Abstract
Studies have documented the potential antitumor activities of oridonin, a compound extracted from medicinal herbs. However, whether oridonin can be used in the selected setting of hematology/oncology remains obscure. Here, we reported that oridonin induced apoptosis of t(8;21) acute myeloid leukemic (AML) cells. Intriguingly, the t(8;21) product AML1-ETO (AE) fusion protein, which plays a critical role in leukemogenesis, was degraded with generation of a catabolic fragment, while the expression pattern of AE target genes investigated could be reprogrammed. The ectopic expression of AE enhanced the apoptotic effect of oridonin in U937 cells. Preincubation with caspase inhibitors blocked oridonin-triggered cleavage of AE, while substitution of Ala for Asp at residues 188 in ETO moiety of the fusion abrogated AE degradation. Furthermore, oridonin prolonged lifespan of C57 mice bearing truncated AE-expressing leukemic cells without suppression of bone marrow or reduction of body weight of animals, and exerted synergic effects while combined with cytosine arabinoside. Oridonin also inhibited tumor growth in nude mice inoculated with t(8;21)-harboring Kasumi-1 cells. These results suggest that oridonin may be a potential antileukemia agent that targets AE oncoprotein at residue D188 with low adverse effect, and may be helpful for the treatment of patients with t(8;21) AML.
Introduction
The Isodon plant, Rabdosia rubescens (RR), and its extracts, were shown in China to be able to suppress disease progress, reduce tumor burden, alleviate syndrome, and prolong survival in patients with esophageal, gastric carcinoma or liver cancer.1-5 Of interest, other Isodon plants including Isodon japonicus Hara (IJ) and I trichocarpus (IT) were also applied as home remedies for similar disorders in Japan and Korea.6 Oridonin (Figure 1A), a bitter tetracycline diterpenoid compound, was isolated from RR, IJ, and IT separately,7,8 suggesting oridonin should be an essential antitumor component of Isodon plants. Studies showed that oridonin induced apoptosis in a variety of cancer cells including those from prostate, breast, non–small cell lung cancers, acute leukemia (NB4, HL-60 cells), glioblastoma multiforme, and human melanoma cells.9-12 Oridonin could also increase lifespan of mice bearing Ehrlich ascites or P388 lymphocytic leukemia.13,14 However, though studies showed that caspase-3 (casp-3), casp-8, P53, Bcl-2/Bax, cytochrome c (cyt C),10,15,16 and nuclear factor kappa B (NFκB)17 were involved in apoptosis induced by oridonin, mechanisms underlying the antitumor activity of oridonin remain largely unknown, and whether oridonin can find clinical application still needs more investigation.
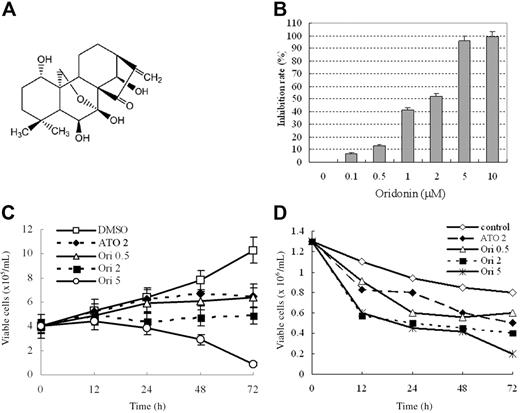
Oridonin inhibits cell growth and proliferation of t(8;21)-bearing leukemic cells. (A) Chemical structure of oridonin. (B) Effects of oridonin on Kasumi-1 cell proliferation detected by using a CCK-8 Kit. (C) Effects of oridonin on Kasumi-1 cell growth. (D) Effects of oridonin on primary leukemic cells isolated from a relapsed patient with t(8;21) leukemia. ATO indicates arsenic trioxide. Error bars represent SD of experiments.
Oridonin inhibits cell growth and proliferation of t(8;21)-bearing leukemic cells. (A) Chemical structure of oridonin. (B) Effects of oridonin on Kasumi-1 cell proliferation detected by using a CCK-8 Kit. (C) Effects of oridonin on Kasumi-1 cell growth. (D) Effects of oridonin on primary leukemic cells isolated from a relapsed patient with t(8;21) leukemia. ATO indicates arsenic trioxide. Error bars represent SD of experiments.
Genetic abnormalities have been shown to play a key role in leukemogenesis,18 and treatment strategies interfering with oncoproteins involved in leukemia pathogenesis have been reported to have high therapeutic efficacy with low adverse effects. The BCR-ABL–targeting STI-571, in the treatment of chronic myeloid leukemia (CML),19 and the PML-RARα–targeting agents all-trans retinoic acid (ATRA) and arsenic trioxide (ATO), in taming acute promyelocytic leukemia (APL),20,21 can serve as paradigms. Hence molecular target–based therapies should be developed for leukemias with poor prognosis. The AML1-ETO (AE) fusion gene is the result of translocation t(8;21)(q22;q22), which is seen in 40% to 80% of M2-type acute myeloid leukemia (AML M2) and 12% to 20% of all cases of AML.22,23 The AE fusion protein recruits the nuclear receptor corepressor (NCoR)–mammalian Sin3 (mSin3)–histone deacetylase (HDAC) complex,24,25 inhibits transcription of AML1 target genes22,24 including interleukin-3 (IL3),26 activates transcription of apoptotic antagonist Bcl-2,27 up-regulates protein tyrosine kinase C-KIT,28 induces the expression of granulocyte colony-stimulating factor receptor (G-CSFR) as well as myeloperoxidase (MPO),29 and blocks transactivation of the GM-CSF promoter.30 The AE oncoprotein enhances self-renewal of hematopoietic stem/progenitor cells, blocks hematopoietic differentiation, disturbs normal cell proliferation,31 and immortalizes murine hematopoietic progenitors.32,33 Although reports suggest that additional mutations are required to cooperate with AE to cause murinefull-blown leukemia,32,34 stem cells expressing AE induce a myeloproliferative disorder35 or distinct myeloid developmental abnormalities in mice,36 while deletion of a C-terminal NCoR/SMRT-interacting region strongly induces leukemia development.37 Recently, a novel isoform of AE transcript, AML1-ETO9a (AE9a), which included an extra exon 9a of the ETO gene and encoded a C-terminally–truncated AE protein, was identified from human t(8;21) AML and was shown to rapidly induce leukemia in a mouse retroviral transduction-transplantation model.38 These data demonstrate that AE plays a critical role in pathogenesis of t(8;21) leukemia, and AE-targeting agents might be helpful for the treatment of t(8;21) AML.
Clinically, aggressive cytosine arabinoside (Ara-C)–based chemotherapy is the standard protocol for t(8;21) AML, and t(8;21) was shown to be a favorable prognostic factor for AML.39 However, others demonstrated that the median survival time of patients with t(8;21) AML was less than 2 years, with a 5-year survival rate of no more than 40%.28,40-44 To further improve the clinical outcome and to provide therapeutic options for t(8;21) AML patients, investigational therapy should be developed. Here, we reported that oridonin not only induced apoptosis in AE-bearing leukemic cells by activating intrinsic apoptotic pathway and triggered a casp-3–mediated degradation of AE at D188, but also showed significant antileukemia efficacies without obvious side effects in 2 murine models for human t(8;21) AML, providing the rationale for potential clinical study to evaluate oridonin as a drug against t(8;21) leukemia.
Materials and methods
Oridonin
Oridonin with a purity of up to 99.5% was a generous gift from Mr Wei Jiang (Zhao Wei Technology and Development, Shanghai, China) and Prof Handong Sun (Kunming Institute of Botany, Chinese Academy of Sciences). A stock solution (10−2 M) was prepared by dissolving oridonin in DMSO (Sigma, St Louis, MO) and stored at −20°C.
Cell culture, cell viability, analysis of externalization of phosphatidylserine (PS), cell cycle, and mitochondrial transmembrane potential (MTP, ΔΨm)
The Kasumi-1, U937, and ponasterone A (PA; Invitrogen, Frederick, MD)–inducible AE-expressing U937 (U937-A/E) cells were cultured as described previously.28 The diagnosis of AML M2 with t(8;21) was established according to methods described previously.28 Primary leukemic cells harvested from bone marrow samples of de novo or relapsed patients with t(8;21) AML were cultured as described28 ; informed consent from these patients was provided in accordance with the Declaration of Helsinki. Approval was obtained from the institutional review board of Shanghai Jiao Tong University School of Medicine. Kasumi-1 cells at exponential growth phase were coincubated with oridonin in the absence or presence of pan-caspase inhibitor z-VAD-fmk (Becton Dickinson [BD], Mountain View, CA), casp-3 inhibitor z-DQMD-fmk (Sigma), or proteasome inhibitors (lactacystin [Lact], MG-132, and proteasome inhibitor I [PSI]; Calbiochem, San Diego, CA) at indicated concentrations. The fresh cells isolated were treated with oridonin at indicated concentrations. U937-A/E cells were coincubated with PA in the presence or absence of oridonin.
Cell proliferation was analyzed using a Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan).28 Cell viability was estimated by trypan blue dye exclusion. Cell morphology was assessed by Wright-Giemsa staining of cells prepared by cytospin centrifugation. The externalization of PS was evaluated by annexin V (AV) detection using an AV–fluorescein isothiocyanate (FITC) kit (Clontech, BD, Palo Alto, CA); cell cycle analysis and MTP assessment were performed as described.45
Terminal deoxynucleotidyl transferase (TdT) in situ cell death detection (TdT-mediated dUTP nick end labeling, TUNEL) and DNA fragmentation
Apoptosis in individual cells was detected using an in situ cell death detection kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions and analyzed under a fluorescence microscope. For assays of DNA fragmentation, cells were collected and lysed. RNase was added to the lysate and incubated for 30 minutes at 37°C. Proteinase K and SDS were added, followed by incubation at 50°C for 16 hours. DNA was extracted, precipitated, and electrophoresed. The stained gel was visualized by transillumination with ultraviolet (UV) light and photographed.
Analysis of the expression of AE target genes at mRNA and protein levels
Total RNA was purified from cells using a Trizol Reagent (Gibco/BRL, Grand Island, NY). First-strand cDNA was synthesized using 2 μg RNA and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI). The polymerase chain reaction (PCR) amplification was conducted. For quantitative reverse-transcription (RT)–PCR, a SYBR Green PCR Master mix (Applied Biosystems, Foster City, CA) was used, and β-actin was coamplified as an endogenous control to standardize the amount of the sample RNA added to the reaction.28 The primers used are listed in Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article.
To detect the expression of G-CSFR and MPO at protein level, 1 × 106 Kasumi-1 cells were washed with PBS and incubated with FITC-conjugated antihuman G-CSFR or MPO antibody (Beckman Coulter, San Diego, CA) according to the manufacturer's instructions. After being washed with PBS, the cells were resuspended in PBS and analyzed by flow cytometry.
Chromatin immunoprecipitation (ChIP)
ChIP was conducted using the ChIP-IT Kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions with antibodies against the ETO (Santa Cruz Biotechnology, Santa Cruz, CA), acetyl–histone H3, and HDAC1 (Upstate, Lake Placid, NY). Briefly, cells treated or untreated with oridonin were fixed using formaldehyde. The DNA was sheared into small, uniform fragments using sonication, and specific protein/DNA complexes were immunoprecipitated using an antibody directed against the DNA-binding protein of interest. Following immunoprecipitation, cross-linking was reversed, the proteins were removed by treatment with proteinase K, and the DNA was purified and analyzed by semiquantitative RT-PCR using primers listed in Table S1.
Plasmid, mutagenesis, and transfection
To explore the exact cleavage site on AE, the full-length AE-containing pFLAG-CMV4-AE plasmid was used, and residues Asp (D) at 171, 188, and 192 were substituted by Ala (A) generated by using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. After being sequenced, 20 μg pFLAG-CMV4-AE or mutant pFLAG-CMV4-AE were mixed with U937 cells and transferred to electroporation cuvettes with a 0.4-cm gap (Bio-Rad, Hercules, CA). Electroporations were performed using a Gene-Pulser II (Bio-Rad) at 280 V and 1050 μF. The samples were then transferred to complete RPMI 1640 medium and incubated at 37°C in 5% CO2. Twelve hours later, the cells were treated with oridonin, and the effect of oridonin on mutant AE was analyzed by Western blot. U937 cells transfected with mutant AE were also coincubated with 1000 μg/mL G418 (Chemicon, Temecula, CA) for 30 days to select cells stably expressing mutant AE. The cells were then treated with oridonin and analyzed.
Western blot analysis
Cell pellets were lysed, and protein extracts were quantitated and loaded on 8% to 12% sodium dodecyl sulfate–polyacrylamide gel, electrophoresed, and transferred to a nitrocellulose membrane. The membrane was incubated with primary antibody, washed, and incubated with horseradish peroxidase (HRP)–conjugated secondary antibody (Santa Cruz). Detection was performed by using a chemiluminescent Western detection kit (Cell Signaling, Beverly, MA). The change in cytoplasmic cyt C was detected using a Cytochrome C Releasing Apoptosis Assay Kit (BioVision, Palo Alto, CA) according to the manufacturer's instructions.
Murine models and oridonin treatment
All mice used in this study were bred and maintained in a specific pathogen-free environment. C57 or FVB/NJ mice (6 to 8 weeks old) were irradiated by a high-energy linear accelerator (6 MV; Siemens Primus; Siemens, New York, NY) at a dose of 450 cGy. Twenty-four hours later, C57 mice were intravenously injected with 3 × 106 leukemic cells expressing a truncated AE (AEtr),37 and FVB/NJ mice were injected with 2 × 105 cells expressing PML-RARα46 via tail vein. When the mice became moribund, they were killed, and splenic cells were isolated and inoculated into secondary recipients for passage. Xenograft model in nude mice was established by subcutaneous inoculation of 4 × 107 Kasumi-1 cells into the right flank.
Five days after leukemic cell transplantation, the C57 mice were treated intraperitoneally with vehicle (1% PluronicF68), low-dose Ara-C (25 mg/kg per day for 5 days; Ara-C-L), high-dose Ara-C (25 mg/kg per day for 10 days; Ara-C-H), oridonin (2.5, 7.5, or 15 mg/kg per day, 5 days per week for 2 weeks), or oridonin (7.5 mg/kg per day) plus Ara-C-L. The FVB/NJ mice were treated with oridonin at 7.5 or 15 mg/kg per day, or ATRA (Sigma) at 10 mg/kg per day as a positive control. The nude mice received oridonin treatment (7.5 or 15 mg/kg per day) when tumor was measurable. Caliper measurements of the longest perpendicular tumor diameters were performed every day to estimate the tumor volume, using the following formula: 4π/3 × (width/2)2 × (length/2), representing the 3-dimensional volume of an ellipse.47 Animals were killed when their tumors reached 2 cm or when the mice became moribund. Survival was evaluated from the first day of treatment until death. TUNEL assay was performed to detect in situ apoptosis on tissue section using a DeadEnd Colorimetric TUNEL System (Promega) according to the manufacturer's instructions.
Statistical analysis
The lifespan of mice was analyzed by Kaplan-Meier methods, while the tumor volume was analyzed with one-way ANOVA and independent-sample t test. P values less than .05 were considered statistically significant and were derived from 2-sided statistical tests. All statistical analysis was performed using the software SPSS 12.0 for Windows (Chicago, IL).
Results
Oridonin inhibits growth and induces apoptosis in t(8;21) leukemic cells
We investigated the effects of oridonin on t(8;21)-bearing cells. We found that oridonin at 0.5 to 5.0 μM inhibited proliferation and growth of Kasumi-1 cells (Figure 1B-C). In particular, the rate of inhibition was 13%, 41%, 52%, 95%, and 99% for 0.5, 1, 2, 5, and 10 μM, respectively. Therefore, oridonin concentrations of 0.5, 2, and 5 μM were used to perform the majority of the experiments. Unlike Kasumi-1 cells, primary t(8;21) leukemic cells showed little proliferation, and the cell viability was gradually decreased when no drug was added. Oridonin reduced the number of viable cells in samples from relapsed or refractory patients (3 cases) and de novo cases (2 cases), exemplified by cells from a patient with resistance to high-dose Ara-C (Figure 1D). This inhibition was greater at concentration of 5 μM. We also tested the effects of oridonin on NB4, HL-60, U937, and K562 cell lines, and found that the IC50s of oridonin for these cells were 8 to 20 μM, much higher than that for Kasumi-1 cells (1.33 μM). For fresh leukemic cells, oridonin at 5 μM induced apoptosis in a much smaller proportion of cells in samples from patients with M3, M4, and M2 types of AML without t(8;21) and CML than that in cells with t(8;21) (Table S2).
Examination on t(8;21)-bearing leukemic cells upon oridonin treatment revealed the appearance of morphologic characteristics of apoptosis, such as shrinking cytoplasm, condensed chromatin, and nuclear fragmentation with intact cell membrane (Figure 2A).Externalized PS, revealed by AV staining, was significantly increased in Kasumi-1 cells treated with oridonin (Figure 2B). After treatment with oridonin at 2 μM for 48 hours, TdT labeling showed a proportion of Kasumi-1 cells experiencing DNA fragmentation (Figure 2C). The sub-G1 cells also increased while Kasumi-1 cells were treated with oridonin at 2 μM for 24 hours (Figure 2D). DNA electrophoresis using samples isolated from oridonin (2 μM)–treated Kasumi-1 cells confirmed DNA fragmentation (Figure 2E). These results demonstrated that oridonin induced apoptosis in Kasumi-1 cells. However, oridonin did not induce differentiation of Kasumi-1 cells, as determined by assessment of cell morphology, nitroblue tetrazolium (NBT) reduction assay, and detection of differentiation antigens CD11b and CD14 (data not shown).
Apoptotic effects of oridonin on t(8;21) leukemic cells. (A) Morphologic changes in Kasumi-1 (top panel) and fresh leukemic (bottom panel) cells untreated or treated with oridonin. Stainings were analyzed using an Olympus BX51 research microscope equipped with a 100×/1.30 numerical aperture (NA) oil objective (Olympus, Tokyo, Japan). Images were processed using Adobe Photoshop CS (Adobe Systems, San Jose, CA). Original magnification, ×1000. (B) Expression of annexin V on Kasumi-1 cells treated with oridonin at indicated concentrations. (C) In situ TdT labeling of Kasumi-1 cells. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. Images were visualized and processed using the equipment described for panel A. (D) Cell cycle analysis showing sub-G1 content in cells treated with 2 μM oridonin. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (E) DNA fragmentation in Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (F) PI-negative Rh123-positive (PI [−] Rh123 [+]) cells are decreased upon oridonin treatment; in particular, the inhibition was greater using oridonin treatment at 5 μM for 48 hours. (G) Effects of oridonin on casp-3, PARP, Bcl-2, survivin, NFκB, and Fas-L. β-Actin is used as a loading control. Error bars represent SD of experiments.
Apoptotic effects of oridonin on t(8;21) leukemic cells. (A) Morphologic changes in Kasumi-1 (top panel) and fresh leukemic (bottom panel) cells untreated or treated with oridonin. Stainings were analyzed using an Olympus BX51 research microscope equipped with a 100×/1.30 numerical aperture (NA) oil objective (Olympus, Tokyo, Japan). Images were processed using Adobe Photoshop CS (Adobe Systems, San Jose, CA). Original magnification, ×1000. (B) Expression of annexin V on Kasumi-1 cells treated with oridonin at indicated concentrations. (C) In situ TdT labeling of Kasumi-1 cells. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. Images were visualized and processed using the equipment described for panel A. (D) Cell cycle analysis showing sub-G1 content in cells treated with 2 μM oridonin. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (E) DNA fragmentation in Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (F) PI-negative Rh123-positive (PI [−] Rh123 [+]) cells are decreased upon oridonin treatment; in particular, the inhibition was greater using oridonin treatment at 5 μM for 48 hours. (G) Effects of oridonin on casp-3, PARP, Bcl-2, survivin, NFκB, and Fas-L. β-Actin is used as a loading control. Error bars represent SD of experiments.
Collapse of ΔΨm, activation of casp-3, and down-regulation of Bcl-2
To evaluate the effects of oridonin on ΔΨm and to determine whether cells with a low ΔΨm also lose plasma membrane integrity, we double stained the Kasumi-1 cells with propidium iodide (PI) and rhodamine 123 (Rh123), a lipophilic cation that is taken up by mitochondria in proportion to the ΔΨm. The results showed that untreated living cells were PI negative and strongly stained by Rh123 (PI−, ΔΨm high) (Figure 2F). Upon treatment with oridonin at 0.5, 2, or 5 μM, a fraction of PI-negative and low–Rh123-staining (PI−, ΔΨm low) Kasumi-1 cells appeared in a dose- and time-dependent manner; in particular, the inhibition was greater using oridonin at 5 μM for 48 hours (Figure 2F), suggesting that oridonin decreased the ΔΨm without altering plasma membrane permeability.
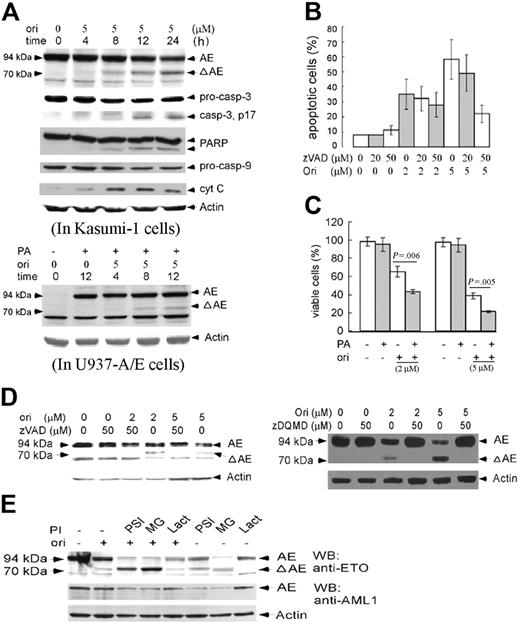
We tested the change of casp-3 in oridonin-induced apoptosis of Kasumi-1 cells and found that casp-3 precursor (pro–casp-3) was decreased with concentration of 5 μM oridonin with generation of an active form, accompanied by a cleavage of its substrate, poly ADP-ribose polymerase (PARP) (Figures 2G and 3A). The pro–casp-9 was also decreased, while the expression of cytoplasmic cyt C was increased (Figure 3A). Moreover, pretreatment with pan-caspase inhibitor z-VAD.fmk and casp-3 inhibitor z-DQMD-fmk significantly reduced the percentage of AV-positive Kasumi-1 cells (Figure 3B). These results suggested that caspases played a critical role in oridonin-induced apoptosis of Kasumi-1 cells. Oridonin treatment at 5 μM also led to down-regulation of Bcl-2, while the expression of Fas-ligand (Fas-L) and survivin was not modulated (Figure 2G).
Effects of oridonin on AE fusion protein and the possible mechanisms. (A) In Kasumi-1 cells, oridonin causes a degradation of AE with generation of an approximately 70-kDa catabolic fragment. In this expression, Western blot assay was done using an anti-ETO antibody whose epitope maps at the C-terminal of ETO. Effects of oridonin on casp-3, PARP, casp-9, and cytoplasmic cyt C are also shown. In U937-A/E cells, oridonin treatment also causes degradation of AE (lower panel, using the anti-ETO antibody). (B) Effects of pan-caspase inhibitor on oridonin-induced apoptosis. (C) The presence of PA, which induces the expression of AE, enhances apoptotic effect of oridonin on U937-A/E cells, as revealed by reduction of viable cells upon oridonin treatment. (D) Pretreatment with pan-caspase inhibitor z-VAD.fmk (left panel) and casp-3 inhibitor z-DQMD-fmk (right panel) prevents AE from proteolysis. (E) Pretreatment with 3 proteasome inhibitors, PSI, Lact, and MG-132, cannot prevent oridonin-induced degradation of AE. Error bars represent SD of experiments.
Effects of oridonin on AE fusion protein and the possible mechanisms. (A) In Kasumi-1 cells, oridonin causes a degradation of AE with generation of an approximately 70-kDa catabolic fragment. In this expression, Western blot assay was done using an anti-ETO antibody whose epitope maps at the C-terminal of ETO. Effects of oridonin on casp-3, PARP, casp-9, and cytoplasmic cyt C are also shown. In U937-A/E cells, oridonin treatment also causes degradation of AE (lower panel, using the anti-ETO antibody). (B) Effects of pan-caspase inhibitor on oridonin-induced apoptosis. (C) The presence of PA, which induces the expression of AE, enhances apoptotic effect of oridonin on U937-A/E cells, as revealed by reduction of viable cells upon oridonin treatment. (D) Pretreatment with pan-caspase inhibitor z-VAD.fmk (left panel) and casp-3 inhibitor z-DQMD-fmk (right panel) prevents AE from proteolysis. (E) Pretreatment with 3 proteasome inhibitors, PSI, Lact, and MG-132, cannot prevent oridonin-induced degradation of AE. Error bars represent SD of experiments.
Oridonin induces degradation of AE oncoprotein with D188 as a possible cleavage site for casp-3
Western blot analysis using an anti-ETO antibody whose epitope maps at the C-terminus of ETO was performed to explore whether oridonin treatment could modulate the expression of AE oncoprotein. We found that oridonin triggered a degradation of the 94-kDa AE with generation of an approximately 70-kDa fragment (ΔAE; Figure 3A upper panel). Detection using an anti-AML1 antibody (maps the Runx homology domain) confirmed the degradation of AE, while the 70-kDa cleavage fragment was not detected (data not shown), indicating the ΔAE should contain the C-terminal of ETO moiety. In U937-A/E cells, the PA-induced expression of AE was also degraded by oridonin treatment with generation of ΔAE (Figure 3A lower panel). Indeed, treatment with PA resulted in high-level expression of AE but not apoptosis of U937-A/E cells (Figure 3C). However, under coincubation with PA, oridonin treatment significantly decreased AV-negative, PI-negative proportion (viable cells) of U937-A/E cells (Figure 3C), suggesting that the expression of AE might enhance the cellular apoptotic response to oridonin.
Pretreatment with z-VAD.fmk not only suppressed oridonin-induced apoptosis (Figure 3B), but also abrogated catabolism of AE and generation of ΔAE (Figure 3D left panel). Furthermore, pretreatment with casp-3–specific inhibitor z-DQMD-fmk almost completely blocked AE degradation and ΔAE generation (Figure 3D right panel). However, pretreatment with proteasome inhibitors, PSI, MG-132, and Lact, could not block oridonin-induced AE degradation (Figure 3E). These results suggested that oridonin-induced degradation of AE was possibly mediated by casp-3.
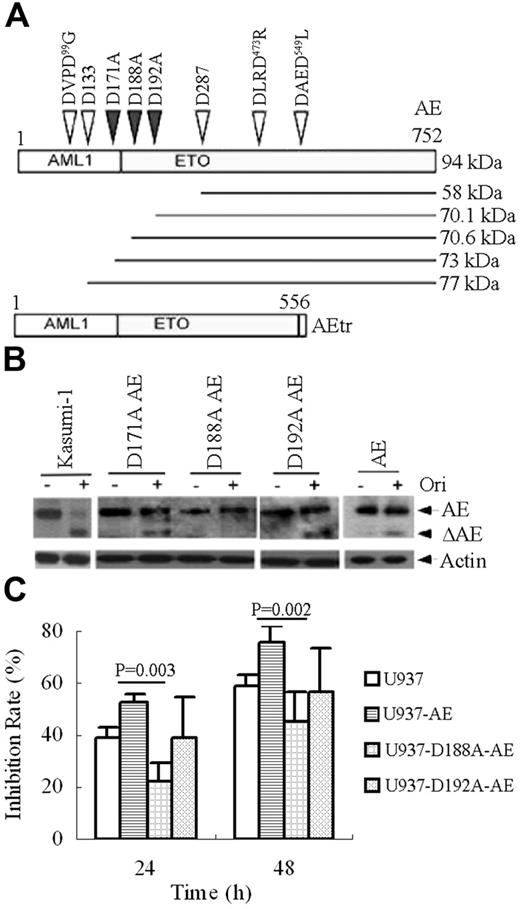
Casp-3 prefers a DXXD-like motif–containing substrate with some variants including XXXD-X.48,49 By analyzing the amino acid sequence, we found 3 DXXD-like motifs, DVPD99G, DLRD473R, and DAED549L, in AE amino acid sequence. However, cleavage at any one of the 3 motifs could not generate a 70-kDa ΔAE containing the C-terminal of ETO. In addition, only 3 Asp's existed in the region between D133 and D287 (Figure 4A). Hence, we hypothesized that the cleavage site(s) should be a XXXD-like, and might possibly reside on D171, D188, or D192 of AE oncoprotein. We thus performed site-directed mutagenesis to generate the substitution of A for D171, D188, and D192 (denoted as D171A, D188A, and D192A, respectively; Figure 4A). The mutant AEs were transfected into U937 cells, which were then treated with oridonin. The results showed that upon oridonin treatment, the wild type, D171A-AE, and D192A-AE could be degraded with generation of ΔAE. However, oridonin treatment could not cause catabolism of AE or generation of ΔAE in D188A-AE mutant (Figure 4B), suggesting D188 to be the cleavage site. Of interest, we found that U937 cells stably expressing D188A-AE showed less sensitivity to oridonin compared with U937 cells expressing wild type or D192A-AE (Figure 4C).
Substitution of Ala for Asp at 188 abrogates AE catabolism and confers oridonin resistance to some extent in U937 cells. (A) Schematics for mutations introduced into the AE fusion protein. (B) D188A mutant in AE abrogates AE degradation or ΔAE generation upon oridonin treatment. Wild type, D171A-AE, D188A-AE, and D192A-AE are transfected into U937 cells that are then treated without or with oridonin. Western blot is performed using an anti-ETO antibody to analyze the effects of oridonin on AE mutants. (C) Oridonin induces a much lower inhibition rate of U937 cells expressing D188A-AE compared with U937 cells expressing wild type or D192A-AE, suggesting D188A-AE confers oridonin resistance to U937 cells. In this experiment, U937 cells expressing AE, D188A-AE, and D192A-AE are treated without or with oridonin, and trypan blue dye exclusion assay is done. Inhibition rate is determined by viable cells in oridonin treatment group compared with those in vehicle control group. Error bars represent SD of experiments.
Substitution of Ala for Asp at 188 abrogates AE catabolism and confers oridonin resistance to some extent in U937 cells. (A) Schematics for mutations introduced into the AE fusion protein. (B) D188A mutant in AE abrogates AE degradation or ΔAE generation upon oridonin treatment. Wild type, D171A-AE, D188A-AE, and D192A-AE are transfected into U937 cells that are then treated without or with oridonin. Western blot is performed using an anti-ETO antibody to analyze the effects of oridonin on AE mutants. (C) Oridonin induces a much lower inhibition rate of U937 cells expressing D188A-AE compared with U937 cells expressing wild type or D192A-AE, suggesting D188A-AE confers oridonin resistance to U937 cells. In this experiment, U937 cells expressing AE, D188A-AE, and D192A-AE are treated without or with oridonin, and trypan blue dye exclusion assay is done. Inhibition rate is determined by viable cells in oridonin treatment group compared with those in vehicle control group. Error bars represent SD of experiments.
Reprogramming of AE targets
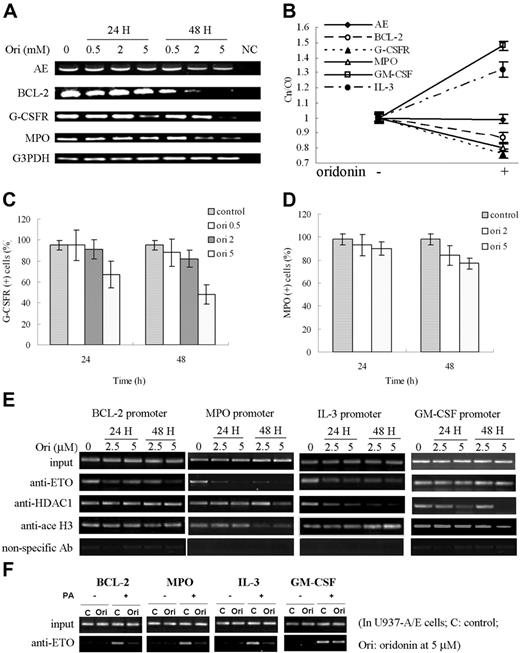
Whether degradation of AE could lead to reprogramming of the expression of its target genes (eg, BCL2, GCSFR, MPO, IL3, and GMCSF) was tested. Bcl-2 was shown to be down-regulated at protein level (Figure 2G). At mRNA level, oridonin treatment at 5 μM suppressed the expression of Bcl-2, G-CSFR, and MPO in Kasumi-1 cells as detected by semiquantitative RT-PCR, while AE was not significantly altered (Figure 5A). The expression of these genes was further analyzed by real-time RT-PCR, and the copy number of the gene transcripts (Cn) at each time point was compared with that of control (C0) (Cn/C0). We found that upon oridonin treatment, the mRNA level of AE was not perturbed significantly. While BCL2, GCSFR, and MPO exhibited down-regulated transcript levels, IL3 and GMCSF were up-regulated (Figure 5B). By analysis using direct immunofluorescence assay detected by flow cytometry, we found that upon oridonin treatment, G-CSFR and MPO were also down-regulated (Figure 5C-D).
Effects of oridonin on AE targets. (A) Semiquantitative RT-PCR analysis shows a down-regulation of Bcl-2, G-CSFR, and MPO in Kasumi-1 cells treated with oridonin. (B) Quantitative real-time RT-PCR analysis of expression of AE target genes. (C-D) Flow cytometry analysis shows a diminution in expression of G-CSFR (C) and MPO (D) on Kasumi-1 cells treated with oridonin. (E-F) Pharmacologic modulation of AE, HDAC1, and acetyl histone H3 on promoters of BCL-2, MPO, IL-3, and GM-CSF. ChIP assay was done in Kasumi-1 (E) and U937-A/E (F) cells treated with oridonin at indicated concentrations using anti-ETO, HDAC1, and acetyl histone H3 antibodies. Error bars represent SD of experiments.
Effects of oridonin on AE targets. (A) Semiquantitative RT-PCR analysis shows a down-regulation of Bcl-2, G-CSFR, and MPO in Kasumi-1 cells treated with oridonin. (B) Quantitative real-time RT-PCR analysis of expression of AE target genes. (C-D) Flow cytometry analysis shows a diminution in expression of G-CSFR (C) and MPO (D) on Kasumi-1 cells treated with oridonin. (E-F) Pharmacologic modulation of AE, HDAC1, and acetyl histone H3 on promoters of BCL-2, MPO, IL-3, and GM-CSF. ChIP assay was done in Kasumi-1 (E) and U937-A/E (F) cells treated with oridonin at indicated concentrations using anti-ETO, HDAC1, and acetyl histone H3 antibodies. Error bars represent SD of experiments.
The reversible acetylation of histones by HDACs and acetyltransferases (HATs) plays a fundamental role in gene transcription. We tested whether oridonin might reverse epigenetic alterations. BCL2 could bind to AML1 and AE through its DNA-binding sequence for AML1 (TGTGGT).27 Using ChIP assay, we found that in Kasumi-1 cells treated with oridonin, the binding activity of AE with BCL2 promoter was decreased (Figure 5E). This was confirmed in U937-A/E cells (Figure 5F). However, the interaction betweenHDAC1 and acetyl histone H3 with BCL2 promoter was not altered significantly (Figure 5E). Oridonin treatment also caused dissociation of MPO promoter from AE and acetyl histone H3, IL3 promoters from AE and HDAC1, and GMCSF promoters from HDAC1, and enhanced binding of IL3 promoter to acetyl histone H3 (Figure 5E-F).
Oridonin prolongs survival and inhibits tumor growth in t(8;21) but not t(15;17) leukemia murine models
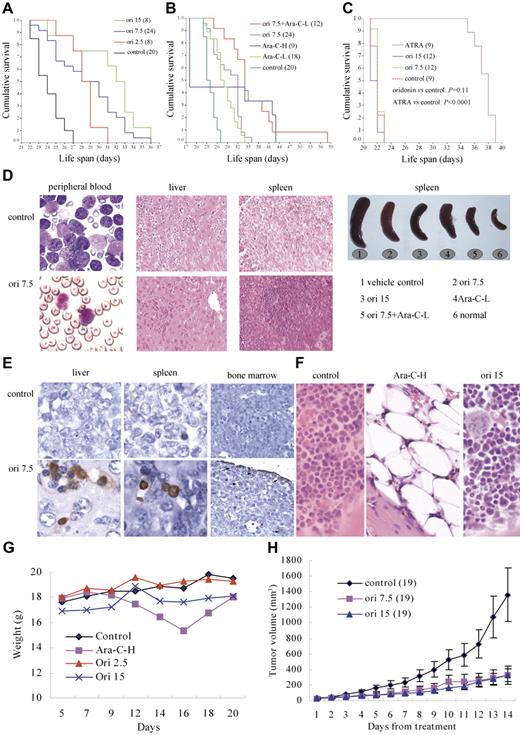
Since a truncated form of AE lacking its C-terminal 200 aa (AEtr; Figure 4A) strongly induced leukemia development,37 and the D188 cleavage site was retained in the truncated form, we evaluated the efficacy of oridonin on this AEtr murine model. C57 mice receiving 3 × 106 fresh leukemic cells isolated from spleen of AEtr AML mice were treated with oridonin 5 days after leukemic cell transplantation. The results showed that oridonin prolonged survival time of mice by 21.7% to 43.5% (median, 23 days for control mice versus 28, 30, and 33 days for mice treated with oridonin at 2.5, 7.5, and 15 mg/kg, respectively; P < .001; Figure 6A). In mice treated with vehicle alone, splenomegaly was observed, while histologic examination revealed extensive infiltrations of immature blast cells in hematopoietic organs (peripheral blood, spleen, and bone marrow) as well as liver, which destroyed the normal architectures of these tissues. Oridonin significantly reduced disseminated disease and prevented destruction of tissue architectures (Figure 6D). TUNEL assay revealed that oridonin induced apoptosis of leukemic cells in liver, spleen, and bone marrow of mice (Figure 6E). Ara-C-L exerted therapeutic efficacy similar to that of oridonin at 2.5 mg/kg (Figure 6B). In mice treated with Ara-C-H, 5 mice died on day 18 due to severe bone marrow suppression (Figure 6B,F), while the remaining 4 mice survived for 34 to 43 days. Treatment with Ara-C-H also led to significant reduction of body weight (Figure 6G). However, oridonin caused neither bone marrow suppression nor weight loss (Figure 6F-G), indicating oridonin to be a relatively safe agent. Combined use of oridonin at 7.5 mg/kg per day and Ara-C-L further prolonged survival of mice (P = .001 compared with Ara-C-L and Ori 7.5 mg/kg per day treatment groups; Figure 6B), with median survival of 34.1 days (49.6% higher than that of control mice), and 4 (33%) mice survived more than 39 days (increased 69.6% of survival time than that of control mice).
In vivo efficacy of oridonin on t(8;21) leukemia. (A) Oridonin prolongs lifespan of C57 mice bearing AEtr-expressing leukemic cells. (B) Oridonin (7.5 mg/kg per day) plus Ara-C (25 mg/kg per day for 5 days, Ara-C-L) further improves survival of the mice. (C) However, oridonin treatment cannot prolong lifespan of FVB/NJ mice bearing PML-RARα–expressing leukemic cells. (D) Oridonin reduces leukemic cell infiltration in C57 mice. (E) TUNEL assay is performed on specimen from C57 mice. The results show that oridonin induces apoptosis of leukemic cells in vivo. (F) Marrow pathological section stained with hematoxylin-eosin (HE) of mice treated with Ara-C and oridonin. (G) Changes in body weight of mice treated with oridonin and Ara-C. (H) Oridonin inhibits tumor growth in nude mice inoculated with Kasumi-1 cells. The numbers in parentheses represent the number of animals used in each group of this study. Error bars in H represent SD. Stainings were analyzed using an Olympus BX51 research microscope equipped with either a 100×/1.30 NA or a 40×/0.75 NA oil objective, and images were processed using Adobe Photoshop CS.
In vivo efficacy of oridonin on t(8;21) leukemia. (A) Oridonin prolongs lifespan of C57 mice bearing AEtr-expressing leukemic cells. (B) Oridonin (7.5 mg/kg per day) plus Ara-C (25 mg/kg per day for 5 days, Ara-C-L) further improves survival of the mice. (C) However, oridonin treatment cannot prolong lifespan of FVB/NJ mice bearing PML-RARα–expressing leukemic cells. (D) Oridonin reduces leukemic cell infiltration in C57 mice. (E) TUNEL assay is performed on specimen from C57 mice. The results show that oridonin induces apoptosis of leukemic cells in vivo. (F) Marrow pathological section stained with hematoxylin-eosin (HE) of mice treated with Ara-C and oridonin. (G) Changes in body weight of mice treated with oridonin and Ara-C. (H) Oridonin inhibits tumor growth in nude mice inoculated with Kasumi-1 cells. The numbers in parentheses represent the number of animals used in each group of this study. Error bars in H represent SD. Stainings were analyzed using an Olympus BX51 research microscope equipped with either a 100×/1.30 NA or a 40×/0.75 NA oil objective, and images were processed using Adobe Photoshop CS.
In nude mice inoculated subcutaneously with 4 × 107 Kasumi-1 cells, all animals developed a measurable tumor after a mean of 5 days (range, 3 to 7 days). Oridonin at 7.5 and 15 mg/kg significantly inhibited tumor growth compared with control mice (both P = .001; Figure 6H). However, oridonin at 7.5 and 15 mg/kg per day did not prolong survival of FVB/NJ mice injected with PML-RARα–expressing cells (P = .11; Figure 6C), while ATRA at 10 mg/kg per day did show significant therapeutic efficacy (P < .001). Taken together, these results demonstrated that oridonin has significant in vivo antileukemia efficacy for t(8;21) AML.
Discussion
Which types of hematologic malignancies could represent the most sensitive ones to oridonin treatment and whether oridonin could be used in the clinic setting remain unclear. Here, we showed that oridonin triggered apoptosis in more than 50% of t(8;21) leukemic cells in vitro at concentration of 2 μM or higher accompanied by degradation of AE oncoprotein, and showed significant antileukemia efficacies with low adverse effects in vivo. These data suggest possible beneficial effects for patients with t(8;21) AML.
The extrinsic and intrinsic apoptotic pathways that ultimately lead to activation of effector caspases (casp-3, casp-2, and casp-7) have been characterized. The extrinsic pathway is initiated by ligation of death receptors (CD95/Fas, TNF receptor, and TRAIL receptor) to stimulate activator caspases (casp-8 and casp-10), which in turn cleave and activate effector caspases.48,49 The intrinsic pathway requires disruption of the mitochondrial membrane and release of proteins, and disruption of this pathway is extremely common in cancer cells.50 We found that oridonin disrupted the ΔΨm of Kasumi-1 cells without altering plasma membrane permeability (Figure 2F), down-regulated Bcl-2, (Figure 2G), activated casp-9, and caused the release of cyt C to cytoplasm (Figure 3A), suggesting the mitochondrial permeability transition pore is opened, the functions of mitochondria are impaired, and machineries of death are unleashed. However, oridonin did not modulate the expression of Fas-L or survivin, which is a member of inhibitor of apoptosis protein (IAP) family (Figure 2G). At protein level, oridonin did not regulate the NFκB expression (Figure 2G), yet modulation of NFκB might also play a role in oridonin-induced apoptosis, because oridonin was shown to interfere with the DNA-binding activity of NFκB to its response DNA sequence, and affect the translocation of NF-κB from the cytoplasm to nuclei without altering IκB-α phosphorylation and degradation.17 Although changes of TNF, TRAIL, casp-8, and casp-10 were not determined, it was unlikely that the extrinsic apoptotic pathway played a major role in oridonin-induced apoptosis. The decrease of the pro–casp-3 and increase of the active p17 subunit, as well as the proteolysis of PARP (Figures 2G and 3A), indicated that casp-3 was activated. Thus, oridonin may trigger apoptosis by insulting mitochondria and activating the intrinsic apoptosis pathway, which results in activation of effector caspases.
The AE oncoprotein is critical for leukemogenesis of t(8;21) AML. Intriguingly, oridonin treatment caused degradation of AE with generation of an approximately 70-kDa fragment (Figure 3A). In U937-A/E cells, the PA-induced expression of AE enhanced the apoptotic effects of oridonin, whereas oridonin also induced degradation of AE in an identical manner to that in Kasumi-1 cells (Figure 3A lower panel). On the other hand, the transcription of AE was not significantly altered (Figure 5A-B). These results demonstrated a cleavage of AE induced by oridonin. Consequently, degradation of AE might cause reprogramming of its target genes. Our results seem to confirm this notion. As shown in Figures 2G and 5A-B, BCL-2 was down-regulated; this might result from cleavage of AE, which led to transcriptional inactivation (Figure 5A-B), and activation of intrinsic apoptotic pathway might also play a role. Previous studies showed that murine homolog of human AML1, PEBP2/CBF, regulated murine Mpo,51,52 and AML1-binding site was found in human MPO,53 suggesting that AE might suppress MPO transcription and AE catabolism might lead to up-regulation of MPO. However, oridonin treatment caused down-regulation of MPO (Figure 5A-B), suggesting that other molecules as well as chromatin conformation could be involved in the regulation of MPO gene expression, and this could be a complex scenario. Using ChIP assay, we found that in Kasumi-1 and U937-A/E cells treated with oridonin, MPO promoter was dissociated from AE, and dissociated slightly from HDAC1 (Figure 5E-F). Theoretically, these phenomena seemed to be able to facilitate MPO transcription, but the binding of MPO promoter to acetyl histone H3 decreased markedly during the 48-hour treatment time course (Figure 5E second panel). This event might silence MPO, corroborating a report showing the up-regulation of MPO by AE mediated indirectly through up-regulation of C/EBPϵ.29 The mechanisms for GCSFR down-regulation by oridonin treatment (Figure 5A-C) might be similar to these. On the other hand, IL3 has a consensus TGTGGT and a similar TGTGGG sequence in its promoter, and thus can be activated by AML1 and repressed by AE.26 As shown in Figure 5E-F, oridonin dissociated IL3 promoter from AE and HDAC1, and slightly enhanced binding to acetyl histone H3. These events can explain the up-regulation of IL3 (Figure 5B). GMCSF is another TGTGGT-containing AML1 target gene suppressed by AE.30 Upon oridonin treatment of Kasumi-1 cells, the expression of GMCSF was increased (Figure 5B). Unlike IL3, GMCSF promoter was dissociated from AE slightly; however, when Kasumi-1 cells were treated with oridonin at 5 μM for 48 hours, GMCSF promoter was completely dissociated from HDAC1, suggesting that release of transcription suppressor may contribute mainly to up-regulation of GMCSF, and recruitment of HDAC1 suppressor complex might also be involved in GMCSF inactivation by AE. Since the regulation network of gene expression is complex, in which regulators as well as status of DNA methylation, histone acetylation, methylation, and other modifications play critical roles, these results provide only a first step in understanding the chemical biology of oridonin on t(8;21) leukemic cells.
Degradation of protein through hydrolysis of peptide bonds is a complex process. In APL, degradation of the PML-RARα fusion protein by ATRA and ATO has been shown to be mediated by proteasome and/or caspases.54,55 We tested the roles that caspases and proteasome can play in AE degradation. We found that pan-caspase inhibitor and casp-3 inhibitor not only blocked oridonin-induced apoptosis in Kasumi-1 cells, but also abrogated AE degradation (Figure 3B,D). However, proteasome inhibitors used in this study could not obstruct AE catabolism (Figure 3E). Indeed, the proline-rich AML1 protein was shown to be degraded through the ubiquitin-proteasome system, while proteasome inhibitors were able to protect AML1 from proteolytic degradation.56 Since casp-3 is the executioner of apoptosis induced by oridonin, it is likely that AE degradation was mediated by casp-3. Because AE was cleaved with generation of a 70-kDa fragment containing the carboxyl terminal of ETO, we speculated the cleavage site(s) should be localized in D171, D188, or D192 at AML1 moiety or N-terminal of ETO moiety (Figure 4A). To test this hypothesis, D171A, D188A, and D192A mutants were generated and mutant AE was transfected into U937 cells that were then treated with oridonin. We found that the wild type, D171A-AE, and D192A-AE could be cleaved by oridonin treatment with generation of ΔAE. However, oridonin could cause neither catabolism of AE nor generation of ΔAE in D188A-AE (Figure 4B), suggesting the D188 was the cleavage site. Consequently, at cellular level D188A-AE conferred oridonin resistance to U937 cells (Figure 4C). Hence, the PD188S in AE represents a novel cleavage site for casp-3. An intriguing question is raised here: can mutation in D188 perturb the leukemogenic activity of AE? Additionally, Yan et al37 reported that deletion of an AE C-terminal NCoR/SMRT-interacting region strongly induced leukemia development. Whether ΔAE acts as a dominant-negative form for the full-length AE and thereby facilitates oridonin-induced apoptosis could be interesting questions for future work.
The in vivo efficacy of oridonin on t(8;21) leukemia had been demonstrated in 2 different murine models. In C57 mice bearing AEtr-expressing leukemic cells, oridonin prolonged survival and reduced infiltration of leukemia (Figure 6A,D). TUNEL assay showed that oridonin also induced apoptosis of leukemic cells in vivo (Figure 6E). Moreover, severe bone marrow suppression and body weight loss were not observed in mice treated with oridonin compared with high-dose Ara-C treatment (Figure 6F-G), suggesting oridonin to be a relatively safe agent. Combined use of oridonin at 7.5 mg/kg per day and Ara-C-L further prolonged survival of mice (Figure 6B). In nude mice inoculated with Kasumi-1 cells, oridonin significantly inhibited tumor growth (Figure 6H). These data suggest that oridonin could be effective in treating human t(8;21) leukemia.
Authorship
Contribution: G.-B.Z. designed research, performed research, analyzed data, and wrote the paper; H.K. and L.W. performed research and analyzed data; L.G., P.L., J.X., F.-X.Z., and X.-Q.W. performed research; Z.-X.S., J.C., and L.-J.G. analyzed data; M.Y. performed research; D.-E.Z. contributed vital new reagents and analytical tools; S.-J.C., Z.-Y.W., and Z.C. designed research, analyzed data, and wrote the paper. G.-B.Z., H.K., L.W., and L.G. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zhu Chen, Shanghai Institute of Hematology, Rui Jin Hospital Affiliated to Shanghai JiaoTong University School of Medicine, 197 Ruijin Rd II, Shanghai 200025, China; e-mail: zchen@stn.sh.cn; Guang-Biao Zhou, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou Science Park, Guangzhou 510663, China; e-mail: zhou_guangbiao@gibh.org.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported in part by the National Natural Science Foundation of China (30400590), China Postdoctoral Science Foundation (2004036353), the Chinese National High Tech Program (863), the National Key Program for Basic Research (973), Cancer Research Foundation of Shanghai Second Medical University (Z200502), the K. C. Wong Education Foundation, Hong Kong, and the National Institutes of Health (CA96735).
The authors thank Prof M. Lübbert at University of Freiburg Medical Center, Germany, for providing the U937-A/E cells, and G. Q. Chen and L. H. Ma at Shanghai Institute of Hematology for their help and support.

![Figure 2. Apoptotic effects of oridonin on t(8;21) leukemic cells. (A) Morphologic changes in Kasumi-1 (top panel) and fresh leukemic (bottom panel) cells untreated or treated with oridonin. Stainings were analyzed using an Olympus BX51 research microscope equipped with a 100×/1.30 numerical aperture (NA) oil objective (Olympus, Tokyo, Japan). Images were processed using Adobe Photoshop CS (Adobe Systems, San Jose, CA). Original magnification, ×1000. (B) Expression of annexin V on Kasumi-1 cells treated with oridonin at indicated concentrations. (C) In situ TdT labeling of Kasumi-1 cells. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. Images were visualized and processed using the equipment described for panel A. (D) Cell cycle analysis showing sub-G1 content in cells treated with 2 μM oridonin. (Top panel) control; (bottom panel) Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (E) DNA fragmentation in Kasumi-1 cells treated with 2 μM oridonin for 48 hours. (F) PI-negative Rh123-positive (PI [−] Rh123 [+]) cells are decreased upon oridonin treatment; in particular, the inhibition was greater using oridonin treatment at 5 μM for 48 hours. (G) Effects of oridonin on casp-3, PARP, Bcl-2, survivin, NFκB, and Fas-L. β-Actin is used as a loading control. Error bars represent SD of experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/8/10.1182_blood-2006-06-032250/4/m_zh80080711410002.jpeg?Expires=1769469503&Signature=nfs-dQh7DkjlkhG~WS29RKyyvxWZum1YXn5~JGenh3zZmmEhgrudP3mCip5MhG9Dj5i1kJOuY8ne50Q2kYVSshUZ3NEjMGl1l~BGzfmOhHy-Gde~9tQpqxRZAppHf9bfuKeIYlc3PU1D-NNMV8sUFj7bd3wsxVHBRfZmBuHV7ahS7NFkhpY8ExRYVtxbsSFLi-vn-AP1TZXfNHQZXoeUwLcS3R4yCahUn7KCa3915j9FGdsSLa9Qn1CZUrVx~gbaeUbh-zLMvNdHAU3Jvn8mfGeWBkRx8~3XvtV5XbdG6hI415Qx3He4CAbTXFAZL12f-pkepmVb8IUd3Z0j6r26Cg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal