in sorted CD4SP thymocytes from Gli2\#916;N2 transgenic and wild-type (WT) littermate thymi. We found an approximately 20-fold increase in Gli1 transcription in transgenic thymocytes, confirming thymocyte-autonomous activation of the downstream transcriptional changes induced by Hh signaling (Figure 1D).
Hh signaling in the Gli2\#916;N2 transgenic influences the ratio of CD4/CD8 SP cells
We analyzed lymphocyte populations in Gli2\#916;N2 transgenic mice. Thymus, lymph nodes, and spleen were grossly normal, with no significant differences in lymphocyte numbers in thymus or spleen between Gli2\#916;N2 transgenic and WT littermates (Figure 2A). Analysis of CD4 and CD8 expression revealed a statistically significant reduction in the proportion of CD4SP cells and an increase in the proportion of CD8SP cells in the Gli2\#916;N2 transgenic thymus, relative to littermates. Typically, 6.8% of thymocytes were CD4SP and 2.6% were CD8SP in the Gli2\#916;N2 transgenic, compared with 9.9% and 1.7%, respectively, in WT littermates (Figure 2B), and the average ratio of CD4SP/CD8SP cells was reduced from 3.7 in WT to 2.3 in the Gli2\#916;N2 transgenic mice (Figure 2C). We also observed a significant increase in the percentage of DP cells in the Gli2\#916;N2 transgenic thymus, compared with littermates (Figure 2B). To determine if the change in CD4/CD8 ratio was genuinely the result of changes in the proportions of mature T cells rather than an increase in pre-DP immature CD8SP cells (ISPs), we analyzed CD3 and HSA expression. CD3 and HSA expression in the SP populations of Gli2\#916;N2 transgenic and WT littermate thymi were identical (Figure 2D-E), excluding an increase in the ISP population. In both spleen and lymph nodes, we found a reduction in the CD4/CD8 T-cell ratio and the overall percentage of CD4 and CD8 T cells (Figure 2B).
Phenotype of Gli2\#916;N2 transgenic mice. (A) Relative cell number (calculated relative to the mean of WT littermates) in the thymus and spleen of WT (gray bars, n = 6) and Gli2\#916;N2 transgenic (\#9633;, n = 6). (B) CD4 and CD8 profiles in the Gli2\#916;N2 transgenic mouse. Thymocytes from Gli2\#916;N2 transgenic and WT littermates were analyzed for CD4 and CD8 expression in the thymus (top panels), spleen (middle panels), and lymph node (bottom panels). Differences between the percentage of populations between transgenic and littermate were statistically significant as given: (thymus) CD4SP, P = .005; DP, P = .014; (spleen) CD4, P = .002; CD8, P = .001; and (lymph node) CD4, P = .005; CD8, P = .041. (C) Bar chart to show change in CD4/CD8 ratio in thymus (WT [n = 6], Gli2\#916;N2 [n = 6], P = .032). (D) CD3 expression on CD4 (top histograms) and CD8 (bottom histograms) SP thymocytes. (E) HSA expression on CD4 (top) and CD8 (bottom) SP thymocytes. (F) CD2 expression on DP (top), CD4SP (middle), and CD8SP (bottom) cells in the Gli2\#916;N2 transgenic (right panels) and the WT (left panels). (G) CD5 expression on CD8SP cells in the Gli2\#916;N2 transgenic thymus (gray line) compared with the WT (black line). The bar chart shows mean MFI for WT (\#9641;) and Gli2\#916;N2 transgenic (\#9633;). (H) CD69 expression on DP (top), CD4 (middle), and CD8 (bottom) SP thymocytes. Numbers in dot plots or histograms indicate the percentage of cells falling in the quadrant, region, or marker. Error bars show standard error (SE). Data are representative of at least 3 experiments.
Phenotype of Gli2\#916;N2 transgenic mice. (A) Relative cell number (calculated relative to the mean of WT littermates) in the thymus and spleen of WT (gray bars, n = 6) and Gli2\#916;N2 transgenic (\#9633;, n = 6). (B) CD4 and CD8 profiles in the Gli2\#916;N2 transgenic mouse. Thymocytes from Gli2\#916;N2 transgenic and WT littermates were analyzed for CD4 and CD8 expression in the thymus (top panels), spleen (middle panels), and lymph node (bottom panels). Differences between the percentage of populations between transgenic and littermate were statistically significant as given: (thymus) CD4SP, P = .005; DP, P = .014; (spleen) CD4, P = .002; CD8, P = .001; and (lymph node) CD4, P = .005; CD8, P = .041. (C) Bar chart to show change in CD4/CD8 ratio in thymus (WT [n = 6], Gli2\#916;N2 [n = 6], P = .032). (D) CD3 expression on CD4 (top histograms) and CD8 (bottom histograms) SP thymocytes. (E) HSA expression on CD4 (top) and CD8 (bottom) SP thymocytes. (F) CD2 expression on DP (top), CD4SP (middle), and CD8SP (bottom) cells in the Gli2\#916;N2 transgenic (right panels) and the WT (left panels). (G) CD5 expression on CD8SP cells in the Gli2\#916;N2 transgenic thymus (gray line) compared with the WT (black line). The bar chart shows mean MFI for WT (\#9641;) and Gli2\#916;N2 transgenic (\#9633;). (H) CD69 expression on DP (top), CD4 (middle), and CD8 (bottom) SP thymocytes. Numbers in dot plots or histograms indicate the percentage of cells falling in the quadrant, region, or marker. Error bars show standard error (SE). Data are representative of at least 3 experiments.
Strength of TCR signaling can influence CD4/CD8 lineage commitment in the thymus, with a strong TCR signal favoring differentiation to CD4SP.31\#8211;34 We therefore analyzed expression of cell surface molecules associated with TCR signaling in thymocytes.27 Expression of CD235 was significantly increased on the DP population in the Gli2\#916;N2 transgenic, from 59% in WT to 83% of DP cells in the Gli2\#916;N2 transgenic (Figure 2F), but was unaffected on SP populations. Expression of CD5, a molecule linked to attenuation of the TCR signal,36 was lower on DP and SP populations in the Gli2\#916;N2 transgenic compared with WT littermates (Figure 2G). In the CD8SP population, 72% stained brightly in WT compared with 45% in the Gli2\#916;N2 transgenic, and mean fluorescence intensity (MFI) was reduced from 113.9 in the WT to 80.9 in the Gli2\#916;N2 transgenic (Figure 2G). Expression of CD69, a marker associated with repertoire selection and T-cell activation, was higher on CD8SP thymocytes in the Gli2\#916;N2 transgenic, with, in a typical experiment, 74% staining positive compared with 60% in the WT (Figure 2H).
Increased differentiation to CD4SP in the Shh\#8722;/\#8722; thymus
Given that activation of the Hh signaling pathway in developing thymocytes reduced the ratio of CD4/CD8 SP thymocytes, we asked if removal of the Shh signal would have the opposing effect. Shh\#8722;/\#8722; embryos die before birth24 and have defects in DN thymocyte proliferation and differentiation, but in FTOC they accumulate a normal proportion of DP cells.5 We therefore cultured E14.5 Shh\#8722;/\#8722; and littermate thymi and analyzed the production of CD4SP cells. After 3 and 6 days in culture, the Shh\#8722;/\#8722; thymi contained a significantly greater proportion of CD4SP cells than those of littermates, indicating that Shh negatively regulates the transition from DP to CD4SP cell (Figure 3A-C). In E14.5+6-day FTOC, only 30% of SP cells express high cell surface CD3.5 Therefore to confirm that the increase in the CD4SP population was due to the accumulation of mature T cells, we also analyzed differentiation to SP cells in E17.5 Shh\#8722;/\#8722; and littermate FTOC after 7 days. On E17.5, the thymus contained approximately 70% DP cells and there was no significant difference in the proportion of DP cells between Shh\#8722;/\#8722; and littermate thymi. Mature SP cells had not arisen, facilitating investigation of the transition from DP to SP. In a typical experiment, the ratio of CD4/CD8 SP cells was increased from 2.6 in the Shh+/+ to 3.4 in the Shh\#8722;/\#8722; FTOC. The proportion of CD4SP cells was significantly increased from 13% in Shh+/+ FTOC to 21.2% in Shh\#8722;/\#8722; FTOC (Figure 3D). There was no difference in the proportion of CD3high cells in each SP population between Shh\#8722;/\#8722; and Shh+/+ littermate cultures, indicating that the increase in CD4SP cells was genuinely due to the accumulation of mature cells (Figure 3D). Of interest, CD69 expression was increased in the CD4SP Shh\#8722;/\#8722; population (Figure 3D). Therefore, under all culture conditions the Shh\#8722;/\#8722; thymus favored the production of CD4SP over CD8SP cells (Figure 3E).
Increased differentiation to CD4SP in the Shh\#8722;/\#8722; thymus. (A) CD4 and CD8 staining of E14.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 6 days. Mean thymus sizes were 2.3 × 104 for Shh\#8722;/\#8722;, 1.6 × 105 for Shh+/\#8722;, and 1.8 × 105 for Shh+/+. (B-C) Bar charts to show the relative proportion of CD4SP thymocytes in E14.5 Shh\#8722;/\#8722; and littermate FTOC after 3 (B) and 6 (C) days in culture. For each thymus, the proportion of CD4SP cells was calculated relative to the mean proportion in WT littermates. The difference in the relative proportion of CD4SP cells between Shh\#8722;/\#8722; and Shh+/+ thymi was statistically significant: P = .006 for 3-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 5]), P = .022 for 6-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 6]). (D) CD4 and CD8 staining of E17.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 7 days. The average percentage of CD4SPs in the Shh\#8722;/\#8722; was increased 1.4 times compared with littermates (P = .046). The ratio of CD4/CD8 SP cells was 2.6 in Shh+/+ FTOC and 3.4 in Shh\#8722;/\#8722; FTOC. Bar charts to show CD3 and CD69 expression on CD4 and CD8 SP thymocytes in the Shh\#8722;/\#8722; (\#9633;, n = 3) and littermates (\#9641;, n = 14). Mean thymus sizes were 9 × 104 for Shh\#8722;/\#8722;, 1.3 × 105 for Shh+/\#8722;, and 1.9 × 105 for Shh+/+. (E) Bar chart to show mean CD4/CD8 ratio for the different culture conditions. (F) Bar chart to show mean SP/DP ratio for the different culture conditions. Error bars show SE. Data are representative of at least 3 experiments.
Increased differentiation to CD4SP in the Shh\#8722;/\#8722; thymus. (A) CD4 and CD8 staining of E14.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 6 days. Mean thymus sizes were 2.3 × 104 for Shh\#8722;/\#8722;, 1.6 × 105 for Shh+/\#8722;, and 1.8 × 105 for Shh+/+. (B-C) Bar charts to show the relative proportion of CD4SP thymocytes in E14.5 Shh\#8722;/\#8722; and littermate FTOC after 3 (B) and 6 (C) days in culture. For each thymus, the proportion of CD4SP cells was calculated relative to the mean proportion in WT littermates. The difference in the relative proportion of CD4SP cells between Shh\#8722;/\#8722; and Shh+/+ thymi was statistically significant: P = .006 for 3-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 5]), P = .022 for 6-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 6]). (D) CD4 and CD8 staining of E17.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 7 days. The average percentage of CD4SPs in the Shh\#8722;/\#8722; was increased 1.4 times compared with littermates (P = .046). The ratio of CD4/CD8 SP cells was 2.6 in Shh+/+ FTOC and 3.4 in Shh\#8722;/\#8722; FTOC. Bar charts to show CD3 and CD69 expression on CD4 and CD8 SP thymocytes in the Shh\#8722;/\#8722; (\#9633;, n = 3) and littermates (\#9641;, n = 14). Mean thymus sizes were 9 × 104 for Shh\#8722;/\#8722;, 1.3 × 105 for Shh+/\#8722;, and 1.9 × 105 for Shh+/+. (E) Bar chart to show mean CD4/CD8 ratio for the different culture conditions. (F) Bar chart to show mean SP/DP ratio for the different culture conditions. Error bars show SE. Data are representative of at least 3 experiments.
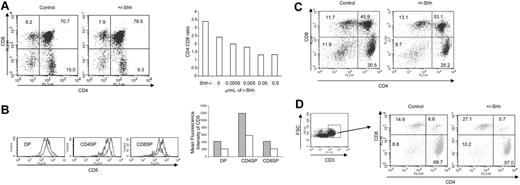
Shh treatment reduced the CD4/CD8 ratio and CD5 expression in thymus explants
Given that the production of mature CD4SP cells was increased in the Shh\#8722;/\#8722; thymus, we asked if treatment of WT FTOC with recombinant Shh (r-Shh) could reduce differentiation to CD4SP. We treated E17.5 FTOC with r-Shh for 7 days. Typically, the percentage of DP cells in r-Shh\#8211;treated cultures increased to 78.5% compared with 70.7% in the control, whereas the proportion of CD4SP cells was decreased from 15% in control to 9.3% in treated cultures. When we compared the ratio of mature (CD3high) CD4/CD8 SP cells (Figure 4A), we found a dose-dependent reduction with maximum inhibition at 0.05 \#956;g/mL r-Shh. CD5 expression was reduced in r-Shh\#8211;treated FTOC relative to controls in DP and SP populations (Figure 4B), consistent with the duller expression observed in the Gli2\#916;N2 transgenic thymus (Figure 2G).
Shh treatment reduced CD5 expression and the CD4/CD8 ratio in thymus explants. (A) CD4 and CD8 profiles of 7-day C57BL/6 E17.5 FTOC control and treated with 0.05 \#956;g/mL r-Shh. The bar chart shows the mean ratio of mature (CD3high) CD4/CD8 SP in 7-day FTOC for Shh\#8722;/\#8722; and C57BL/6 E17.5 cultured with the stated concentration of recombinant Shh (r-Shh). For all treatments, n = 4. Mean cell recovery was as follows: control, 1.87 × 105; 0.5 \#956;g/mL r-Shh, 1.66 × 105; 0.05 \#956;g/mL r-Shh, 2.07 × 105; 0.005 \#956;g/mL r-Shh, 2.1 × 105; and 0.0005 \#956;g/mL r-Shh, 1.54 × 105. (B) CD5 expression on r-Shh\#8211;treated FTOC. Overlaid histograms of CD5 expression gated on the subsets from the FACS plots above for control cultures (gray filled) and r-Shh\#8211;treated cultures (gray line only, no fill). MFIs were (control/+ r-Shh) as follows: DP, 424.5/219.6; CD4SP, 1197.7/586.6; and CD8SP, 425.3/220.8. The bar chart shows MFI for the control (\#9641;) and Shh-treated (\#9633;) cultures. (C) CD4 and CD8 profiles of 10-day C57BL/6 E17.5 FTOC control and treated with 0.5 \#956;g/mL r-Shh. Mean cell recovery was as follows: control, 1.34 × 105; 0.5 \#956;g/mL r-Shh, 1.44 × 105. (D) Profiles of CD4 and CD8, gated on CD3high of 10-day C57BL/6 FTOC control and treated with 0.5 \#956;g/mL r-Shh.
Shh treatment reduced CD5 expression and the CD4/CD8 ratio in thymus explants. (A) CD4 and CD8 profiles of 7-day C57BL/6 E17.5 FTOC control and treated with 0.05 \#956;g/mL r-Shh. The bar chart shows the mean ratio of mature (CD3high) CD4/CD8 SP in 7-day FTOC for Shh\#8722;/\#8722; and C57BL/6 E17.5 cultured with the stated concentration of recombinant Shh (r-Shh). For all treatments, n = 4. Mean cell recovery was as follows: control, 1.87 × 105; 0.5 \#956;g/mL r-Shh, 1.66 × 105; 0.05 \#956;g/mL r-Shh, 2.07 × 105; 0.005 \#956;g/mL r-Shh, 2.1 × 105; and 0.0005 \#956;g/mL r-Shh, 1.54 × 105. (B) CD5 expression on r-Shh\#8211;treated FTOC. Overlaid histograms of CD5 expression gated on the subsets from the FACS plots above for control cultures (gray filled) and r-Shh\#8211;treated cultures (gray line only, no fill). MFIs were (control/+ r-Shh) as follows: DP, 424.5/219.6; CD4SP, 1197.7/586.6; and CD8SP, 425.3/220.8. The bar chart shows MFI for the control (\#9641;) and Shh-treated (\#9633;) cultures. (C) CD4 and CD8 profiles of 10-day C57BL/6 E17.5 FTOC control and treated with 0.5 \#956;g/mL r-Shh. Mean cell recovery was as follows: control, 1.34 × 105; 0.5 \#956;g/mL r-Shh, 1.44 × 105. (D) Profiles of CD4 and CD8, gated on CD3high of 10-day C57BL/6 FTOC control and treated with 0.5 \#956;g/mL r-Shh.
When E17.5 FTOC were cultured for 10 days, Shh treatment again reduced differentiation from DP to CD4SP (Figure 4C). Gating on CD3high cells revealed a relative increase in the proportion of CD8SP cells from 14.9% in the control to 27.1% in Shh-treated cultures, concomitant with the decrease in the CD4SP population. Overall, the ratio of mature (CD3high) CD4/CD8 SP cells was reduced from 4.7 in the control to 2.1 in the Shh-treated cultures.
Taken together, these data show that Shh signaling is involved in regulating differentiation from DP to SP and CD4/CD8 ratio. Absence of Shh increased production of CD4SP cells, but Hh pathway activation, both by thymocyte autonomous Gli2\#916;N2 expression in vivo and by treatment with r-Shh in vitro, decreased cell surface CD5 expression and the production of CD4SP cells.
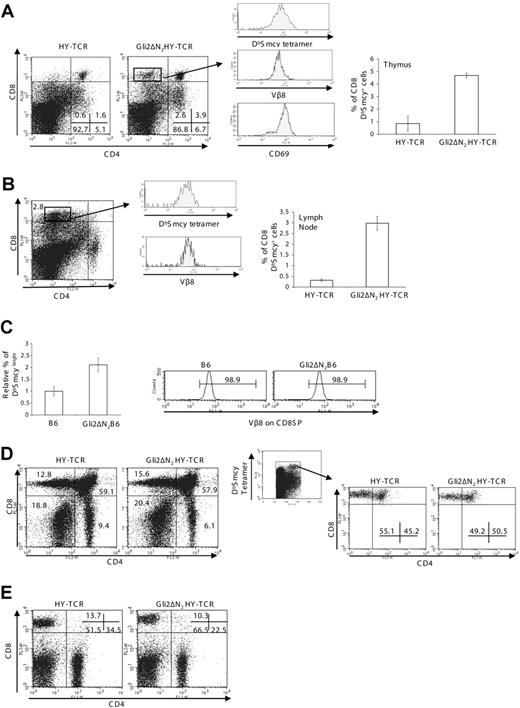
Negative selection is compromised in the Gli2\#916;N2HY-TCR transgenic male thymus
The pattern of expression of CD2, CD5, and CD69 in the Gli2\#916;N2 thymus together with the opposing effects of removal or activation of the Shh signal on differentiation from DP to CD4SP cell suggested that Hh signaling influences the later stages of thymocyte development by a TCR-dependent mechanism, and that activation of the Hh signaling pathway in thymocytes might lower the strength of the TCR signal. Therefore, to assess the effect of Hh signaling on repertoire selection we crossed the Gli2\#916;N2 transgenic with transgenic mice expressing the HY-TCR.22 The HY-TCR recognizes a male-specific peptide from the Smcy gene presented by H-2Db,37 allowing assessment of negative selection in male mice.22 As expected, male HY-TCR mice showed efficient clonal deletion of CD8SP and DP cells (Figure 5A). However, in the Gli2\#916;N2HY-TCR thymus we found an increase in the percentage of DP cells and the presence of a small population of CD8SPs that had escaped deletion (Figure 5A). This population made up 2.5% of thymocytes and was absent in HY-TCR littermates. It expressed high levels of CD8 and CD69, and stained bright with anti-V\#946;8 and DbSmcy tetramer, confirming that it expressed the transgenic male-reactive TCR and had genuinely escaped negative selection (Figure 5A). We found that a small proportion (\#8764; 3%) of peripheral lymph node cells expressed high levels of CD8 and stained brightly with DbSmcy tetramer in the Gli2\#916;N2HY-TCR mice, which were absent in male HY-TCR littermates (Figure 5B)
Gli2\#916;N2 expression influences TCR repertoire selection. (A) Thymus phenotype of male mice with the transgenic HY-TCR. CD4 and CD8 expression in HY-TCR\#8211;positive mice with (Gli2\#916;N2HY-TCR) and HY-TCR littermates without the Gli2\#916;N2 transgene. Mean thymus sizes were 2.4 × 107 for HY-TCR and 2.3 × 107 for Gli2\#916;N2HY-TCR. The histograms show DbSmcy tetramer, V\#946;8, and CD69 staining on CD8SP cells from the Gli2\#916;N2HY-TCR. The bar chart shows the percentage of CD8 DbSmcy-positive cells in the HY-TCR (n = 12) and Gli2\#916;N2HY-TCR (n = 8) thymus (P = < .001). (B) Peripheral phenotype of male mice with the HY transgenic TCR. CD4 and CD8 profile of a Gli2\#916;N2HY-TCR lymph node. The bar chart shows the percentage of CD8 DbSmcy-positive cells in HY-TCR (n = 12) and Gli2\#916;N2HY-TCR (n = 8) lymph node (P = < .001). The histograms show DbSmcy and V\#946;8 expression on CD8 T cells from the Gli2\#916;N2HY-TCR. (C) Lymph node of male mice with the B6.2.16\#946; transgenic TCR. Bar chart to show the relative number of CD8+ DbSmcybright cells in B6.2.16\#946;-positive mice (B6) and B6.2.16\#946;Gli2\#916;N2-positive mice (Gli2\#916;N2B6) (P = .021, n = 4, n = 5, respectively). The histograms show V\#946;8 expression on CD8 T cells. (D) Thymus phenotype of HY-TCR female mice. CD4 and CD8 profiles of HY-TCR and Gli2\#916;N2HY-TCR mice (left dot plots). Mean thymus sizes were 8.7 × 107 for HY-TCR and 8.4 × 107 for Gli2\#916;N2HY-TCR. CD4 and CD8 profiles gated on DbSmcy-positive cells in the HY-TCR and Gli2\#916;N2HY-TCR thymus (right dot plots). The percentages of cells falling in the DbSmcy-positive gate were typically 9.6% in the HY-TCR and 9.4% in the Gli2\#916;N2HY-TCR. (E) Peripheral phenotype of HY-TCR female mice. CD4 and CD8 profiles in HY-TCR and Gli2\#916;N2HY-TCR mice. Numbers in dot plots or histograms indicate the percentage of cells falling in the quadrant, region, or marker. Error bars show SE. Data are representative of at least 3 experiments.
Gli2\#916;N2 expression influences TCR repertoire selection. (A) Thymus phenotype of male mice with the transgenic HY-TCR. CD4 and CD8 expression in HY-TCR\#8211;positive mice with (Gli2\#916;N2HY-TCR) and HY-TCR littermates without the Gli2\#916;N2 transgene. Mean thymus sizes were 2.4 × 107 for HY-TCR and 2.3 × 107 for Gli2\#916;N2HY-TCR. The histograms show DbSmcy tetramer, V\#946;8, and CD69 staining on CD8SP cells from the Gli2\#916;N2HY-TCR. The bar chart shows the percentage of CD8 DbSmcy-positive cells in the HY-TCR (n = 12) and Gli2\#916;N2HY-TCR (n = 8) thymus (P = < .001). (B) Peripheral phenotype of male mice with the HY transgenic TCR. CD4 and CD8 profile of a Gli2\#916;N2HY-TCR lymph node. The bar chart shows the percentage of CD8 DbSmcy-positive cells in HY-TCR (n = 12) and Gli2\#916;N2HY-TCR (n = 8) lymph node (P = < .001). The histograms show DbSmcy and V\#946;8 expression on CD8 T cells from the Gli2\#916;N2HY-TCR. (C) Lymph node of male mice with the B6.2.16\#946; transgenic TCR. Bar chart to show the relative number of CD8+ DbSmcybright cells in B6.2.16\#946;-positive mice (B6) and B6.2.16\#946;Gli2\#916;N2-positive mice (Gli2\#916;N2B6) (P = .021, n = 4, n = 5, respectively). The histograms show V\#946;8 expression on CD8 T cells. (D) Thymus phenotype of HY-TCR female mice. CD4 and CD8 profiles of HY-TCR and Gli2\#916;N2HY-TCR mice (left dot plots). Mean thymus sizes were 8.7 × 107 for HY-TCR and 8.4 × 107 for Gli2\#916;N2HY-TCR. CD4 and CD8 profiles gated on DbSmcy-positive cells in the HY-TCR and Gli2\#916;N2HY-TCR thymus (right dot plots). The percentages of cells falling in the DbSmcy-positive gate were typically 9.6% in the HY-TCR and 9.4% in the Gli2\#916;N2HY-TCR. (E) Peripheral phenotype of HY-TCR female mice. CD4 and CD8 profiles in HY-TCR and Gli2\#916;N2HY-TCR mice. Numbers in dot plots or histograms indicate the percentage of cells falling in the quadrant, region, or marker. Error bars show SE. Data are representative of at least 3 experiments.
In the male HY-TCR transgenic model, negative selection in the thymus can commence at an earlier developmental stage than would occur physiologically because of early expression of the \#945;\#946;TCR in DN cells.38 To confirm that inhibition of clonal deletion was not restricted to early TCR\#945; expression, we analyzed the effect of the Gli2\#916;N2 transgene on negative selection in male TCR\#946; chain\#8211;only transgenic mice, B6.2.16\#946;.23 The B6.2.16\#946; mice express the same TCR\#946; chain as the HY-TCR transgenic, but the TCR\#945; chain is variable and male-reactive DbSmcy-binding T cells are deleted in the male animals. The proportion of CD8SP cells (all of which expressed the transgenic \#946; chain) that stained brightly with DbSmcy tetramer in the lymph node was more than doubled in the presence of the Gli2\#916;N2 transgene (Figure 5C). Thus, clonal deletion of self-reactive thymocytes was also compromised in Gli2\#916;N2B6.2.16\#946; male mice.
Hh signaling influences positive selection in HY-TCR transgenic female mice
In the female Gli2\#916;N2HY-TCR thymus, we found an increased proportion of CD8SPs compared with the control HY-TCR female, together with a decrease in the CD4SP population (Figure 5D), as observed in the non-TCR transgenic Gli2\#916;N2 thymus. On first consideration, it therefore seemed that the Gli2\#916;N2 transgene might actually be enhancing positive selection of the transgenic \#945;\#946;TCR to the CD8 lineage. However, when we gated on DbSmcybright cells (Figure 5D), we consistently found a reduction in the proportion of CD8SP cells and increase in DP cells in the Gli2\#916;N2HY-TCR thymus compared with littermates, indicative of reduced positive selection of the transgenic \#945;\#946;TCR. In a representative experiment the percentage of DbSmcybright cells that was CD8SP was reduced from 55.1% in the control HY-TCR thymus to 49.2% in the Gli2\#916;N2HY-TCR thymus, whereas the total percentage of DbSmcybright thymocytes was unaffected (Figure 5D).
In lymph nodes, the Gli2\#916;N2 transgene caused a reduction in CD4 and CD8 peripheral T cells. The CD8 population was reduced from 14% to 10% and the CD4SP population from 35% to 23% (Figure 5E), suggesting that Gli2\#916;N2 expression also affected peripheral T-cell expansion and homeostasis.
Altered TCR repertoire selection in Shh mutant thymus
To ask if Shh affects TCR repertoire selection in a physiological situation, we introduced the transgenic HY-TCR onto the Shh+/\#8722; background and compared positive selection in young adult female Shh+/\#8722; HY-TCR and their littermate thymi. We found increased positive selection of the transgenic TCR to the CD8 lineage in the Shh+/\#8722; HY-TCR thymus (Figure 6A) with an overall increase in the proportion of CD8SP thymocytes from 11% to 16%. When we gated on DbSmcybright cells (Figure 6A) the proportion of CD8SP cells was consistently increased, typically from 39% in the Shh+/+ littermate to 49% in the Shh+/\#8722; thymus, whereas the proportion of DP cells was decreased from 60% in the Shh+/+ to 49% in the Shh+/\#8722; thymus.
Altered TCR repertoire selection in Shh mutant thymus. (A) Thymus phenotype of 4-week-old Shh+/\#8722; HY-TCR female mice. CD4 and CD8 profiles of HY-TCR and Shh+/\#8722; HY-TCR littermate mice (left dot plots). Mean thymus sizes were 3.7 × 107 for HY-TCR and 4.8 × 107 for Shh+/\#8722; HY-TCR. CD4 and CD8 profiles gated on DbSmcy-positive cells in the HY-TCR and Shh+/\#8722; HY-TCR thymus (right dot plots). (B) Thymus phenotype of E17.5 Shh\#8722;/\#8722; HY-TCR female FTOC cultured for 10 days. CD4 and CD8 profiles of HY-TCR and Shh\#8722;/\#8722; HY-TCR littermate FTOC. Bar chart shows the relative percentage of transgenic (Tg) TCR+ CD8bright SP cells for HY-TCR (n = 3) and Shh\#8722;/\#8722; HY-TCR (n = 3); P = .027. The histograms show V\#946;8 and T3.70 (transgenic TCR\#945; chain) staining on CD8SP cells. (C) Thymus phenotype of 12-day-old Shh+/\#8722; HY-TCR male. CD4 and CD8 profiles of HY-TCR and Shh+/\#8722; HY-TCR littermates. Mean thymus sizes were 2.4 × 107 for HY-TCR and 2.1 × 107 for Shh+/\#8722; HY-TCR. Bar charts show the relative percentage of transgenic (Tg) TCR+ CD8bright SP or DP cells for HY-TCR (n = 3) and Shh+/\#8722; HY-TCR (n = 3); P = .019. The histograms show V\#946;8 and T3.70 expression on CD8SP cells. (D) Thymus phenotype of E19 Shh\#8722;/\#8722; HY-TCR male. CD4 and CD8 profiles of HY-TCR, Shh+/\#8722; HY-TCR, and Shh\#8722;/\#8722; HY-TCR littermates. Mean thymus sizes were 2.5 × 105 for HY-TCR, 2.4 × 105 for Shh+/\#8722; HY-TCR, and 1.1 × 104 for Shh\#8722;/\#8722; HY-TCR. Bar charts show the relative percentage of Tg TCR+ CD8bright SP or DP cells for HY-TCR (n = 4), Shh+/\#8722; HY-TCR (n = 4, P = .034), and Shh\#8722;/\#8722; HY-TCR (n = 3, P = .018). The histograms show DbSmcy and T3.70 staining on CD8SP cells. Numbers in dot plots indicate the percentage of cells falling in the quadrant. Error bars show SE. Data are representative of at least 3 experiments.
Altered TCR repertoire selection in Shh mutant thymus. (A) Thymus phenotype of 4-week-old Shh+/\#8722; HY-TCR female mice. CD4 and CD8 profiles of HY-TCR and Shh+/\#8722; HY-TCR littermate mice (left dot plots). Mean thymus sizes were 3.7 × 107 for HY-TCR and 4.8 × 107 for Shh+/\#8722; HY-TCR. CD4 and CD8 profiles gated on DbSmcy-positive cells in the HY-TCR and Shh+/\#8722; HY-TCR thymus (right dot plots). (B) Thymus phenotype of E17.5 Shh\#8722;/\#8722; HY-TCR female FTOC cultured for 10 days. CD4 and CD8 profiles of HY-TCR and Shh\#8722;/\#8722; HY-TCR littermate FTOC. Bar chart shows the relative percentage of transgenic (Tg) TCR+ CD8bright SP cells for HY-TCR (n = 3) and Shh\#8722;/\#8722; HY-TCR (n = 3); P = .027. The histograms show V\#946;8 and T3.70 (transgenic TCR\#945; chain) staining on CD8SP cells. (C) Thymus phenotype of 12-day-old Shh+/\#8722; HY-TCR male. CD4 and CD8 profiles of HY-TCR and Shh+/\#8722; HY-TCR littermates. Mean thymus sizes were 2.4 × 107 for HY-TCR and 2.1 × 107 for Shh+/\#8722; HY-TCR. Bar charts show the relative percentage of transgenic (Tg) TCR+ CD8bright SP or DP cells for HY-TCR (n = 3) and Shh+/\#8722; HY-TCR (n = 3); P = .019. The histograms show V\#946;8 and T3.70 expression on CD8SP cells. (D) Thymus phenotype of E19 Shh\#8722;/\#8722; HY-TCR male. CD4 and CD8 profiles of HY-TCR, Shh+/\#8722; HY-TCR, and Shh\#8722;/\#8722; HY-TCR littermates. Mean thymus sizes were 2.5 × 105 for HY-TCR, 2.4 × 105 for Shh+/\#8722; HY-TCR, and 1.1 × 104 for Shh\#8722;/\#8722; HY-TCR. Bar charts show the relative percentage of Tg TCR+ CD8bright SP or DP cells for HY-TCR (n = 4), Shh+/\#8722; HY-TCR (n = 4, P = .034), and Shh\#8722;/\#8722; HY-TCR (n = 3, P = .018). The histograms show DbSmcy and T3.70 staining on CD8SP cells. Numbers in dot plots indicate the percentage of cells falling in the quadrant. Error bars show SE. Data are representative of at least 3 experiments.
To analyze positive selection in the complete absence of Shh, we cultured female E17.5 Shh+/+ HY-TCR and Shh\#8722;/\#8722; HY-TCR littermate FTOC for 10 days. The proportion of HY-TCR+ CD8SP cells was increased approximately 3-fold (P = .027), from 8% in the Shh+/+ HY-TCR to 23% in the Shh+/\#8722; HY-TCR thymus (Figure 6B). Thus, in contrast to the Gli2\#916;N2HY-TCR thymus, we found that in both the embryo and the adult, mutation of Shh increased positive selection of the HY-TCR.
In the adult male HY-TCR thymus, clonal deletion is efficient and complete (Figure 5A and Kisielow et al22), and so we could not test if reduction in Shh copy number increased deletion. In the embryo, clonal deletion starts at E18 and a significant proportion of DP and CD8SP cells persists at birth and in the neonate.39,40 We therefore analyzed negative selection in the fetal and neonatal thymus.
In 12-day-old male Shh+/+ HY-TCR thymi, we detected a small population of HY-TCR+ CD8SP (\#8764; 5%) and DP (\#8764; 4%) cells, which were significantly reduced (P = .019) to approximately 3.5% and 2%, respectively, in Shh+/\#8722; HY-TCR littermates, indicating more efficient clonal deletion in the Shh+/\#8722; thymus (Figure 6C). On E19, the proportion of HY-TCR+ CD8SP and DP cells was significantly reduced (P = .018) from 19% and 17%, respectively, in the Shh+/+ thymus to only 12% and 4% in the Shh\#8722;/\#8722; littermate thymus (Figure 6D). The Shh+/\#8722; thymus gave an intermediate phenotype (P = .034). Therefore, we again found that removal of Shh increased the efficiency of TCR repertoire selection, in this case increasing negative selection of the self-reactive transgenic TCR. These data support the hypothesis that under physiological conditions, Shh modulates TCR signal strength in developing thymocytes.
Attenuated T-cell activation in peripheral Gli2\#916;N2 transgenic T cells
Given that activation or reduction of Hh signaling altered the outcome of TCR ligation in thymocytes, we investigated the effect of Gli2\#916;N2 expression on peripheral T-cell activation. We confirmed Gli2\#916;N2 transgene expression and up-regulation of Gli1 transcription by quantitative RT-PCR in sorted CD4 and CD8 T-cell populations (Figure 7A). As expected from previous expression studies,29,30 the transgene was highly expressed in CD4 and CD8 T cells, and not detected in B cells. Transcription of Gli1 was up-regulated in both T-cell populations, confirming functional activity of the transgene. To assay T-cell activation, we stimulated splenocytes with anti-CD3 and anti-CD28 antibodies. After 24 hours, induction of cell surface expression of the early activation marker CD69 was inhibited in both CD4 and CD8 T cells from Gli2\#916;N2 transgenic spleens. In a typical experiment, only 18% of Gli2\#916;N2 transgenic CD4 T cells expressed CD69, compared with more than 60% of WT CD4 T cells (Figure 7B). Expression of the later activation marker CD25 was also decreased in Gli2\#916;N2 transgenic cultures on both CD4 and CD8 T cells, from 47% and 72% in WT CD4 and CD8 T populations, respectively, to 17% and 9% in their Gli2\#916;N2 transgenic counterparts (Figure 7B). These data show that activation of Hh signaling in T cells attenuated T-cell activation.
Gli2\#916;N2 expression impairs TCR-induced activation and proliferation of peripheral T cells and reduces ERK phosphorylation. (A) Quantitative RT-PCR analysis of Gli2\#916;N2 transgene expression in FACS-sorted peripheral T-cell populations (left graph). The right graph shows up-regulation of Gli1 transcription on CD4 and CD8 T cells in the Gli2\#916;N2 (\#9641;) compared with the WT (\#9633;). Error bars represent SD. (B) Histograms showing the early activation marker CD69 (top panels) and the later activation marker CD25, on CD4 and CD8 SP cells after 24 hours in culture stimulated with 0.01 \#956;g/mL of each of anti-CD3 and anti-CD28. Gray fill indicates WT; no fill, Gli2\#916;N2. Numbers indicate the percentage of cells falling within the marker. (In all unstimulated controls, CD69 expression was approximately 5%, and CD25 expression was approximately 8%; data not shown). (C) Bar graphs to show cell divisions using CFSE staining in CD4 cells cultured for 96 hours with 0.01 \#956;g/mL of each of anti-CD3 and anti-CD28 with (middle panel) and without (left panel) 20 IU/mL IL-2. The numbers 0-7 represent the number of divisions for cells falling in the marker. Gray indicates WT; white, Gli2\#916;N2 transgenic. The bar graph on the far right shows the percentage of CFSE-labeled cells falling in the markers for 5 to 8 divisions for the WT and the Gli2\#916;N2 transgenic with and without IL-2. (D) Bar graph shows proliferation for WT and the Gli2\#916;N2 transgenic cultured in 0.1, 0.01, and 0.001 \#956;g/mL of each of anti-CD3 and anti-CD28. (E) Western blot showing the levels of phosphorylated (upper panel) Erk1 and Erk2 kinases in WT and Gli\#916;N2 transgenic splenocytes stimulated with 1 \#956;g/mL of each of anti-CD3 and anti-CD28 for the indicated period of time. For comparison, the total levels of Erk kinases expressed in the cytoplasm of stimulated cells is shown (lower panel). (F) Intracellular phosphorylated ERK (p-Erk) on CD4+ cells. Top left histogram shows the CD4+ gate. The right histograms show intracellular p-Erk levels in unactivated CD4+ (dashed line) and CD4+ activated with 1 \#956;g/mL of each of anti-CD3 and anti-CD28 for 2 minutes (gray line). The bar chart shows the MFI (unactivated/activated) for WT (10.0/19.1) and Gli\#916;N2 (7.4/11.5). (G) Histograms to show basal intracellular p-Erk in WT (top, MFI = 45.2) and Gli\#916;N2 (bottom, MFI = 29.0) thymocytes.
Gli2\#916;N2 expression impairs TCR-induced activation and proliferation of peripheral T cells and reduces ERK phosphorylation. (A) Quantitative RT-PCR analysis of Gli2\#916;N2 transgene expression in FACS-sorted peripheral T-cell populations (left graph). The right graph shows up-regulation of Gli1 transcription on CD4 and CD8 T cells in the Gli2\#916;N2 (\#9641;) compared with the WT (\#9633;). Error bars represent SD. (B) Histograms showing the early activation marker CD69 (top panels) and the later activation marker CD25, on CD4 and CD8 SP cells after 24 hours in culture stimulated with 0.01 \#956;g/mL of each of anti-CD3 and anti-CD28. Gray fill indicates WT; no fill, Gli2\#916;N2. Numbers indicate the percentage of cells falling within the marker. (In all unstimulated controls, CD69 expression was approximately 5%, and CD25 expression was approximately 8%; data not shown). (C) Bar graphs to show cell divisions using CFSE staining in CD4 cells cultured for 96 hours with 0.01 \#956;g/mL of each of anti-CD3 and anti-CD28 with (middle panel) and without (left panel) 20 IU/mL IL-2. The numbers 0-7 represent the number of divisions for cells falling in the marker. Gray indicates WT; white, Gli2\#916;N2 transgenic. The bar graph on the far right shows the percentage of CFSE-labeled cells falling in the markers for 5 to 8 divisions for the WT and the Gli2\#916;N2 transgenic with and without IL-2. (D) Bar graph shows proliferation for WT and the Gli2\#916;N2 transgenic cultured in 0.1, 0.01, and 0.001 \#956;g/mL of each of anti-CD3 and anti-CD28. (E) Western blot showing the levels of phosphorylated (upper panel) Erk1 and Erk2 kinases in WT and Gli\#916;N2 transgenic splenocytes stimulated with 1 \#956;g/mL of each of anti-CD3 and anti-CD28 for the indicated period of time. For comparison, the total levels of Erk kinases expressed in the cytoplasm of stimulated cells is shown (lower panel). (F) Intracellular phosphorylated ERK (p-Erk) on CD4+ cells. Top left histogram shows the CD4+ gate. The right histograms show intracellular p-Erk levels in unactivated CD4+ (dashed line) and CD4+ activated with 1 \#956;g/mL of each of anti-CD3 and anti-CD28 for 2 minutes (gray line). The bar chart shows the MFI (unactivated/activated) for WT (10.0/19.1) and Gli\#916;N2 (7.4/11.5). (G) Histograms to show basal intracellular p-Erk in WT (top, MFI = 45.2) and Gli\#916;N2 (bottom, MFI = 29.0) thymocytes.
Proliferation of activated T cells is compromised by the Gli2\#916;N2 transgene
To assess proliferation, splenocytes were labeled with CFSE and stimulated with anti-CD3 and anti-CD28. The Gli2\#916;N2 transgenic T cells were unable to proliferate as well as their WT counterparts (Figure 7C). After 96 hours, only 20% of Gli2\#916;N2 transgenic CD4 T cells had undergone 4 to 7 divisions compared with more than 50% of WT CD4 T cells (Figure 7C). Addition of IL-2 to the cultures overcame the inhibition of proliferation (Figure 7C). We titrated the strength of activation signal given by anti-CD3 and anti-CD28 antibodies. Although at low antibody concentration proliferation was suboptimal in the WT cultures, we consistently saw approximately 50% fewer CD4 T cells that had undergone 4 or more divisions in the Gli2\#916;N2 transgenic cultures compared with WT cultures (Figure 7D).
Hh signaling reduces TCR signal strength and inhibits TCR-induced ERK phosphorylation
To measure directly the effect of Hh signaling on TCR signals, we stimulated splenocytes from Gli2\#916;N2 transgenic and littermate mice with antibodies against CD3 and CD28 and measured MAPK activation by following the kinetics of ERK phosphorylation by Western blot. Gli2\#916;N2 expression inhibited TCR-induced MAPK activation (Figure 7E). In WT splenocytes, strong activation of ERK2 was observed within 5 minutes of stimulus, and full phosphorylation of ERK1 and ERK2 kinases was detected after 30 minutes. In contrast, activation of ERK2 in Gli2\#916;N2 transgenic splenocytes was much delayed and reduced, and ERK1 failed to activate to normal levels during the experiment. To confirm that this reduction in ERK phosphorylation was T-cell specific, we used a FACS-based assay27 to compare Erk phosphorylation in the CD4+ T-cell population. Gating on CD4+ cells, the MFI of intracellular anti\#8211;phospho-ERK on unactivated cells was slightly lower in the Gli2\#916;N2 compared with WT spleen. On TCR ligation, there was a clear shift in the WT intracellular anti\#8211;phospho-ERK staining, with a doubling in MFI. In contrast, the shift in the Gli2\#916;N2 T cells was negligible (Figure 7F). Thus, expression of the Gli2\#916;N2 transgene inhibited TCR-induced activation by a mechanism upstream of ERK phosphorylation.
Given that basal phospho-ERK levels were reduced in Gli2\#916;N2 peripheral T cells, we also measured ERK phosphorylation in thymocytes (Figure 7G). The MFI of intracellular anti\#8211;phospho-ERK was reduced, typically from 45 in WT to 29 in Gli2\#916;N2 thymocytes. This reduction in basal ERK phosphorylation is consistent with reduction in TCR signal strength during repertoire selection in the Gli2\#916;N2 transgenic.
Discussion
Here, we show that the transcriptional events induced by activation of the Hh signaling pathway in mature peripheral T cells resulted in a reduction of TCR signal strength, profoundly inhibiting the ability of the TCR to transduce signals for activation and proliferation. Likewise, activation of the Hh signaling pathway in thymocytes changed the outcome of TCR ligation at many stages of development: allowing self-reactive T cells to escape clonal deletion; reducing transgenic TCR-mediated positive selection of CD8 T cells; and reducing the ratio of CD4/CD8 SP cells produced in the thymus. In contrast, in the Shh\#8722;/\#8722; thymus the CD4/CD8 ratio was affected in the opposite way, and differentiation from DP to CD4SP was increased. Both positive and negative selection of a transgenic TCR were also increased, demonstrating that Shh does indeed influence TCR repertoire selection and the transition from DP to SP cell in a physiological situation. Taken together, the consequences for thymocyte development of removal of the Shh signal (in the Shh\#8722;/\#8722; thymus), or activation of Hh signaling in vivo (by Gli2\#916;N2 expression) and in vitro (by Shh treatment), all strongly support the hypothesis that Hh signaling in thymocytes modulates TCR signal strength.
Reduction of TCR signal strength by Hh-dependent transcription has many consequences for thymocyte development and peripheral immunity, as the outcome of TCR ligation would depend on the Hh microenvironment of the cell. Hh proteins have been shown to function as morphogens,1,41 produced at localized sources and specifying different cell fates in a concentration-dependent manner. Thymocytes move through distinct areas of the thymus at different stages of their development,28 and therefore will receive different strengths of Hh signal at different stages. Our data suggest that their location in the thymus, relative to the source of Hh, will influence the outcome of TCR ligation (positive or negative selection, CD4/CD8 lineage decisions). Of interest, Shh is expressed by epithelial cells scattered in the medulla and corticomedullary junction, and the thymocyte populations that are located in this part of the thymus (early DN populations and SPs) show highest expression of the Hh-target gene Gli1 (Figure 1). As not all medullary epithelial cells express Shh,4,6 it is possible that those that express Shh are specialized for a particular function in TCR repertoire selection.
In the periphery, Hh signaling could function to change the threshold of T-cell activation and influence TCR-dependent T-cell fate decisions, such as TH1/TH2 differentiation or the induction of anergy. Shh is expressed by follicular dendritic cells,42 but Hh proteins are also expressed in skin, gut, and lung, where they are involved in tissue homeostasis and renewal.1 These tissues as sites of frequent immune challenge by pathogens are also subject to many inflammatory and autoimmune diseases, and Hh expression could function to reduce the initial threshold for T-cell activation. Up-regulation of Hh proteins during renewal or remodeling after infection or tissue damage could further influence T-lymphocyte populations at sites of damage.
Inappropriate activation of the Hh signaling pathway has been implicated in many cancers,2 and expression of Gli2\#916;N2 under the control of the keratin-5 promoter increased the incidence of skin tumors in mice.43 Of interest, we have not observed T-cell tumors in Gli2\#916;N2 transgenic mice, although we have maintained the colony for 3 years (N.J.R., unpublished observation, February 2004 to present).
Inhibition of TCR signaling by Hh-induced transcriptional changes involves compromised MAPKinase activation (Figure 7E-G). It is unclear why addition of IL-2 restores proliferation of activated Gli2\#916;N2 peripheral T cells, but one possibility is that the defect in proliferation is directly due to a decrease in IL2 transcription caused by the reduction in ERK activation.44,45 How Gli2\#916;N2 inhibits TCR-induced ERK phosphorylation remains to be determined, but both TCR and Hh signaling require PKA activity, which may therefore be involved in interactions between the 2 pathways.
It has previously been reported that addition of Shh to partially purified T-cell populations in vitro enhanced the induction of T-cell activation and proliferation by suboptimal concentrations of anti-CD3 and anti-CD28 antibody, whereas addition of anti-Hh antibody to these cultures reduced activation and proliferation.19,20 In contrast, our data indicate that the Hh pathway can inhibit T-cell activation and proliferation. There are several possible explanations for this discrepancy: Gli2\#916;N2 transgene expression was restricted to T cells (Figure 7A), but addition of Shh to partially purified T-cell populations might affect T-cell activation indirectly through another cell type present in the culture. Alternatively, differences in the method or strength of activation of the signaling pathway may lead to differences in the relative timing of complete induction of Hh and TCR signaling.
Recently,8 conditional deletion of Smo from T-lineage cells has failed to reveal any positive or negative influence on anti-CD3 induced T-cell proliferation in vitro. From the data presented here, we predict that Smo-deficient T cells transduce a stronger TCR signal than WT T cells, and therefore that differences in proliferation would be detectable only if a suboptimal anti-CD3 signal were given. In the same study, early Smo deletion in the thymus confirmed that Hh positively signals for survival and proliferation of early DN cells,8 but did not influence later stages of T-cell development. Our experiments show that Hh signaling is involved at the transition from DP to SP cell and in TCR repertoire selection. We observed increased CD4/CD8 ratio and TCR repertoire selection in the Shh\#8722;/\#8722; thymus after backcrossing our Shh+/\#8722; colony onto a pure C57BL/6 background5 and introducing a transgenic TCR, whereas in the conditional Smo study the mice were outbred and effects on TCR repertoire were not examined.8
We have previously shown that addition of Shh to FTOC negatively regulated the transition from DN to DP cell, whereas treatment with anti-Hh antibody enhanced differentiation to DP cell.4 The Gli2\#916;N2 transgenic confirms that Hh can act as a negative regulator of TCR signaling, but inhibition of pre-TCR signaling for differentiation from DN to DP cell was not observed. Similarly, transgenic mice that express dominant-negative components of the MAPKinase cascade under the control of the lck promoter have failed to show reduced differentiation to DP cell, despite the fact that MAPKinase activation is downstream of pre-TCR signaling.10 The reason why use of this transgenic cassette is not ideal for dissecting signaling events downstream of the pre-TCR signal is unclear, but it may reflect incomplete transgene expression in the DN subsets,29,30 or powerful pre-TCR independent mechanisms that allow recovery of the DP population.
In summary, we have shown that that the Hh signaling pathway influences TCR repertoire selection in the thymus and can reduce the strength of the TCR signal in mature peripheral T cells, having profound effects for T-cell development and function. It will be important to test the consequences of Hh-induced reduction in TCR signal strength in peripheral immunity and human disease.
Authorship
Contribution: All authors contributed experimentally and in preparation of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tessa Crompton, Division of Cell and Molecular Biology, Imperial College London, Sir Alexander Fleming Building, South Kensington Campus, London SW7 2AZ, United Kingdom; e-mail: t.crompton@imperial.ac.uk.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked \#8220;advertisement\#8221; in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the MRC, BBSRC, Leukemia Research Fund, and Wellcome Trust
We thank Ross Anders for microinjection, Tony Magee for his support, Derek Davies (CRUK FACS laboratory) for sorting, Philip Beachy for Shh+/\#8722; mice, Hiroshi Sasaki for the Gli\#916;N2 plasmid, and Curis for r-Shh.

![Figure 2. Phenotype of Gli2\#916;N2 transgenic mice. (A) Relative cell number (calculated relative to the mean of WT littermates) in the thymus and spleen of WT (gray bars, n = 6) and Gli2\#916;N2 transgenic (\#9633;, n = 6). (B) CD4 and CD8 profiles in the Gli2\#916;N2 transgenic mouse. Thymocytes from Gli2\#916;N2 transgenic and WT littermates were analyzed for CD4 and CD8 expression in the thymus (top panels), spleen (middle panels), and lymph node (bottom panels). Differences between the percentage of populations between transgenic and littermate were statistically significant as given: (thymus) CD4SP, P = .005; DP, P = .014; (spleen) CD4, P = .002; CD8, P = .001; and (lymph node) CD4, P = .005; CD8, P = .041. (C) Bar chart to show change in CD4/CD8 ratio in thymus (WT [n = 6], Gli2\#916;N2 [n = 6], P = .032). (D) CD3 expression on CD4 (top histograms) and CD8 (bottom histograms) SP thymocytes. (E) HSA expression on CD4 (top) and CD8 (bottom) SP thymocytes. (F) CD2 expression on DP (top), CD4SP (middle), and CD8SP (bottom) cells in the Gli2\#916;N2 transgenic (right panels) and the WT (left panels). (G) CD5 expression on CD8SP cells in the Gli2\#916;N2 transgenic thymus (gray line) compared with the WT (black line). The bar chart shows mean MFI for WT (\#9641;) and Gli2\#916;N2 transgenic (\#9633;). (H) CD69 expression on DP (top), CD4 (middle), and CD8 (bottom) SP thymocytes. Numbers in dot plots or histograms indicate the percentage of cells falling in the quadrant, region, or marker. Error bars show standard error (SE). Data are representative of at least 3 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-07-037655/4/m_zh80090700520002.jpeg?Expires=1767808531&Signature=ufyYY341vCj72i5mFu1BoUIrXbYQWDMCSDOlNnsY9fiNU6B~W7ume4AYrsryR5x~EvAbgATdffilX5r0WD06zizUGSlM4-M7CRfqQYkYeJkrtJ1wl6eKeHO5TT~0toW9G7mgjKn4mRq0s1xK8Cb~amoLVVXZIhp1LjmAhQ-l3tMUS5FUbbjWByLMgyk5yqD4aEVAXDp4TdKIHRvwy~oF9Ith5lvF8YCi8Xo8IvvPQmwS-27JTMCudX7T6LPZCYxRYHFVzYjs1zL4Dp6p6W9MG2ELpDdZyDOuA1p6sv8cXMK8kErI8jWXKi2hknShBBknU7cxSluvLAs4SQOSWSAQgQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Increased differentiation to CD4SP in the Shh\#8722;/\#8722; thymus. (A) CD4 and CD8 staining of E14.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 6 days. Mean thymus sizes were 2.3 × 104 for Shh\#8722;/\#8722;, 1.6 × 105 for Shh+/\#8722;, and 1.8 × 105 for Shh+/+. (B-C) Bar charts to show the relative proportion of CD4SP thymocytes in E14.5 Shh\#8722;/\#8722; and littermate FTOC after 3 (B) and 6 (C) days in culture. For each thymus, the proportion of CD4SP cells was calculated relative to the mean proportion in WT littermates. The difference in the relative proportion of CD4SP cells between Shh\#8722;/\#8722; and Shh+/+ thymi was statistically significant: P = .006 for 3-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 5]), P = .022 for 6-day cultures (WT [n = 7], Shh\#8722;/\#8722; [n = 6]). (D) CD4 and CD8 staining of E17.5 Shh\#8722;/\#8722;, Shh+/\#8722;, and Shh+/+ FTOC littermates, cultured for 7 days. The average percentage of CD4SPs in the Shh\#8722;/\#8722; was increased 1.4 times compared with littermates (P = .046). The ratio of CD4/CD8 SP cells was 2.6 in Shh+/+ FTOC and 3.4 in Shh\#8722;/\#8722; FTOC. Bar charts to show CD3 and CD69 expression on CD4 and CD8 SP thymocytes in the Shh\#8722;/\#8722; (\#9633;, n = 3) and littermates (\#9641;, n = 14). Mean thymus sizes were 9 × 104 for Shh\#8722;/\#8722;, 1.3 × 105 for Shh+/\#8722;, and 1.9 × 105 for Shh+/+. (E) Bar chart to show mean CD4/CD8 ratio for the different culture conditions. (F) Bar chart to show mean SP/DP ratio for the different culture conditions. Error bars show SE. Data are representative of at least 3 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-07-037655/4/m_zh80090700520003.jpeg?Expires=1767808531&Signature=akjm~hghiw2GPEsI~kB0OzJwZK92pa9VoDyrCPYkLGXirwtA4-FUPU56H3JSbQb1rp2gf7BjWt7dAOyc4z4qWF-jtc6-dngy-5shvLsE6NW~tLgBMflU5iraxk3E5lSGgJbU2I90~rHc-WXvSCn9WENYGhOSEOCkHSl9RzcooVUPTJO2nKwatddujCV0TSiJb~2OMzyKLap2zuHDyj-xWNmrtRidfz5T6t-XATQXWcFQqwg56NKU0wazrEXG3U2O3Y0Tf4ShiesX1XtUSgc1ZwZ7GFss0oYdcMPT1EEYqnsSFJjTpt8na4MWr~t2slbFn2KcxI55iWGoixNr5fcekw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal